Abstract
All RecA-like recombinase enzymes catalyze DNA strand exchange as elongated filaments on DNA. Despite numerous biochemical and structural studies of RecA and the related Rad51 and RadA proteins, the unit oligomer(s) responsible for nucleoprotein filament assembly and coordinated filament activity remains undefined. We have created a RecA fused dimer protein and show that it maintains in vivo DNA repair and LexA co-protease activities, as well as in vitro ATPase and DNA strand exchange activities. Our results support the idea that dimeric RecA is an important functional unit both for assembly of nucleoprotein filaments as well as their coordinated activity during the catalysis of homologous recombination.
The bacterial RecA protein is the founding member of a family of enzymes that catalyzes recombination between homologous DNA substrates, a family which includes the phage T4 UvsX, yeast Rad51, archael RadA and human Rad51 proteins (1, 2). To perform this function these recombinase enzymes form a helical filament that assembles cooperatively on a single-stranded region of DNA resulting in the formation of an active nucleoprotein complex that subsequently interacts with a dsDNA substrate to catalyze a search for homology and DNA strand exchange (1, 3, 4). Early biochemical studies of RecA revealed that in the absence of substrate DNA the protein exists in solution as a complex heterogeneous mix of various oligomers (5–8) and later work showed that this mix included monomers, ring-shaped hexamers/heptamers, short filaments ranging in length from 0.03 to 0.15 μm as well as bundles of filaments (9–12).
The RecA nucleoprotein filament assembles in an ATP-dependent, highly cooperative manner (13–16). Kinetic analysis of nucleoprotein filament assembly has shown that RecA polymerization onto ssDNA can be divided into two major steps, i. nucleation, which is rate limiting, and ii. filament extension (17). However, the oligomeric heterogeneity of RecA in these studies precluded identification of a specific RecA oligomer, e.g. monomers or dimers, as being an obligatory component of the assembly process, or whether various larger RecA oligomers, e.g. trimers or hexamers, could participate in filament assembly. It was also found that RecA oligomers formed in the absence of ATP or ssDNA are not directly interconvertible with active nucleoprotein filaments, and that disassembly of the pre-formed self filaments was required prior to filament assembly on DNA (18, 19). Again, however, the oligomeric state of the dissociated RecA could not be determined in these studies. Thus, the identity of the nucleating unit of recombinase filament assembly remains unclear.
In this study we have spliced two copies of the recA gene into a single transcriptional unit and purified a version of the protein we refer to as the RecA-FD (fused dimer). RecA-FD shows wild-type RecA activities in vivo, and catalyzes strand exchange in vitro. Our results show that formation of active RecA nucleoprotein filaments does not require monomeric RecA, and suggest that oligomeric forms of the protein can participate in filament assembly.
EXPERIMENTAL PROCEDURES
Expression Constructs
The gene encoding wild type recA was expressed using a previously described construct, pTRecA420, in which recA is regulated by the tac promoter (20, 21). The gene encoding the fused RecA dimer (recA-FD) was constructed as follows. Two point mutations, Lys6Ala and Arg28Ala, were introduced into the wild type recA gene carried in plasmid pTRecA420 using the QuikChange protocol (Stratagene). This mutant recA gene was PCR amplified using a top strand primer that overlapped the NcoI restriction site (underlined) at the fMet initiation codon (5′-GG AGT GAT GCC ATG GCT ATC GAC G - 3′) and a bottom strand primer that adds a five-residue linker (Gly3Ser2) at the RecA C-terminus, and also contains an NcoI restriction site (underlined; 5′-GCA TGC CAT GGA GCT CCC GCC TCC AAA ATC TTC GTT AGT TTC TGC – 3′). This PCR product was digested with NcoI, ligated into the NcoI site in pTRecA420, and candidates resulting in two fused recA genes carrying the Lys6Ala and Arg28Ala substitutions only in the N-terminal copy were screened by restriction mapping (see Figure 1). Identification of the correct construct was confirmed by DNA sequencing.
Figure 1.
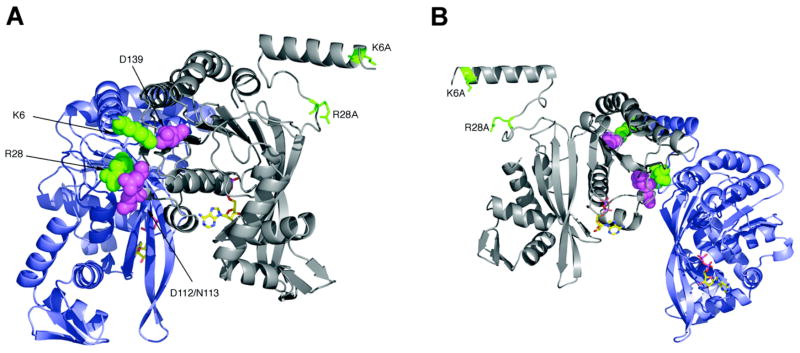
RecA dimer structure. These views show (A) the outside surface and (B) inside surface of a RecA dimer as it exists within the helical RecA protein filament. The α-carbon backbones of the two subunits are colored gray and blue, with side chains of residues Lys6 and Arg28 in green, Asn112, Asp113 and Asp139 in violet, and ADP in atom colors. The Lys6Ala and Arg28Ala in the N-terminal subunit of the fused dimer are indicated. The images were created using PyMOL (DeLano Scientific LLC) and the pdb file 1REA.
Recombinational DNA Repair and LexA Coprotease Activities in vivo
Recombinational DNA repair activity was measured following exposure of cells to varying doses of UV irradiation as previously described (20). LexA coprotease activity was measured using strain DE1663′ and the appearance of red vs. white colonies on MacConkey lactose plates as previously described (22).
Protein Purification
Wild type RecA was purified as previously described (23). RecA-FD was purified using a similar procedure with the following modifications. Cells were lysed by sequential additions of lysozyme (final concentration = 0.2 mg/ml), EDTA (final concentration = 0.5 mM) and Brij-35 (final of 0.5%). The suspension was stirred at 4 °C for 30 min after each addition followed by brief sonication and the resulting lysate was centrifuged for 90 min (Sorval SA-600 at 13,500 rpm). The supernatant was recovered and proteins precipitated by addition of 0.32 g/ml (NH4)2SO4 and incubation overnight at 4 °C. Precipitate was recovered by centrifugation, dissolved in R buffer (20 mM Tris, pH 7.5/10% glycerol/1 mM DTT/0.1 mM EDTA) and dialyzed extensively against the same buffer. Nucleic acids were removed by addition of streptomycin sulfate (final concentration = 0.5%) followed by centrifugation. The supernatant was loaded onto a 40 ml DEAE (DE52) column equilibrated in R buffer/50 mM NH4Cl and proteins were eluted using a 50 – 400 mM NH4Cl gradient. Fractions containing RecA-FD were pooled, precipitated as above, protein was dissolved and dialyzed using R buffer/25 mM NaCl and loaded onto a ssDNA cellulose column (8 ml) equilibrated in the same buffer. The column was washed with 2 column volumes of R buffer/25 mM NaCl and RecA-FD was eluted using 1 column volume of this buffer containing 2 mM ATP. Fractions containing RecA-FD were pooled, precipitated as above, dissolved in R buffer and dialyzed extensively against the same. The concentration was determined spectrophotometrically (ε280 = 0.59 mg−1·ml) and by comparison to standards on SDS gels. Glycerol was added to a final concentration of 25% and samples were quick frozen in liquid nitrogen and stored at −80 °C. Both the wild type RecA and RecA-FD proteins were judged to be > 95% pure by Coomassie-stained SDS polyacrylamide gels. In all RecA-FD preparations, approximately 2–5% of the protein appeared as monomeric RecA (Figure 2). Because protease inhibitors were added to the cells immediately before freezing, and were maintained during the early steps of purification, this likely resulted from intracellular proteolysis near the Gly3Ser2 linker. This contaminating monomeric RecA, which consists of equal amounts of wild type RecA and the Lys6Ala/Arg28Ala mutant, can increase the measured activities of the fused dimer preparation by no more than 5%. However, in no case did any of the activities measured in vivo or in vitro show a decrease approaching 20-fold (see RESULTS). Therefore, the fused dimer itself maintains significant catalytic activity. For all results the concentration of both wild type RecA and the fused dimer refers to monomeric protein.
Figure 2.
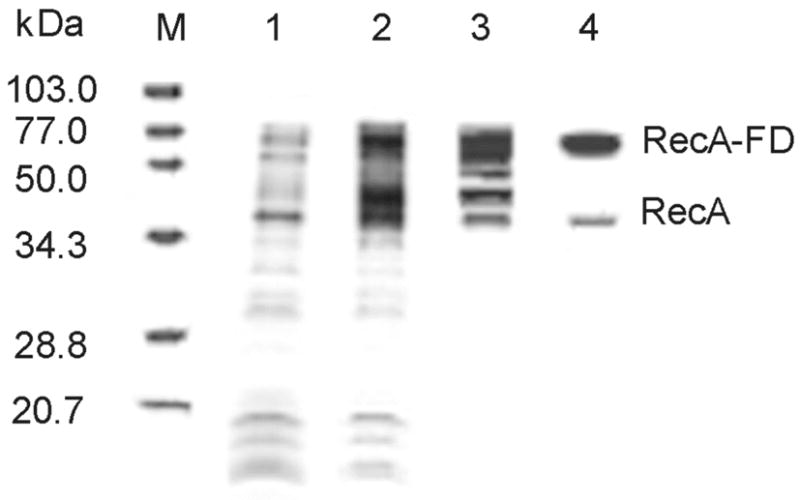
Purification of the RecA-fused dimer protein. SDS-polyacrylamide gel (11%) showing molecular mass markers (lane M), crude extract supernatant before and after induction with IPTG (lanes 1 and 2, respectively), pooled fractions from DE52 column gradient (lane 3) and elution from ssDNA cellulose (lane 4). The final step produced RecA-FD protein at >95% purity with less than 5% contamination of monomeric RecA resulting from intracellular proteolysis (see EXPERIMENTAL PROCEDURES).
ATPase Activity
Hydrolysis of [α-32P] dATP was performed essentially as described (24). Reactions included the following components: 20 mM Tris (pH 7.5), 20 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1.0 mM DTT, 0.5 mM [α-32P] dATP (20 μCi/ml), 2.0 μM RecA or RecA-FD protein and 25 μM ss φX174 DNA (concentration in bases). Percent hydrolysis as a function of time over 40 min was measured by scanning polyethyleneimine chromatography plates using a Molecular Imager FX and QuantityOne software (Bio-Rad).
Gel Shift DNA Binding Assay
Reactions (10 μl) contained 20 mM triethanolamine-HCl (pH 7.5,) 1 mM DTT, 10 mM MgCl2, 0.1 mg/ml BSA, 100 nM 5′ fluorescein-labeled 95 base oligonucleotide (concentration in bases), and the indicated amounts of protein. The oligonucleotide was made using an ABI392 synthesizer and fluorescein was conjugated in the last step of the synthesis using 5′-fluorescein phosphoramidite (Glen Research). The sequence is as follows: 5′-AGA CGA TAG CGA AGG CGT AGC AGA AAC TAA CGA AGA TTT TGG CGG TGG TCT GAA CGA CAT CTT TGA GGC GCA GAA AAT CGA GTG GCA CTA ATA AG - 3′. Reactions were incubated at 37 °C for 30 min followed by addition of 2 ml loading buffer and were loaded onto a 1.0% agarose gel and electrophoresed at 100 mV in 0.5 X TBE buffer with 10 mM MgCl2. DNA was visualized by excitation at 473 nm using a FUJI FILM FLA-5000 Multifunctional Imaging System (FUJI) and Image Reader software (FUJI).
DNA Strand Exchange
Reaction mixtures contained 25 mM Tris Acetate (pH 7.5), 10 mM MgOAc, 5% glycerol, 1 mM DTT, 8 mM phosphocreatine, 10 units/ml creatine kinase, single stranded circular DNA (φX174, 20 μM bases) and 6.0 μM RecA or RecA-FD. Reaction mixtures were preincubated at 37 °C for 10 min, at which point linear duplex DNA substrate (XhoI digested φX174 virion DNA) was added to a final concentration of 26 μM (bases) and incubated an additional 10 min. Strand exchange was initiated by addition of a premixed solution containing 2 μM SSB (US Biologicals) and 3 mM ATP. Samples (10 μl) were taken at indicated times and strand transfer was terminated by addition of 2 μl of a stop buffer containing 5% SDS, 20% glycerol, 60 mM EDTA and proteinase K to a concentration of 1 mg/ml. After incubation at 42 °C for 30 min, samples were analyzed by electrophoresis through a 0.8% agarose gel run in 40 mM Tris acetate and 2 mM EDTA. Gels were processed by staining in Vistra Green fluorescent stain (Amersham Pharmacia, Inc., 1:10,000) for 60 min and then analyzed by Image Reader 1 Laser/1 Image analysis at 473 nm on a phosphorimager (Fuji Films). The gels were quantified using Image Gauge software v. 3.1.
Electron Microscopy
Samples were prepared for electron microscopy as previously described (25). Protein was 2.0 μM, ATPγS was 1.0 mM and φX174 single-stranded circular DNA was 0.5 μM. Reactions with φX174 single-stranded circular DNA included 0.03 μM E. coli single-stranded DNA binding protein. Reactions were spread onto thin carbon films on holey grids (400 mesh), stained with 1% uranyl acetate and visualized using a Philips CM10 electron microscope.
RESULTS
Design of the RecA fused dimer and in vivo activities
Our previous studies of two recA mutants, RecA K6A and RecA R28A, showed that each reduced formation of free protein filaments but formed nucleoproteins on ssDNA. Additionally, each purified mutant protein showed both a lower rate and extent of DNA strand exchange relative to wild type RecA over a 60 min time course (21). Therefore, we originally created the RecA fused dimer carrying the N-terminal K6A and R28A substitutions to investigate the possibility that this protein could be used in structural studies designed to reveal the filament subunit interface in the active form of RecA. While the subunit interface at the dimer junction is identical to wild type RecA (Figure 1), we expected the N-terminal substitutions to prohibit further assembly of RecA filaments. We were surprised, however, to find that the in vivo activities of RecA-FD mimicked those of wild type RecA.Expression of the plasmid-borne recA-FD gene (pTRecA-FD) in the ΔrecA strains MV1190 or DE1663′ rescued the extreme sensitivity of each strain to mitomycin C and UV light to the same extent as expression of the wild type recA gene (pTRecA420; Table I). Additionally, analysis of the LexA co-protease function using a MacConkey lactose plate assay showed the RecA-FD protein to have the same level of DNA damage-inducible activity as wild type RecA (Table I). Thus, the ability of RecA-FD to catalyze LexA cleavage and recombinational DNA repair in vivo appears similar to that of wild type RecA. Therefore, we exploited this protein to further investigate questions regarding the identity of the nucleating protein unit for recombinase filament assembly, as well as possible identification of a functional unit within an active filament.
Table 1.
DNA repair, co-protease, and ATPase activities for wild type RecA and the RecA-FD protein
| protein | UV survival* | MMC survival* | LexA coprotease* | ATPase† |
|---|---|---|---|---|
| RecA | 1.0 | 1.0 | 1.0 | 0.3 (−ssDNA)
18.3 (+ssDNA) |
| RecA-FD | 1.0 | 0.9 | 1.0 | 0.3 (−ssDNA)
18.0 (+ssDNA) |
fractional activity relative to wild type RecA
units are (mol ADP/min·mol enzyme)
Protein purification
The RecA-FD protein was purified using procedures similar to those used for wild type RecA (see EXPERIMENTAL PROCEDURES). All RecA-FD preparations contained 2–5% monomeric RecA, despite the fact that protease inhibitors were present throughout cell harvesting and the purification procedure (Figure 2). However, as explained below (see DISCUSSION), this small level of monomeric RecA, which likely arises from intracellular proteolysis in the region of the Gly3Ser2 linker, could in no way account for the in vivo or in vitro activities observed for the RecA-FD protein.
ATPase Activity
Purified wild type RecA and the RecA-FD proteins showed similar levels of both basal and ssDNA-dependent ATPase activity (Table I). Turnover numbers for both wild type RecA and RecA-FD in the absence and presence of φX174 ssDNA were calculated as 0.3 and 18.3 mol ADP·min−1·mol RecA−1, respectively.
Nucleoprotein Filament Formation and DNA Binding
The ability of the RecA-FD protein to form an active nucleoprotein filament was assessed by electron microscopy and gel shift DNA binding assays. Electron micrographs of both wild type RecA and RecA-FD in the absence of DNA show that both proteins form a mixed population of different sized oligomers (Figure 3, panels A and C). Similar results have been observed previously by several groups using either electron microscopy or various hydrodynamic methods (9–12). In the presence of φX174 ssDNA both proteins form cooperatively assembled nucleoprotein filaments that show a striated helical pattern typical of RecA-DNA complexes (Figure 3, panels B and D). Despite the similar appearance of the nucleoprotein filaments, gel shift DNA binding assays reveal that the affinity of RecA-FD for a 95 base oligonucleotide is reduced compared to wild type RecA. The gel in Figure 4 is representative of 4 different experiments run under similar conditions, and in each case we estimated half maximal DNA binding to be approximately 0.6 μM for wild type RecA and 1.5 μM for RecA-FD. Therefore, despite being able to form wild type-like nucleoprotein filaments the ssDNA binding affinity of RecA-FD is reduced approximately 2.5-fold.
Figure 3.
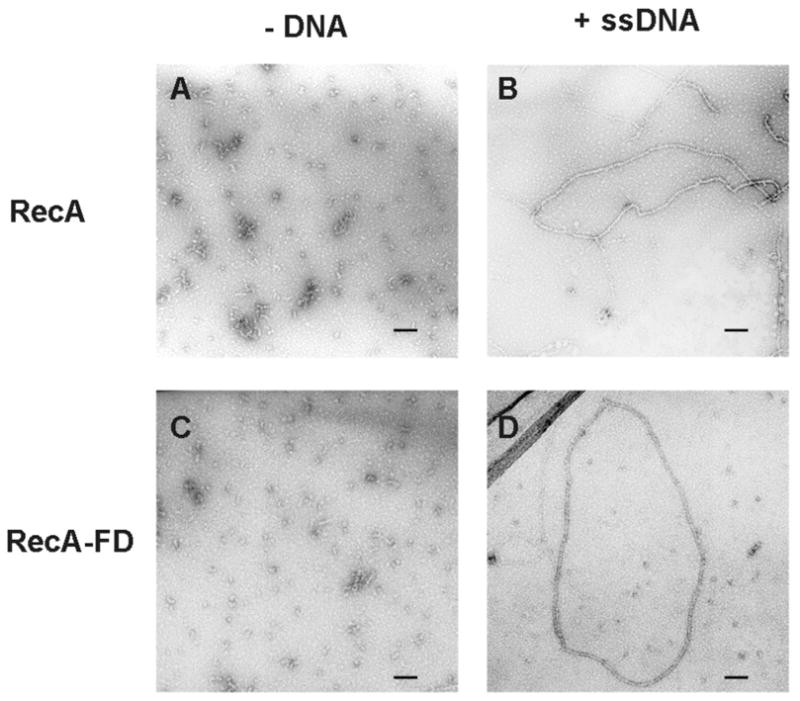
Electron micrographs of RecA and RecA-FD proteins with and without ssDNA. Wild type RecA (A, B) and RecA-FD protein (C, D) were incubated as described in EXPERIMENTAL PROCEDURES in the presence of ATPγS and the absence of φX174 phage ssDNA (A, C) or in the presence of both ATPγS and φX174 phage ssDNA (B, D). Black bar in each panel equals 0.1 μM.
Figure 4.
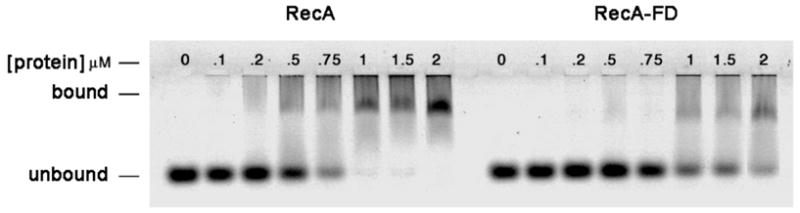
Gel shift DNA binding assay of RecA and RecA-FD proteins. Proteins were incubated at the indicated concentrations with a 5′ fluorescein-labeled 95 base oligonucleotide, and free and protein-bound DNAs were separated using agarose gel electrophoresis as described (see EXPERIMENTAL PROCEDURES).
DNA Strand Exchange
DNA strand exchange assays were performed using single-stranded circular and homologous linearized duplex φX174 DNAs. Using wild type RecA, joint molecule intermediates are seen at 5 min along with a small amount of the final reaction product, nicked circular DNA (Figure 5). The reaction goes to completion between approximately 30 to 45 min, in agreement with previous studies (21, 26). By comparison, joint molecule intermediates are not observed until 15 min in reactions containing the RecA-FD protein, and discernable amounts of product appear at approximately 45 min. The reaction is still progressing at 90 min at which time approximately 50% of the substrate duplex DNA has been converted to nicked circle products. Averages obtained from three separate experiments showed that the initial rate of strand exchange measured by substrate uptake was approximately 4-fold slower for the RecA-FD protein whereas the rate of appearance of final product (nicked circles) was approximately 8-fold slower.
Figure 5.
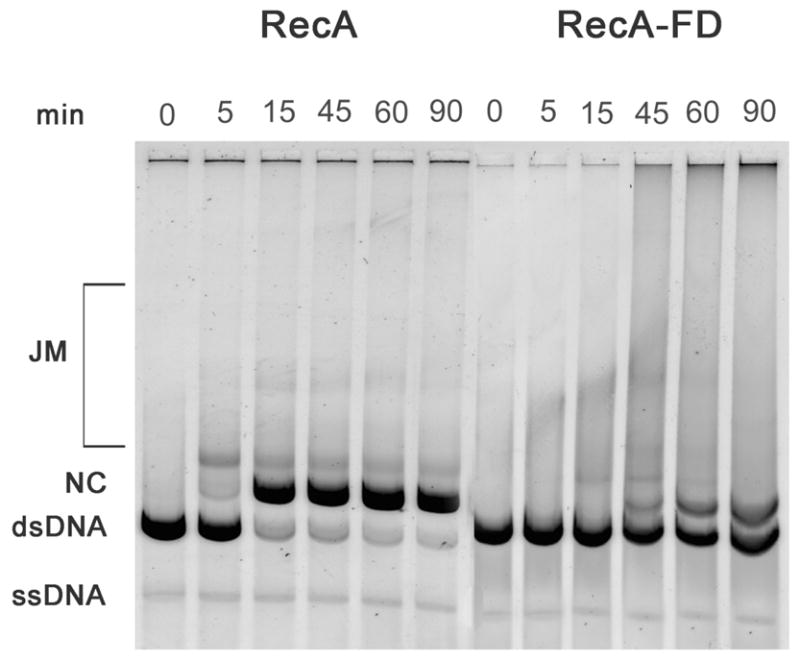
DNA strand exchange catalyzed by the RecA and RecA-FD proteins. Strand exchange reactions were performed as described (see EXPERIMENTAL PROCEDURES) and include 20 μM φX174 ss- and dsDNAs (concentration in bases), 6.0 μM RecA or RecA-FD proteins, 2.0 μM SSB and 3.0 mM ATP. Samples were removed at the indicated times and substrates and products were separated by agarose gel electrophoresis. Products of the reaction include joint molecule intermediates (JM) and final product nicked circles (NC).
DISCUSSION
In this study we show that a covalently fused dimer of the RecA protein is functional both in vivo and in vitro for activities associated with wild type RecA, e.g. repair of UV-mediated DNA damage, cleavage of the LexA repressor, DNA-dependent ATPase and DNA strand exchange. Measurements of DNA repair and co-protease activities carried out in vivo show that the RecA-FD protein is as active as wild type RecA. Biochemical assays show similar ATP turnover numbers for RecA-FD and wild type RecA, while the ssDNA binding affinity is reduced approximately 2-fold for the RecA-FD protein. DNA strand exchange assays show that the activity of RecA-FD is approximately 5–fold slower than that of wild type RecA, a result that may be in part attributed to the decrease in DNA binding affinity for the RecA-FD protein. We note that previous studies of the RecA K6A and RecA R28A mutant proteins show a decreased rate and extent of DNA strand exchange relative to wild type RecA (21). Therefore, the decrease in strand exchange activity of the RecA-FD protein carrying both substitutions is most likely due to the mutations themselves rather than any effect of the dimer fusion. We also note that different preparations of RecA-FD protein typically have a low level of monomeric RecA, between 2–5% of the total purified protein. Therefore, the observed activities clearly do not result from residual monomeric RecA, rather all activities measured reflect those catalyzed by the RecA-FD protein.
Many studies have shown that the oligomeric state of free RecA protein is exquisitely sensitive to a variety of solution conditions including protein concentration, ionic strength and temperature, and that at the protein concentrations typically used in DNA strand exchange assays RecA exists as a complex mix of various oligomeric forms (9–12, 27–30). The fact that the RecA-FD protein functions both in vivo and in vitro demonstrates that dimeric RecA can serve as the nucleating unit for filament assembly onto ssDNA. Biochemical and structural evidence support the idea that inactive oligomers of RecA must disassemble prior to the assembly of active nucleoprotein filaments (18, 19, 31–33), and our data now suggest that disassembly does not necessarily need to progress all the way to monomeric RecA. At the RecA concentrations present in cells both before DNA damage and following DNA damage-mediated induction of recA expression, it is likely that most RecA exists in some oligomeric form. This is supported by analyses performed by Ruigrok and DiCapua (28) in which they estimated that aggregates of RecA could account for greater than 70% of the total RecA in the cell. In fact, recent studies now provide a direct demonstration that greater than 50% of cellular RecA exists in clusters not associated with DNA (34). While solution conditions have been manipulated to show that monomeric RecA can indeed initiate nucleoprotein filament formation (35), the ability of dimeric RecA to serve as a nucleating unit of filament assembly on ssDNA may be a cellular necessity given that, as described above, significant amounts of monomeric RecA are not likely to be present especially following induction of recA expression.
The idea that there is functional coupling between subunits in an active recombinase nucleoprotein filament has been well established by numerous studies (17, 36). The cooperative binding of RecA and Rad51 to DNA demonstrates coordinated subunit interactions during nucleoprotein filament assembly (13, 15, 16), and earlier modeling of DNA binding data supported the idea that various oligomeric forms of RecA could interact directly with DNA (37). Additionally, recent studies describe a coordinated wave of ATP hydrolysis that propagates through a RecA filament that is likely linked to the enzyme’s motor function responsible for catalysis of DNA strand exchange (38). Although the number of subunits that form an interacting unit within a recombinase could not be identified in any of these studies, there is intriguing structural evidence supporting the idea that recombinase dimers may contribute significantly to the underlying functional coordination within a recombinase filament. Electron microscopic analyses of T. aquaticus RecA in the absence of DNA revealed that a large population of hexameric rings was divided between those with 6-fold symmetry and those with 3-fold symmetry, with the latter observed to be trimers of dimers (39). While at that time it was not clear whether this 3-fold symmetry would be maintained to any degree within an active recombinase filament, new structural data strongly supports this idea. In the X-ray structure of yeast Rad51 Conway et al. (40) find that alternate subunit interfaces are different and suggest that a dimer may be the functional unit within the active recombinase filament.
Our results clearly demonstrate that RecA monomers are not an obligatory protein unit required for nucleoprotein filament assembly. Together with the published data discussed above, our data support a model in which dimeric RecA can serve as a nucleating unit for assembly of an active nucleoprotein filament, and may also serve as a functional unit within the filament. Of course, this does not preclude monomers as also participating in these roles, and given the heterogeneous complexity of the RecA oligomeric population both in vitro and in vivo, it may be that more than one type of oligomer, e.g. both monomers and dimers, can carry out both functions.
Acknowledgments
We are grateful to Ms. Tuba Bas, Ms. Ellen Nalivaika and Dr. J.P. Verderese for their technical contributions, and to members of the Knight lab for helpful discussions and comments on the manuscript.
This work was supported by NIH grant GM44772 (K.L.K.).
Footnotes
Abbreviations: RecA-FD, RecA fused dimer; PCR, polymerase chain reaction; UV, ultraviolet; EDTA, ethylenediamine tetra-acetic acid; Tris, tris (hydroxymethyl) aminomethane; DTT, dithiothreitol
References
- 1.McGrew DA, Knight KL. Molecular design and functional organization of the RecA protein. Crit Rev Biochem Mol Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- 2.Bell CE. Structure and mechanism of Escherichia coli RecA ATPase. Mol Microbiol. 2005;58:358–366. doi: 10.1111/j.1365-2958.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 3.Radding CM. Helical RecA nucleoprotein filaments mediate homologous pairing and strand exchange. Biochim Biophys Acta. 1989;1008:131–145. doi: 10.1016/0167-4781(80)90001-9. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa T, Wabiko H, Tsurimoto T, Horii T, Masukata H, Ogawa H. Characteristics of purified recA protein and the regulation of its synthesis in vivo. Cold Spring Harb Symp Quant Biol 43 Pt. 1979;2:909–915. doi: 10.1101/sqb.1979.043.01.099. [DOI] [PubMed] [Google Scholar]
- 6.McEntee K, Weinstock GM, Lehman IR. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J Biol Chem. 1981;256:8835–8844. [PubMed] [Google Scholar]
- 7.Flory J, Radding CM. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982;28:747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 8.Cotterill SM, Satterthwait AC, Fersht AR. recA protein from Escherichia coli. a very rapid and simple purification procedure: binding of adenosine 5′-triphosphate and adenosine 5′-diphosphate by the homogeneous protein. Biochemistry. 1982;21:4332–4337. doi: 10.1021/bi00261a023. [DOI] [PubMed] [Google Scholar]
- 9.Brenner SL, Zlotnick A, Griffith JD. RecA protein self-assembly. Multiple discrete aggregation states. J Mol Biol. 1988;204:959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- 10.Brenner SL, Zlotnick A, Stafford WFr. RecA protein self-assembly. II. Analytical equilibrium ultracentrifugation studies of the entropy-driven self-association of RecA. J Mol Biol. 1990;216:949–964. doi: 10.1016/S0022-2836(99)80013-8. [DOI] [PubMed] [Google Scholar]
- 11.Wilson DH, Benight AS. Kinetic analysis of the pre-equilibrium steps in the self-assembly of RecA protein from Escherichia coli. J Biol Chem. 1990;265:7351–7359. [PubMed] [Google Scholar]
- 12.DiCapua E, Schnarr M, Ruigrok RW, Lindner P, Timmins PA. Complexes of RecA protein in solution. A study by small angle neutron scattering. J Mol Biol. 1990;214:557–570. doi: 10.1016/0022-2836(90)90198-U. [DOI] [PubMed] [Google Scholar]
- 13.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 14.Thresher RJ, Christiansen G, Griffith JD. Assembly of presynaptic filaments. Factors affecting the assembly of RecA protein onto single-stranded DNA. J Mol Biol. 1988;201:101–113. doi: 10.1016/0022-2836(88)90442-1. [DOI] [PubMed] [Google Scholar]
- 15.De Zutter JK, Knight KL. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J Mol Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- 16.Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein. III. Modulation of DNA binding by adenosine nucleotides. J Biol Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 17.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 18.Lee JW, Cox MM. Inhibition of recA protein promoted ATP hydrolysis. 1. ATP gamma S and ADP are antagonistic inhibitors. Biochemistry. 1990;29:7666–7676. doi: 10.1021/bi00485a016. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 20.Logan KM, Knight KL. Mutagenesis of the P-loop motif in the ATP binding site of the RecA protein from Escherichia coli. J Mol Biol. 1993;232:1048–1059. doi: 10.1006/jmbi.1993.1459. [DOI] [PubMed] [Google Scholar]
- 21.Eldin S, Forget AL, Lindenmuth DM, Logan KM, Knight KL. Mutations in the N-terminal region of RecA that disrupt the stability of free protein oligomers but not RecA-DNA complexes. J Mol Biol. 2000;299:91–101. doi: 10.1006/jmbi.2000.3721. [DOI] [PubMed] [Google Scholar]
- 22.Nastri HG, Knight KL. Identification of residues in the L1 region of the RecA protein which are important to recombination or coprotease activities. J Biol Chem. 1994;269:26311–26322. [PubMed] [Google Scholar]
- 23.Konola JT, Nastri HG, Logan KM, Knight KL. Mutations at Pro67 in the RecA protein P-loop motif differentially modify coprotease function and separate coprotease from recombination activities. J Biol Chem. 1995;270:8411–8419. doi: 10.1074/jbc.270.15.8411. [DOI] [PubMed] [Google Scholar]
- 24.Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Steady state kinetic analysis of ATP hydrolysis. J Biol Chem. 1981;256:8845–8849. [PubMed] [Google Scholar]
- 25.Logan KM, Forget AL, Verderese JP, Knight KL. ATP-mediated changes in cross-subunit interactions in the RecA protein. Biochemistry. 2001;40:11382–11389. doi: 10.1021/bi011081u. [DOI] [PubMed] [Google Scholar]
- 26.Nayak S, Bryant FR. Differential rates of NTP hydrolysis by the mutant [S69G]RecA protein. Evidence for a coupling of NTP turnover to DNA strand exchange. J Biol Chem. 1999;274:25979–25982. doi: 10.1074/jbc.274.37.25979. [DOI] [PubMed] [Google Scholar]
- 27.Heuser J, Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J Mol Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 28.Ruigrok RW, DiCapua E. On the polymerization state of recA in the absence of DNA. Biochimie. 1991;73:191–198. doi: 10.1016/0300-9084(91)90202-c. [DOI] [PubMed] [Google Scholar]
- 29.Budzynski DM, Gao X, Benight AS. Isolation, characterization, and magnesium-induced self-association kinetics of discrete aggregates of RecA protein from Escherichia coli. Biopolymers. 1996;38:471–491. doi: 10.1002/(sici)1097-0282(199604)38:4<471::aid-bip4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 30.Skiba MC, Logan KM, Knight KL. Intersubunit proximity of residues in the RecA protein as shown by engineered disulfide cross-links. Biochemistry. 1999;38:11933–11941. doi: 10.1021/bi991118z. [DOI] [PubMed] [Google Scholar]
- 31.Morrical SW, Cox MM. Light scattering studies of the recA protein of Escherichia coli: relationship between free recA filaments and the recA X ssDNA complex. Biochemistry. 1985;24:760–767. doi: 10.1021/bi00324a034. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW, Cox MM. Inhibition of recA protein promoted ATP hydrolysis. 2. Longitudinal assembly and disassembly of recA protein filaments mediated by ATP and ADP. Biochemistry. 1990;29:7677–7683. doi: 10.1021/bi00485a017. [DOI] [PubMed] [Google Scholar]
- 33.Roca AI, Singleton SF. Direct evaluation of a mechanism for activation of the RecA nucleoprotein filament. J Am Chem Soc. 2003;125:15366–15375. doi: 10.1021/ja0270165. [DOI] [PubMed] [Google Scholar]
- 34.Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- 35.Masui R, Mikawa T, Kato R, Kuramitsu S. Characterization of the oligomeric states of RecA protein: monomeric RecA protein can form a nucleoprotein filament. Biochemistry. 1998;37:14788–14797. doi: 10.1021/bi981296c. [DOI] [PubMed] [Google Scholar]
- 36.Cox MM. The bacterial RecA protein as a motor protein. Annu Rev Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi M. Analysis of DNA-RecA protein interactions involving the protein self-association reaction. J Biol Chem. 1989;264:288–295. [PubMed] [Google Scholar]
- 38.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Egelman EH. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Biol. 1997;4(2):101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 40.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]