Abstract
Immunoperoxidase histochemistry is a widespread method of assessing expression of biomolecules in tissue samples. Accurate assessment of the expression levels of genes is critical for the management of disease, particularly as therapy targeted to specific molecules becomes more widespread. Determining the quality of preservation of macromolecules in tissue is important to avoid false negative and false positive results. In this review we discuss (1) issues of sensitivity (false negativity) and specificity (false positivity) of immunohistochemical stains, (2) approaches to better understanding differences in immunostains done by different laboratories (including the recently proposed MISFISHIE specification for tissue localization studies), and (3) approaches to assessing the quality of preservation of macromolecules in tissue, particularly in small biopsy samples.
Keywords: Quality control, Immunoperoxidase, Quantum dots, Analyte, Sensitivity, Specificity, MISFISHIE
Introduction
Tissue specimens are a source of invaluable information for not only the diagnosis of disease, but also, to ever greater extent, for the management of disease. Historically, tissue biopsies, analyzed by hematoxylin and eosin stains that are often supplemented by additional special stains, have provided information for the basic classification of disease and characterization of neoplasms. The development of immunohistochemistry has provided a basis for the molecular characterization of tumors using antibodies to localize molecules expressed by tumor cells. As targeted therapy using agents, most of which are antibody derivatives or small drug molecules specific to designated molecules, have been incorporated into clinical management, the molecular characterization of tumors has become increasingly important. The differential expression of molecules by tumor cells provides a basis for both prognosis and for the selection of targeted therapies (Ross et al. 2004). Adding further impetus to selecting a therapy to which a specific neoplasm is sensitive is the risk of significant side effects. For example, ≥5% of patients receiving Trastuzumab as anti-her2 therapy for treating breast cancer have significant cardiac toxicity, resulting in potentially irreversible impaired cardiac pump function (Telli et al. 2007). This unanticipated side effect is consequent to suppression of her2-dependent cell survival pathways. Loss of these pathways permits the cardiotoxic effect of anthracyclines, which are administered concomitant with Trastuzumab, to become more readily manifest (Crone et al. 2002).
For these reasons determining the molecular phenotype of a tumor is an increasingly more important laboratory test. Essential to accurate characterization of the molecular phenotype of a tumor is determining the quality of preservation of macromolecules in tissue. In this short review I will focus on tumors, the identification and characterization of tumor tissue biomarkers by immunohistochemistry, and parameters that affect the preservation and detection of these macromolecules. In current practice immunoperoxidase histochemistry is the method that is most widely used to determine the molecular phenotype of tumor cells. That immunohistochemical methods are so useful is consequent to (1) the fact that cell and/or tissue architecture is preserved so that the cell types expressing the gene product(s) can be identified, and (2) the sensitivity of immunostains, i.e. single tumor cells can serve as the basis for characterizing a tumor.
Correspondingly, issues of characterizing, controlling, and optimizing the quality of tissues samples has become paramount to minimize the frequency with which “false negative” and false positive results from molecular analysis of tumors by immunohistochemical methods occurs. At a basic level, assessment of biomolecule expression in tissue deals with sensitivity of the assay (if the molecule is present, will the assay detect it?) and specificity (if the molecule is present might the assay erroneously report its presence?). Our focus will be on the three-step indirect immunoperoxidase stain, which is currently the most widely used method for assessing gene product expression in tissue.
Sensitivity
With respect to sensitivity—is the molecule present or absent—let us consider the following sources of decreased sensitivity that can result in a false negative immunohistochemical stain.
The epitopes to which antibodies are directed are too few to be detected by light microscopy.
The epitopes are not available to bind the specific antibodies despite presence of the molecule due to conformation features. This is exemplified by differential staining using different antibodies to MUC1 and to endothelin.
Epitopes can only be made available to binding the antibody after an antigen retrieval technique, using either proteolytic enzymes or heating the tissue section in a buffer.
The primary antibodies have binding constants that are too low to be retained in the section during the multiple steps of tissue processing. For example, antibodies used in ELISA assays are of lower affinity than those used in immunohistochemical stains since the former type of reaction is a competitive assay and the latter reaction depends upon high affinity binding to survive the multiple steps of processing the tissue section.
The molecule may be within a compartment of the cell that is not accessible to the immunohistochemical reagents. Although this limitation is typically not a problem when using sections of tissue, immunostains of intact cells using whole immunoglobulin molecules that may be conjugated to other molecules such as biotin, might not be able to penetrate to the compartment of the cell in which the antigen is located.
Degradation of the molecule: The rate of loss of immunoreactivity varies with the molecule. In one study, estrogen receptor immunoreactivity could be detected in sections from paraffin blocks that had been stored for up to 60 years (Camp et al. 2000). Conversely, in sections that were not paraffin-coated and stored in a nitrogen environment but that were exposed to ambient atmosphere at room temperature, immunoreactivity of keratin, estrogen receptor, p53 and Ki-67 was lost within 3 months (DiVito et al. 2004; Jacobs et al. 1996).
Tissue processing: The multiple steps of tissue processing affect antigens differently. In one study, UCHL1 and vimentin were most susceptible to the duration and pH of fixation and to the type of fixative. Conversely, L26 was most sensitive to changes in tissue processing (Williams et al. 1997). Another study reported that the frequency of KI-67-positive tumor cells was dependent, in part, on the nature and duration of steps in the immunohistochemistry procedure (Mengel et al. 2002).
Understanding the chemistry of what happens to molecules and their epitopes in tissue is important for developing better methods for assessing the quality of protein in tissue. For example, heating of tissue sections, typically by microwave, increases the detectability of most antigens (Cuevas et al. 1994). Although the mechanism by which epitopes become exposed is unknown, a series of immunohistochemical experiments using antibodies to nuclear, cytoplasmic, cell membrane, and extracellular antigens applied at a range of different pH’s and of salt concentrations provides evidence that epitopes are variably exposed (in a molecule-specific manner) by electrostatic forces that may prevent refolding to a non-immunoreactive conformation (Emoto et al. 2005).
Formaldehyde fixation itself has chemical complexities that are probably not widely known. Understanding the chemistry of formaldehyde fixation is a step in trying to predict the effect of tissue processing on immunoreactivity of a given molecule. During formaldehyde fixation an equilibrium between formaldehyde, as carbonyl formaldehyde, and methylene glycol is established. The formation of the glycol explains why formaldehyde penetrates rapidly (as methylene glycol) and fixes slowly (as carbonyl formaldehyde). For example, 16-µ thick sections maximize incorporation of covalently linked formaldehyde only after 24 h at 37°C and after 48 h at 25°C (Fox et al. 1985). Consequently, endogenous proteases exposed to fixative for less than 24 h at 37°C may still be active and capable of degrading tissue biomarker macromolecules.
Thus, understanding the details of these two parameters of tissue processing—epitope exposure and formaldehyde fixation—begins to provide a basis for establishing conditions that optimize the quality of macromolecule preservation in tissue.
Specificity
Non-specificity of an immunohistochemical stain may lead to a false positive stain, resulting in a report that the molecule is expressed in that tissue, whereas, in reality, it is not. We can subcategorize sources of non-specific reaction product into immunologic and non-immunologic causes. With respect to immunologic sources of non-specific reactions:
The epitope may be sufficiently similar to the epitope of completely unrelated molecules, due to a combination of binding affinity and structural homology, as to result in high-affinity binding of the primary antibody to an unrelated and unintended molecule.
The epitope may be a non-protein moiety that is a post-translational modification common to functionally unrelated molecules.
Non-immunologic causes leading to a false positive stain include the following:
Leakage of antigen into different tissue compartments. This varies with the molecule. In a post-mortem study of the effect of autolysis on antigen localization, alpha-1-antichymotrypsin, alpha-2-macroglobulin, and fibronectin were mislocalized, whereas lysozyme was not (Fieguth et al. 1997).
Expression of endogenous peroxidase in cells at a sufficiently high level to produce a signal independent of the immunolocalization reaction. Cells that express a particularly high-level of peroxidase-like activity include red blood cells and neutrophils. This source of “false positivity” can often be obviated by pre-incubation of the sections in hydrogen peroxide.
Expression of biotin at sufficiently high levels as to bind the avidin–biotin–peroxidase complex of an immunoperoxidase procedure. Renal tubular epithelial cells and hepatocytes, which contain high concentrations of biotin, can produce a reaction product that is misinterpreted as a positive immunoperoxidase stain when biotin conjugated reagents are used. Methods to suppress endogenous biotin activity may not by fully effective in preventing this source of false positivity.
Idiopathic causes: False positivity can also be associated with necrosis and with other curious artifacts, such as nuclear staining using antibodies to the cytoskeletal protein vimentin. There is no ready explanation for these phenomena.
A frequent source of “false positivity” results from overexposure of the tissue to the immunoperoxidase reaction. An example is the edge effect where tissue lifts from the slide and both sides of the section are exposed to the immunoperoxidase reaction.
Even if all sources of non-immunologic false positivity can be dealt with, the question of whether specificity can ever be proven remains (Swaab et al. 1977). Competition with or pre-absorption of the antibody with excess peptide controls for the antigen, demonstrating that localization of the antibody is consistent with binding to that antigen. However, failure of pre-absorption to abolish immunoreactivity is not necessarily evidence that the antibody binding to the sought protein has not occurred since the conformation of the protein in tissue might be different than that of the free peptide. Control for antibody specificity is provided, in part, by immunoprecipitation and demonstration that the precipitated protein has the biochemical characteristics of the sought protein.
Quality of tissue
From the perspective of assessing and controlling the quality of tissue, let us first consider tissue handling. Historically, the “quality” of a sample has been based on the microscopic assessment of such features as necrosis, crush artifact, and biologic phenomena that might affect gene expression such as hemorrhage and inflammation. However, there is not a good correlation between such histological features as necrosis and retention of molecules that can be detected by immunohistochemistry. For example, cells types can be identified and characterized by immunostains in tissue that at the light microscopic level is totally necrotic. Our ability to predict retention of immunoreactivity in such a situation is imperfect. Whether or not necrotic tissue contains epitopes that are immunoreactive is unpredictable.
Historically, the quality of RNA and DNA has been assessed in samples of tissue large enough to analyze by, respectively, Northern and Southern blots. Molecular amplification techniques have permitted characterization of nucleic acids extracted from such small samples as needle biopsies of tissue using reverse transcriptase polymerase chain reaction. Instruments such as the Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) can assess the quality of nucleic acids based on degree of fragmentation. As little as 1 µl of solution can be analyzed for expression of as little as 1 ng of nucleic acid.
The lability of macromolecules to certain conditions of tissue handling has been characterized at the nucleic acid level. Gene expression profiles have been characterized for warm ischemia time (during surgery when the prostate has been devitalized by clamping vessels but not yet removed from the patient for processing) (Lin et al. 2006) and for autolysis (while the prostate sits on a bench at room temperature) (Dash et al. 2002). These profiles can be used to “correct” expression profiles of tissue samples. Immunohistochemical assessment of expression of a set of antigens in tissue let sit at 37° C for a range of times (0, 4, 8, 12, 24, 48, and 72 h) yields thematically similar findings. Although Leu2a immunoreactivity was preserved for only 12 h, CLA and UCHL-1 immunoreactivity was retained for at least 72 h (Pelstring et al. 1991).
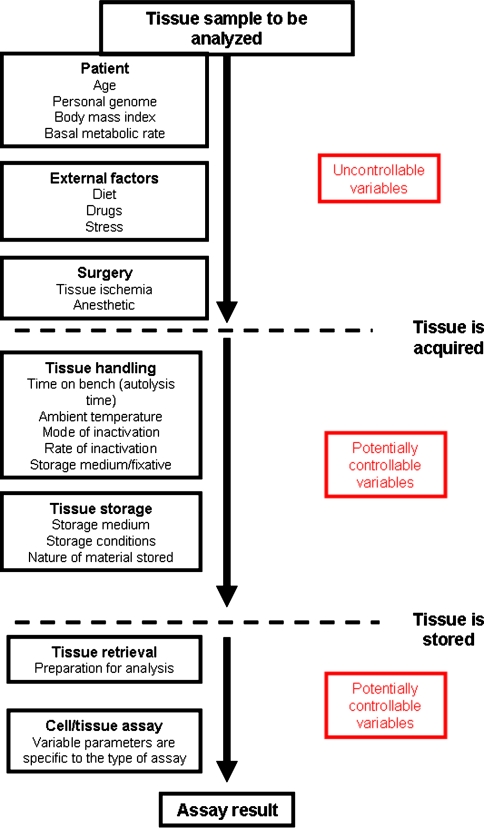
However, these ischemia and autolysis profiles analyze only two of numerous steps in tissue processing (Fig. 1). Furthermore, the quality of RNA may not accurately reflect levels of protein expression since expression levels of mRNA and of the corresponding protein translation product may differ, for some genes up to 5-fold (Pascal et al. 2008). Thus, these profiles do not provide a particularly good basis for assessing the quality of protein in tissue. The assessment of the quality of proteins in tissues, particularly very small samples, is a significant challenge since there is no current method to amplify proteins for analysis. Furthermore, since expression levels of protein differ widely in tissue, there is no protein that can be used as a marker for the overall quality of protein in tissue. That said, there are specific gene products that have been used as markers of extent of preservation/fixation in buffered formalin—p27 (De Marzo et al. 2002) and vimentin (using an antibody to an epitope of vimentin that is partially susceptible to formaldehyde fixation) (Battifora 1991). These potential markers of tissue preservation have not seen general use.
Fig. 1.
Schematic of steps in tissue processing with examples of potential sources of variance in gene expression levels. These steps are classified into those steps over which the investigator has no control, i.e. either the events occurred prior to receipt of tissue or obtaining the tissue was obtained in a setting where clinical needs were paramount, or those that the investigator can potentially modify
Development of the MISFISHIE standard
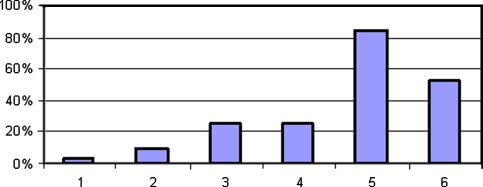
What does seem important as an initial step in determining the overall quality of macromolecules in tissue studies is identification of the critical steps that influence gene expression. To explicitly identify these steps we have proposed a specification for all tissue localization experiments—by immunohistochemical and in-situ hybridization methods (see http://www.scgap.systemsbiology.net/standards/misfishie/ and http://www.mged.sourceforge.net/misfishie/) (Deutsch et al. 2008). Termed MISFISHIE (an abbreviation of Minimum Information Specification For In Situ Hybridization and Immunohistochemistry Experiments), and developed by a large group of colleagues working in different fields of biomedicine, the specification stipulates the steps of all such experiments that should be documented and characterized in detail (Fig. 1). The principle of this specification is to ensure that the minimum information that a researcher at a different lab needs to reproduce or evaluate the experiment is provided. MISFISHIE does not specify the data format or in any manner limit the nature of the experiment. MISFISHIE merely stipulates what information should be communicated. Ideally these principles are being used by authors and are looked for by reviewers and editors in publications of such experiments. However, this is not uniformly the case. A significant number of publications in a wide range of journals fail to satisfy MISFISHIE standards by not providing sufficient detail to have confidence that the experiment can be repeated, as we found in a retrospective review of 30 published articles (Fig. 2). Since the conclusions of so many current immunohistochemical studies are based on interpretations of images, and since image interpretation is a source of observer variability, one section of MISFISHIE requests that all images be made available for independent interpretation (Pascal et al. 2007). Although this expectation is currently onerous, given the effort necessary to obtain the images and the disc storage required to save the images, we think that making images available for independent review and interpretation will be a long-term benefit to biomedical science.
Fig. 2.
Percentage of 30 randomly selected papers that would not have satisfied the MISFISHIE specification, by MISFISHIE category (key: 1 Experimental Design; 2 Biomaterials and Treatment, i.e. tissue processing, fixation medium, storage conditions; 3 Reporters, i.e. antibody source, clone, concentration; 4 Staining, i.e. antigen retrieval method, staining protocol; 5 Imaging Data, i.e. images of immunostains; 6 Image Characterization, i.e. algorithm for reporting and analyzing the immunostaining images, criteria for positivity
By applying MISFISHIE rules to experiments all potential sources of variance in tissue based experiments will, in principle, be revealed. Although documenting all steps of immunohistochemical experiments provides a basis for targeting those parts of the procedure that are the source of greatest variance, implementation of quality control procedures is the next step. Using on slide positive and negative control tissues (Mengel et al. 2005) and parallel immunostains of tissue microarrays that contain a variety of control tissues (Hsu et al. 2002) are ways to increase confidence that an immunohistochemical finding is accurate.
Standardization
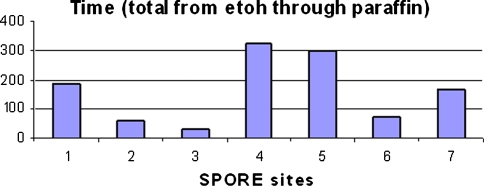
Standardization of procedures in different labs would increase the reliability of an immunohistochemical procedure. A step in addressing standardization of tissue processing is to document how different labs process their samples. A component of an ongoing study by the NIH-funded Prostate Cancer SPORE’s was documenting the timing for different steps of tissue processing at the SPORE sites. Data recorded by the tissue processors illustrate the range in time for processing tissue from ethanol into paraffin—0 to 310 min (Fig. 3). This sequence is but one set of intermediate steps in the tissue processing cycle. The effect of this range of processing time on antigen immunoreactivity is being assessed. Unfortunately, documentation of the time required for the entire sequence of tissue processing is virtually never done due to the imprecision and impracticality of being able to accurately record the time required for many steps, such as time the tissue sits at ambient temperature on the bench before fixation and time in formalin before initiating tissue processing.
Fig. 3.
Time to process blocks of formaldehyde-fixed, tissue by tissue processors at seven different institutions (NIH-funded Prostate Cancer SPORE’s) from the first ethanol dehydration step through paraffin infiltration (Fine SW, Trock B, Reuter VE, Ayala G, Cheville JC, Fearn P, Jenkins RB, Knudsen BS, Loda M, Netto GJ, Said J, Shah RB, Simko J, Troncoso P, True LD, Yang XJ, Rubin MA, DeMarzo AM (2007) Effects of tissue processing on biomarker analysis in prostate needle biopsies: a multi-institutional study. Annual Meeting of US–Canadian Academy of Pathology
Although desirable, standardization of routine tissue handling procedures and of tissue localization methods does not appear imminent due to clinical and practical considerations, which differ by laboratory, and the absence of compelling evidence that such standardization will be of value. What would be of value would be developing a metric to assess the quality of preservation of macromolecules in fixed tissue. Methods are being applied that measure integrity of nucleic acids in fixed tissue (Jewell et al. 2002). Although assessment of protein integrity is a greater challenge, mass spectrometry is being used as a protein integrity assessment tool (Shi et al. 2006).
“Quantification” by immunoperoxidase histochemistry
The conditions necessary to ensure that tissue quality is sufficient to minimize the possibility of false negative and false positive immunohistochemical results pale at the challenge posed by the goal of obtaining reliable and reproducible quantifications of molecules in sections of tissue. To truly quantify levels of gene expression by immunohistochemistry entails developing a standard curve for each antigen and each immunohistochemical method. Based on a standard curve, where known (either absolute or relative) numbers of molecules per unit of tissue (or, ideally, per average cell), is determined at multiple concentrations over the range of anticipated expression level in the tissue of interest, the expression level of the molecule of interest can be calculated. However, with only rare exceptions, expression levels of gene products are “quantified” by immunohistochemical methods and published without reference to a standard curve for that gene produce in that tissue. In a study that exemplifies the use of a standard curve, levels of a gene product were measured by correlating the optical density of immunoperoxidase stained sections to the amount of the same protein, which was extracted and measured by radioimmunoassay from an adjacent, histologically homologous tissue sample (Gross and Rothfeld 1985). Although developing such reference standards is an expensive and laborious process, it is the only way to ensure that the optical density of an immunoperoxidase stained section reflects gene expression level. The challenge to using immunohistochemical stains to quantify molecules is that sources of variance are numerous (True 1988). And, yet, the issue is critical. As just one example, the concentration of biomarker antibody appears to dramatically affect the apparent relationship between her2 expression and prognosis of patients with breast cancer (McCabe et al. 2005).
Despite these caveats, many investigators have “quantified” immunostains by assigning numbers to ranges of staining intensity and summarizing the results as single value numbers. For example, estrogen receptor (ER) immunostains have been reported as continuous variables, termed by some the H Score (McClelland et al. 1990; McCarty et al. 1985). An assumption made in these calculations is that these numerical values represent a continuous variable. However, that assumption has not been validated. For example, there is no evidence that a breast cancer expressing ER with an H score of 200 has twice the per tumor cell ER content as a breast cancer with an H score of 100. The use of letters instead of numbers would make explicit the fact that interpretations of immunostains provide categorical, not continuous, variables.
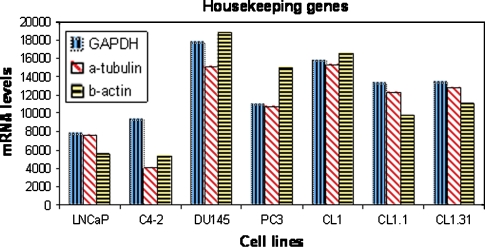
With respect to quantification of gene products, housekeeping genes have traditionally been used as reference levels for the quantification of nucleic acids (de Kok et al. 2005). In principle, the products of housekeeping genes can serve as metrics for the quality of preservation of the nucleic acids. However, there appears to be no single housekeeping gene that can be relied upon to be expressed by the range of cells in a given tissue that might be usable as a standard to assess quality of preservation of nucleic acids. For example, GAPDH is an androgen regulated gene which, due to the androgen regulation nature of the gene, is variably expressed in tissue that may be subjected to different androgen concentrations. There may well not be a single, or, even, set of housekeeping genes that remains invariant throughout different conditions (Thellin et al. 1999; Jain et al. 2006). This observation is true even for such a pure cell population as those of cell lines (Fig. 4).
Fig. 4.
Levels of mRNA (in relative units) of three putative housekeeping genes by seven prostate cancer cell lines. Note the wide range of expression levels of each gene (unpublished work by Mengchu Wu, Ilsa Coleman, and Peter Nelson)
Consequently, there appears to be little immediate prospect that there are housekeeping proteins or sets of proteins that can serve as a basis for assessing protein quality in tissue. Furthermore, based on all the sources of variance of immunohistochemical stains, immunohistochemistry seems an unlikely method to assess protein integrity in tissue. To cite one more example of how methodology can have a profound affect on extent of immunohistochemical reactivity, different antigen retrieval techniques have a variable effect. More cells in tissue sections that were heated prior to immunostaining expressed vimentin immunoreactivity than did those in sections that were treated with proteolytic enzymes as the antigen retrieval method (Kahveci et al. 2003; Hazelbag et al. 1995).
Heterogeneity
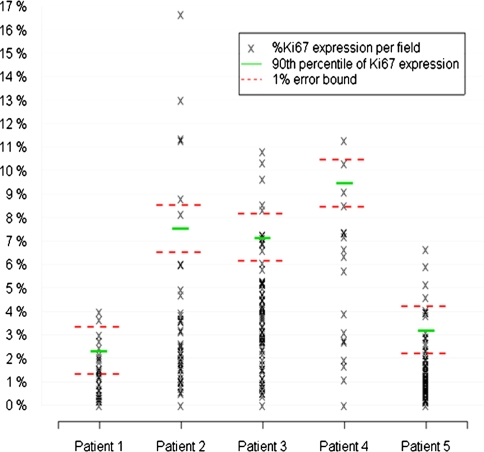
One aspect of analyzing tissue samples that can be a major challenge for accurate molecular phenotyping of tumors is the heterogeneity of tumor cell phenotypes within a given tumor. For example, assessing breast cancer tissue for expression of her2 or of estrogen receptor in a sample of “insufficient” size may not accurately reflect the molecular status of a tumor (Moeder et al. 2007; Chung et al. 2007). The extent of heterogeneity and the challenge posed to accurate sampling varies with both uniformity and frequency of expression of the molecule. Obtaining a sample that is representative of the status of that macromolecule in the tumor is particularly challenging for molecules expressed at low frequencies by cells that are not uniformly distributed throughout a tumor. In a study of expression of one such antigen, we tabulated the frequency of expression of Ki-67 (Fig. 5). The fact that in one case, which exhibited the greatest range of Ki-67-positivity (patient 2), 40 (of 100 possible) microscopic fields would need to have been analyzed to find the 90th percentile of KI-67 expression emphasizes the challenge of dealing with heterogeneity and in developing a strategy to sample the most clinically relevant part of a tumor. As an aside, extent of heterogeneity of gene expression might reflect tumor biology. For example, tumor cells in breast carcinomas that vary in extent of expression of estrogen receptor appear to be more likely to fail systemic therapy than those tumors that exhibit more uniform expression of the receptor (Sklarew et al. 1990).
Fig. 5.
The percentage of prostate carcinoma cells that are Ki67-positive was counted in 100 sequential 1-mm diameter optical fields in a section of cancer from the radical prostatectomy from each of 5 patients. Note the wide range of Ki67-positivity in each section (Roudier MP, Hawley S, Etzioni R, Luthringer D, True LD (2003) Heterogeneity of Ki-67 expression in prostate cancer. Implications for tissue microarray design. Annual Meeting of US-Canadian Academy of Pathology, March 2003)
Thus, developing a strategy for sampling a tumor for a specific gene product is an important goal. One approach would be to sample the specimen for the specific analyte until the variance of the running mean of the measured value does not exceed a pre-determined value, such as 5% (Dunnill 1985). However, this strategy may be inadequate for gene products expressed in very small samples at low levels, such as Ki-67 in prostate carcinoma. Sampling algorithms that report the error range need to be developed for such clinical scenarios.
Future possibilities
Since, at least for the foreseeable future, tissue specimens in the form of biopsies will continue to be the source of tumor cells to be analyzed for presence and expression level of specific molecules, better methods of assessing quality of preservation of the macromolecules will need to be developed. There are several possible solutions to this challenge, such as:
Sets of housekeeping proteins that are specific to the type of tissue will be developed. Given the realization that there appears to be no single or, even, set of housekeeping genes that can serve as a universal metric for tissue quality (as discussed above), tumor-specific sets of proteins may have to be developed. Complementing this development would have to be methods to measure proteins in samples as small as needle biopsies, which have an estimated mass of no more than 0.000054 g.
A mass spectrometry method to assay the quality of protein in a tissue may provide a better method than an immunohistochemical stain. However, this is speculative. Mass spectrometry profiles of formalin-fixed, paraffin-embedded samples would have to be developed. And, due to the variably fragmented nature of proteins in these samples, mass spectrometry may be inadequate to assess degree of protein integrity.
A better assay tool than immunoperoxidase stains may be quantum dot-based immunohistochemical stains (Smith et al. 2006; True and Gao 2007). Quantum dot-based reagents provide several advantages. As light emitting markers they have a wider dynamic range than the optical density of immunoperoxidase stains. Since the emitted light is intense, quantum dots can be conjugated directly to the primary antibodies, minimizing the number of steps and, consequently, decreasing the sources of variance. Finally, due to their narrow emission spectra, quantum dots of different sizes (and, hence, having different emission spectra) can be conjugated to antibodies and multiplexed to immunohistochemically characterize a single tissue section.
Even with these possibilities, optimism must be tempered with acknowledgement that the functional state of molecules often depends of post-translational modifications of proteins. For example, the phosphorylated state of a protein is often the active form of the protein. However, assaying the extent of phosphorylation of a protein in a sample of tissue is subject to many additional sources of variance. Antibodies may not be immunoreactive to fixed tissue, though they are reactive with the soluble protein. And, the state of phosphorylation may both be transient or, alternatively, may be artifactually induced by the process of handling tissue (Mandell 2003).
To conclude, the challenges are multiple and large. But, the need for better metrics to assess the quality of protein and of other macromolecules in tissue is urgent and timely.
Acknowledgments
In addition to the co-authors of manuscripts cited in the text and in figure legends, particularly Dr. Alvin Liu and Dr. Eric Deutsch for their role in developing the MISFISHIE specification, discussions over many years with Dr. Fre Bosman have provided me continual intellectual stimulus for this topic.
References
- Battifora H (1991) Assessment of antigen damage in immunohistochemistry. The vimentin internal control. Am J Clin Pathol 96:669–671 [DOI] [PubMed]
- Camp RL, Charette LA, Rimm DL (2000) Validation of tissue microarray technology in breast carcinoma. Lab Invest 80:1943–1949 [DOI] [PubMed]
- Chung GG, Zerkowski MP, Ghosh S et al (2007) Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest 87:662–669 [DOI] [PubMed]
- Crone SA, Zhao YY, Fan L et al (2002) ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med 8:459–465 [DOI] [PubMed]
- Cuevas EC, Bateman AC, Wilkins BS et al (1994) Microwave antigen retrieval in immunocytochemistry: a study of 80 antibodies. J Clin Pathol 47:448–452 [DOI] [PMC free article] [PubMed]
- Dash A, Maine IP, Varambally S et al (2002) Changes in differential gene expression because of warm ischemia time of radical prostatectomy specimens. Am J Pathol 161:1743–1748 [DOI] [PMC free article] [PubMed]
- de Kok JB, Roelofs RW, Giesendorf BA et al (2005) Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 85:154–159 [DOI] [PubMed]
- De Marzo AM, Fedor HH, Gage WR et al (2002) Inadequate formalin fixation decreases reliability of p27 immunohistochemical staining: probing optimal fixation time using high-density tissue microarrays. Hum Pathol 33:756–760 [DOI] [PubMed]
- Deutsch EW, Ball CA, Berman JJ et al (2008) Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE). Nat Biotechnol 26:305–312 [DOI] [PMC free article] [PubMed]
- DiVito KA, Charette LA, Rimm DL et al (2004) Long-term preservation of antigenicity on tissue microarrays. Lab Invest 84:1071–1078 [DOI] [PubMed]
- Dunnill MS (1985) Some statistical aspects of sampling in morphometry. Anal Quant Cytol Histol 7:250–255 [PubMed]
- Emoto K, Yamashita S, Okada Y (2005) Mechanisms of heat-induced antigen retrieval: does pH or ionic strength of the solution play a role for refolding antigens? J Histochem Cytochem 53:1311–1321 [DOI] [PubMed]
- Fieguth A, Kleemann WJ, von Wasielewski R et al (1997) Influence of postmortem changes on immunohistochemical reactions in skin. Int J Legal Med 110:18–21 [DOI] [PubMed]
- Fox CH, Johnson FB, Whiting J et al (1985) Formaldehyde fixation. J Histochem Cytochem 33:845–853 [DOI] [PubMed]
- Gross DS, Rothfeld JM (1985) Quantitative immunocytochemistry of hypothalamic and pituitary hormones: validation of an automated, computerized image analysis system. J Histochem Cytochem 33:11–20 [DOI] [PubMed]
- Hazelbag HM, van den Broek LJ, van Dorst EB et al (1995) Immunostaining of chain-specific keratins on formalin-fixed, paraffin-embedded tissues: a comparison of various antigen retrieval systems using microwave heating and proteolytic pre-treatments. J Histochem Cytochem 43:429–437 [DOI] [PubMed]
- Hsu FD, Nielsen TO, Alkushi A et al (2002) Tissue microarrays are an effective quality assurance tool for diagnostic immunohistochemistry. Mod Pathol 15:1374–1380 [DOI] [PubMed]
- Jacobs TW, Prioleau JE, Stillman IE et al (1996) Loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer. J Natl Cancer Inst 88:1054–1059 [DOI] [PubMed]
- Jain M, Nijhawan A, Tyagi AK et al (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345(2):646–651 [DOI] [PubMed]
- Jewell SD, Srinivasan M, McCart LM et al (2002) Analysis of the molecular quality of human tissues: an experience from the Cooperative Human Tissue Network. Am J Clin Pathol 118:733–741 [DOI] [PubMed]
- Kahveci Z, Minbay FZ, Noyan S et al (2003) A comparison of microwave heating and proteolytic pretreatment antigen retrieval techniques in formalin fixed, paraffin embedded tissues. Biotech Histochem 78:119–128 [DOI] [PubMed]
- Lin DW, Coleman IM, Hawley S et al (2006) Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol 24:3763–3770 [DOI] [PubMed]
- Mandell JW (2003) Phosphorylation state-specific antibodies: applications in investigative and diagnostic pathology. Am J Pathol 163:1687–1698 [DOI] [PMC free article] [PubMed]
- McCabe A, Dolled-Filhart M, Camp RL et al (2005) Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst 97:1808–1815 [DOI] [PubMed]
- McCarty KS Jr, Miller LS, Cox EB et al (1985) Estrogen receptor analyses Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109:716–721 [PubMed]
- McClelland RA, Finlay P, Walker KJ et al (1990) Automated quantitation of immunocytochemically localized estrogen receptors in human breast cancer. Cancer Res 50:3545–3550 [PubMed]
- Mengel M, von Wasielewski R, Wiese B et al (2002) Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J Pathol 198:292–299 [DOI] [PubMed]
- Mengel M, Hebel K, Kreipe H et al (2005) Standardized on-slide control for quality assurance in the immunohistochemical assessment of therapeutic target molecules in breast cancer. Breast J 11:34–40 [DOI] [PubMed]
- Moeder CB, Giltnane JM, Harigopal M et al (2007) Quantitative justification of the change from 10% to 30% for human epidermal growth factor receptor 2 scoring in the American Society of Clinical Oncology/College of American Pathologists guidelines: tumor heterogeneity in breast cancer and its implications for tissue microarray based assessment of outcome. J Clin Oncol 25:5418–5425 [DOI] [PubMed]
- Pascal LE, Deutsch EW, Campbell DS et al (2007) The urologic epithelial stem cell database (UESC)—a web tool for cell type-specific gene expression and immunohistochemistry images of the prostate and bladder. BMC Urol 7:19 [DOI] [PMC free article] [PubMed]
- Pascal LE, True LD, Campbell DS et al (2008) Correlation of mRNA and protein levels: cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics 9:246 [DOI] [PMC free article] [PubMed]
- Pelstring RJ, Allred DC, Esther RJ et al (1991) Differential antigen preservation during tissue autolysis. Hum Pathol 22:237–241 [DOI] [PubMed]
- Ross JS, Schenkein DP, Pietrusko R et al (2004) Targeted therapies for cancer 2004. Am J Clin Pathol 122:598–609 [DOI] [PubMed]
- Shi SR, Liu C, Balgley BM et al (2006) Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem 54:739–743 [DOI] [PubMed]
- Sklarew RJ, Bodmer SC, Pertschuk LP (1990) Quantitative imaging of immunocytochemical (PAP) estrogen receptor staining patterns in breast cancer sections. Cytometry 11:359–378 [DOI] [PubMed]
- Smith AM, Dave S, Nie S et al (2006) Multicolor quantum dots for molecular diagnostics of cancer. Expert Rev Mol Diagn 6:231–244 [DOI] [PubMed]
- Swaab DF, Pool CW, Van Leeuwen FW (1977) Can specificity ever be proved in immunocytochemical staining. J Histochem Cytochem 25:388–391 [DOI] [PubMed]
- Telli ML, Hunt SA, Carlson RW et al (2007) Trastuzumab-related cardiotoxicity: calling into question the concept of reversibility. J Clin Oncol 25:3525–3533 [DOI] [PubMed]
- Thellin O, Zorzi W, Lakaye B et al (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295 [DOI] [PubMed]
- True LD (1988) Quantitative immunohistochemistry: a new tool for surgical pathology? Am J Clin Pathol 90:324–325 [DOI] [PubMed]
- True LD, Gao X (2007) Quantum dots for molecular pathology: their time has arrived. J Mol Diagn 9:7–11 [DOI] [PMC free article] [PubMed]
- Williams JH, Mepham BL, Wright DH (1997) Tissue preparation for immunocytochemistry. J Clin Pathol 50:422–428 [DOI] [PMC free article] [PubMed]