Abstract
A new bisintercalating anthracycline (WP762) has been designed, in which monomeric units of daunorubicin have been linked through their amino groups on the daunosamine moieties using an m-xylenyl linker. Differential scanning calorimetry and UV melting experiments were used to measure the ultratight binding of WP762 to DNA. The binding constant for the interaction of WP762 with herring sperm DNA was determined to be 7.3 (±0.2) × 1012 M−1 at 20°C. The large favorable binding free energy of −17.3 kcal mol−1 was found to result from a large negative enthalpic contribution of −33.8 kcal mol−1 and an opposing entropic term (−TΔS = +16.5 kcal mol−1). A comparative molecular modeling study rationalized the increased binding by the m-xylenyl linker of WP762 positioning in the DNA minor groove compared to the p-xylenyl linker found in WP631, the first bis-anthracycline of this type. The cytotoxicity of WP762 was compared to that of other anthracyclines in Jurkat T lymphocytes. These studies, together with an analysis of the cell-cycle traverse in the presence of WP762, suggest that in these cells the new drug is more cytotoxic than the structurally related WP631.
INTRODUCTION
The anthracycline antibiotics, which include daunorubicin and doxorubicin (Adriamycin), have broad utility in the treatment of several malignancies.1–4 Although there have been intense efforts to synthesize more efficacious anthracyclines with fewer toxic side effects, these efforts have not succeeded, and the parent compounds currently remain the best chemotherapeutic agents of this class.2,4 Anthracyclines bind tightly to DNA, and are useful in modular drug design.4–7 Daunorubicin binding to DNA has been thoroughly characterized8–10 and the information obtained has provided a foundation for the rational design of new bisanthracyclines targeted toward DNA.5,7,11 The logic of the design of WP631 (Figure 1), a new bisanthracycline composed of two daunorubicin monomers connected via a p-xylenyl linker, was based on the high-resolution crystallographic structure of two separate daunorubicin monomers bound to a DNA oligonucleotide.10 In these daunorubicin-DNA complexes, each drug molecule was intercalated at either end of a hexanucleotide, with the daunosamine moieties of the two drug molecules pointing toward one another, lining the minor groove.10 This arrangement brings the reactive amine substituents of each drug molecule to within 7 Å of one another. Cross-linking these amines by means of a p-xylenyl linker yielded WP631. Biophysical and crystallographic studies have demonstrated on a detailed molecular level that this molecular design strategy was a success.5,7,12,13
Figure 1.
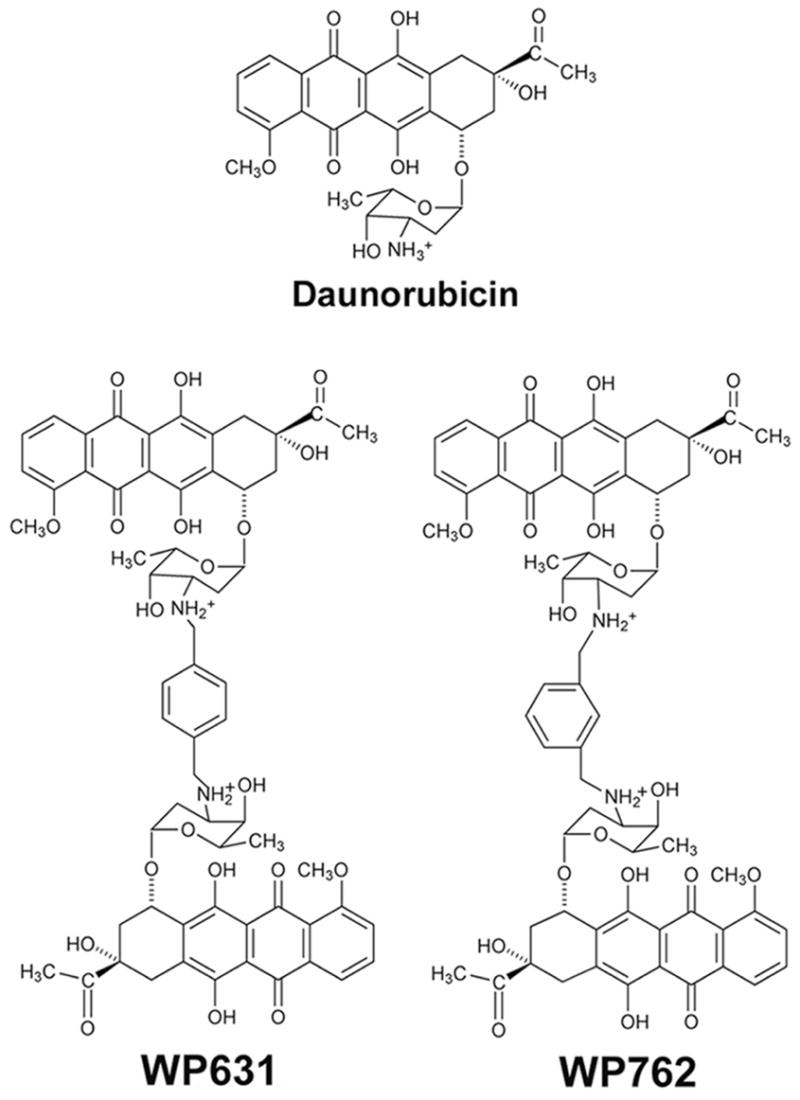
Chemical formulae of daunorubicin and bisantracyclines WP631 and WP762.
From the results obtained with WP631, it was suggested that the design of bisanthracyclines might be refined to increase DNA affinity even further.13 Structural data indicated that the fit of the p-xylenyl linker in the minor groove was not optimal, and could be improved.13,14 Therefore, a linker was designed to be of the appropriate length (approximately 7 Å) but that might fit better into the minor groove of DNA with less steric hindrance. The new WP762 (Figure 1) differs from WP631 in that the linker is m-xylenyl instead of p-xylenyl. The change in the orientation of the two sugars affects the precise orientation as well as the distance between the two chromophores (from roughly 7 Å in WP631 to 6 Å in WP762), which was hypothesized to influence the strength of binding. The design, synthesis, and characterization of WP762, including an initial analysis of its biological properties, are described in this paper, and are compared to both daunorubicin and WP631.
RESULTS
Initial attempts to determine the binding constant of WP762 indicated that it binds to DNA extremely tightly. Since daunorubicin binds to DNA with a binding constant of ~107 M−1 under the ionic conditions used in our experiments,12 a binding constant of ~1014 M−1 is expected for a bisintercalator like WP762. It is very difficult to determine such a large binding constant by traditional spectrophotometric methods since it becomes impossible to work at ligand concentrations near the reciprocal of the association constant and still obtain a reliable signal. Therefore, UV melting studies (Figure 2) and differential scanning calorimetry (Figure 3) were used to determine the binding constant for the interaction of WP762 with herring sperm DNA in BPE buffer (16 mM total Na+ concentration). Figure 2 shows the melting of DNA in the presence of increasing molar ratios of WP762. At less than saturating concentrations, melting curves are complex and multiphasic, due to the ligand redistribution during the melting process. These experiments establish the drug concentrations needed for saturation, where the change in Tm becomes constant and the redistribution is absent. In the absence of WP762, the Tm of herring sperm DNA was found to be 68.5 °C. In the presence of 10 μM WP762, a concentration sufficient to saturate the DNA lattice, the Tm was 98.0 °C. The enthalpy of DNA melting in the absence of ligand (ΔHm) was determined by differential scanning calorimetry to be 6.8 (±0.2) kcal mol−1 (bp) (Figure 3). These values were used to determine the DNA binding constant at the melting temperature by using equation 1 (in which the ligand site size (n) was considered to be 6 base pairs, based on the crystal structure of the structurally related WP631),13 yielding Ktm = 3.8 × 107 M−1. Correction of this value to lower temperature requires knowledge of the binding enthalpy, which we determined by DSC. Figure 3 shows the results of DSC experiments using herring sperm DNA in the presence and absence of saturating amounts of WP762. By Hess’s law, these data may be used to determine the enthalpy of WP762 binding to DNA. The equilibria to be considered, along with the experimentally determined enthalpy values, are as follows:
Figure 2.
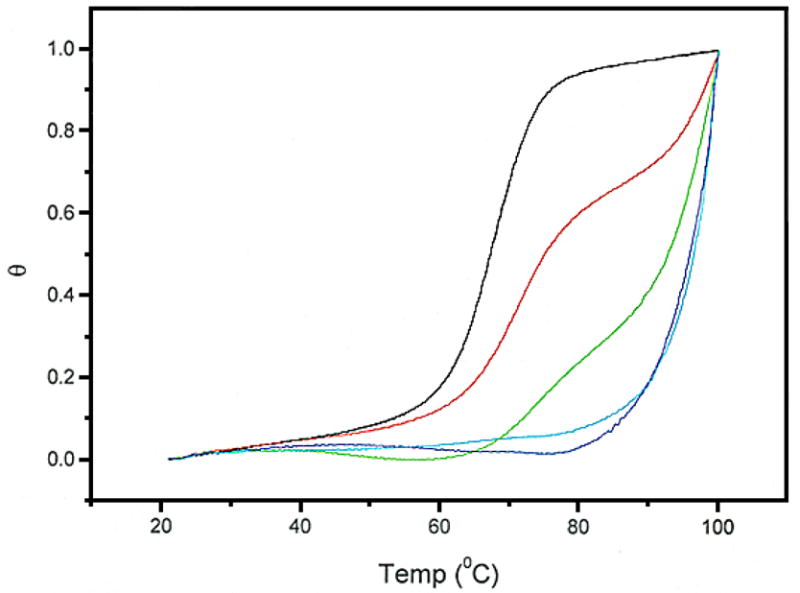
UV melting curves for the denaturation of herring sperm DNA (20 μM bp) in the presence of increasing concentrations of WP762. WP762 concentrations were (from left to right): 0, 1, 2, 10, and 20 μM.
Figure 3.
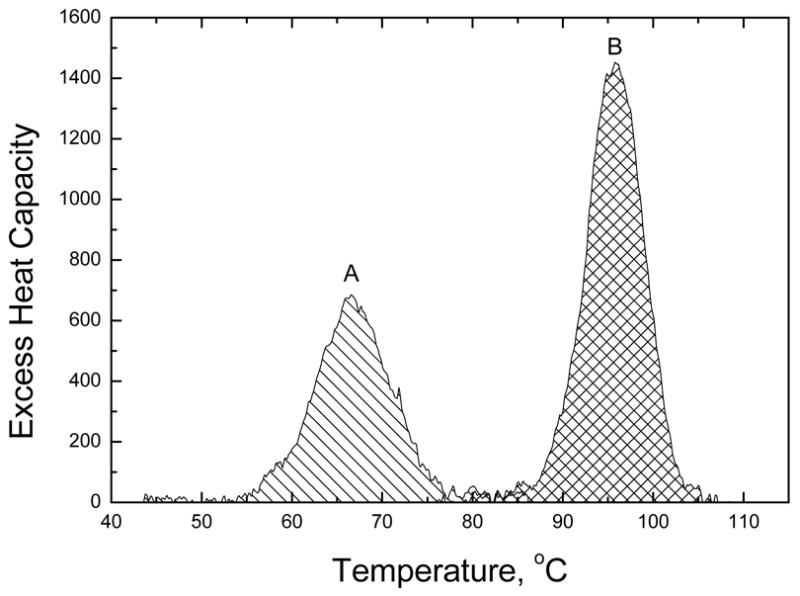
Results of DSC experiments. Melting curves are shown for herring sperm DNA at 1 mM (bp) concentration (A) and for an identical DNA solution with saturating amounts (0.16 mol antibiotic/mol bp) of WP762 added (B). The areas under the peaks provide direct estimates of the enthalpy for melting of the DNA duplex (A) and the WP762-DNA complex (B).
Combining these two reactions, the binding reaction and its enthalpy may be obtained:
The enthalpy ΔH3 must be corrected for the amount of WP762 bound to the DNA determined spectrophotometrically, and the sign changed, to obtain the binding enthalpy (ΔHb):
From three independent DSC determinations using separately prepared samples, a value for ΔHb of −33.6 (±2.8) kcal mol−1 was obtained for the association of WP762 with DNA. For the monomeric daunorubicin the same experimental procedure had yielded ΔHb = −10.5 kcal mol−1, while for the structurally related WP631 the ΔHb was −30.2 kcal mol−1 (Table 1). In all these cases, enthalpy values refer to the temperature at the respective Tm. Because of limited solubility and avid self-association, ITC cannot be used to measure ΔHb for WP762 at the temperature of reference (20°C), so we were forced to neglect any possible heat capacity changes when calculating the binding of this bisintercalator.
Table 1.
Comparison of thermodynamic parameters for daunorubicin, WP631 and WP762 binding to herring sperm DNAa.
| Kobs (M−1) | ΔGobs (kcal mol−1) | SK | ΔGpe (kcal mol−1) | ΔGt (kcal mol−1) | ΔGcalc (kcal mol−1)c | ΔH (kcal mol−1) | −TΔS (kcal mol−1) | |
|---|---|---|---|---|---|---|---|---|
| Daunorubicinb | 1.6 × 107 | − 9.7 | 0.9 | − 2.0 | − 7.7 | −7.7 | − 10.5 | 0.8 |
| WP631b | 2.7 × 1011 | − 15.3 | 1.6 | − 3.9 | − 11.4 | −11.9 | − 30.2 | 14.9 |
| WP762 | 7.3 × 1012 | − 17.3 | 1.5 | − 3.7 | − 13.6 | −12.6 | − 33.8 | 16.6 |
Kobs (M−1) is the binding constant for the interaction of the ligand with salmon testes DNA in 18 mM Na+ at 20°C. ΔGobs is the binding free energy calculated from ΔGobs = −RTlnK. SK is the slope of the plot of logKobs versus log[Na +] shown in Fig. 4. ΔGpe and Δ Gt are the polyelectrolyte and non-electrostatic contributions to the binding free energy (ΔGpe = SKRTln[Na+]) evaluated at 16 mM Na+. Enthalpy values were determined by DSC (Fig. 3).
Data adapted from.12
Once the binding enthalpy was determined, the DNA binding constant of WP762 at 20°C was calculated to be 7.33 (±0.2) × 1012 M−1 by application of the standard van’t Hoff equation (eq. 2). The same procedure had also been used previously to determine the binding constant for daunorubicin and WP631, using herring sperm DNA (Table 1).12 Knowledge of the binding constant and binding enthalpy allowed us to construct the complete thermodynamic profiles for the binding of WP762 to herring sperm DNA and to compare them with those obtained by daunorubicin and WP631 (Table 1). The free energy is obtained from the standard relation ΔG = −RT lnK, and the entropy is calculated with the equation: ΔS = −(ΔG − ΔH)/T.
Because WP762 is a dication, its binding to DNA is thermodynamically linked to Na+ binding to DNA, and as a result, its DNA binding constant will depend on the total Na+ concentration (Figure 4). Polyelectrolyte theories based on Manning’s counterion condensation model describe the process and provide a basis for interpreting the data.15,16 For WP762, SK (the slope of the least-squares fit of its data in Figure 4) was found to be 1.54, which is in accord with the theoretical predictions by Record et al. for dication binding to DNA.15 In Figure 4, the slopes of the different least-squares fits of the data (SK) are equivalent to the number of the counterions released upon binding of a ligand with net charge Z (eq. 3). For WP631 and daunorubicin, we had found that 1.63 and 0.85 counterions were respectively released from herring sperm DNA upon binding of these ligands.12 From these values of Z, the ‘actual’ charge on each drug is as follows: +1.75 for WP762, +1.85 for WP631, and +0.96 for daunorubicin, as estimated using Record’s polyelectrolyte theory.15,16 These values correspond quite well to the two positive charges carried by WP762 and WP631, and the one positive charge carried by daunorubicin at neutral pH (Figure 1).
Figure 4.
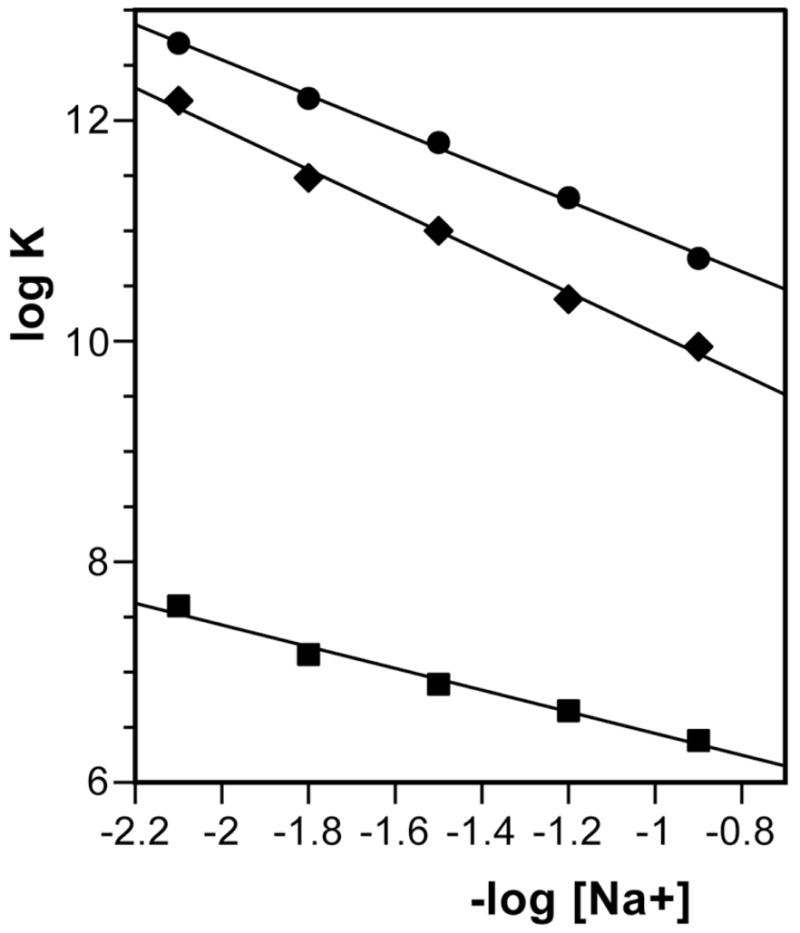
Salt dependence of WP762 binding constant at 20°C, compared to daunorubicin and WP631. Data are presented according to the Record’s theory.15 WP762 (circles), WP631 (diamonds), daunorubicin (squares). Data for daunorubicin and W631 were adapted from.12 The linear least-squares fit of the data yields a slope (SK) of 1.54 for WP762. From this value, it was estimated that the charge of WP762 is +1.75 at neutral pH.
Knowledge of the binding constant and the binding enthalpy allowed us to construct the complete thermodynamic profiles for the binding of WP762 to DNA and dissect the observed binding free energy into some component parts. The binding constant for WP762 decreases with increasing salt concentrations, due to counter ion release that accompanies the binding of a charged ligand to DNA. From the dependence of the binding constant on salt concentration, the observed binding free energy may be partitioned into two contributions:
where ΔGt is the non-polyelectrolyte contribution to the binding free energy and ΔGpe is the polyelectrolyte contribution. The latter term may be calculated from the experimental results by using the equation:
where ZΨ is the slope (SK) of the corresponding line in Figure 4.
The magnitude of ΔGt provides a measure of that portion of the binding free energy contributed by the non-polyelectrolyte forces that stabilize the DNA-drug complex (i.e., hydrogen bonding, van der Waals interactions, hydrophobic transfer, etc). ΔGpe is the free energy contribution arising from coupled polyelectrolyte effects, the most important of which is the release of the condensed counter ions from the DNA helix upon binding of the charged ligand.15
Table 1 summarizes the comparative energetics for the binding of WP762, WP631 and daunorubicin. For all of the anthracyclines analyzed, ΔGt is large in magnitude, indicating that non-polyelectrolyte forces play a significant role in stabilization of their DNA complexes. The ΔGt value of WP762 is almost twice that of daunorubicin and about 2 kcal mol−1 higher than for WP631. We caution that while the thermal denaturation methods used here to obtain thermodynamic profiles are the most rigorous possible under the circumstance, the methods are not without limitations and possible error. The analysis of melting data assumes homogeneous binding sites along the DNA lattice, and further assumes that heat capacity changes for both DNA melting and ligand binding are negligible. Neglect of heat capacity changes could introduce error into our estimates of thermodynamic values at 20 °C. These possible difficulties were discussed more fully in our previous publication on WP631 binding.12
Differences between the binding of WP631 and WP762 can be visualized through the comparison of their structures when bound to DNA in a comparative molecular modeling study. A view of the average structures of both of the WP631 and WP762 complexes with DNA, obtained from molecular dynamics simulations, indicates that there is a positional shift with regard to the location of the xylenyl linker, as well as a slight positional shift of both of the daunosamine sugars (Figure 5: A and B). This positional shift is seemingly responsible for much of the increase in binding energy of WP762 versus WP631. A more comprehensive look into the nature of these interactions can be seen in Figure 6(A and B), showing a HINT 18–22 interaction map of the average structure of WP631 and WP762. Figure 6C and 6D show a close-up view of the linker region. The green regions represent areas of favorable hydrophobic or van der Waal’s interactions, while the blue regions represent areas of favorable electrostatic interactions between the polar functional groups. In both maps, we see significant electrostatic and hydrophobic (van der Waal’s) interactions surrounding the intercalating chromophores of both WP631 and WP762. We also see significantly more favorable van der Waal’s interactions between the daunosamine sugar rings of WP762 and the sugars of the wall of the DNA minor groove, compared to those in WP631. Additionally, the more favorable positioning of the m-xylenyl linker of WP762, allows for greater van der Waal’s interactions of the linker with the the sugars of the wall of the DNA minor groove (Figure 6A). There is also a notable widening of the DNA minor groove to accommodate drug binding in both the WP631 (7.9 Å) and WP762 (8.6 Å) complexes, as compared to the minor groove width of an unbound DNA molecule (5.9 Å), calculated by the program Curves.17 Interestingly, the linker region of the minor groove of the DNA-WP762 complex is ~0.7 Å narrower than the DNA-WP631 complex. This could result from the additional van der Waal’s interactions between the m-xylenyl linker and the sugar wall of the minor groove, compared to the p-xylenyl linker.
Figure 5.
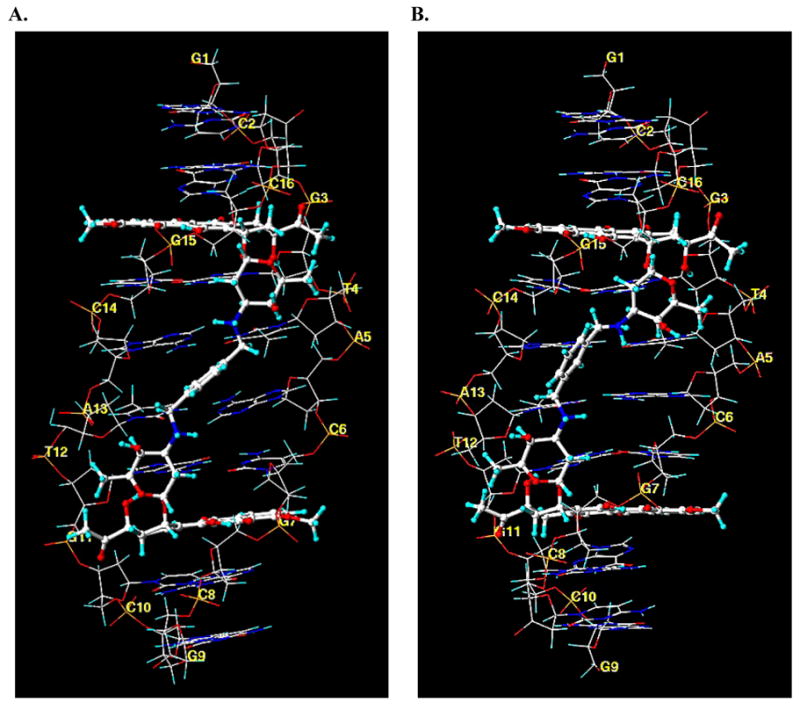
Molecular models of the average structures of WP631 (A) and WP762 (B), indicating the DNA base pairs and the position of intercalation of each compound.
Figure 6.
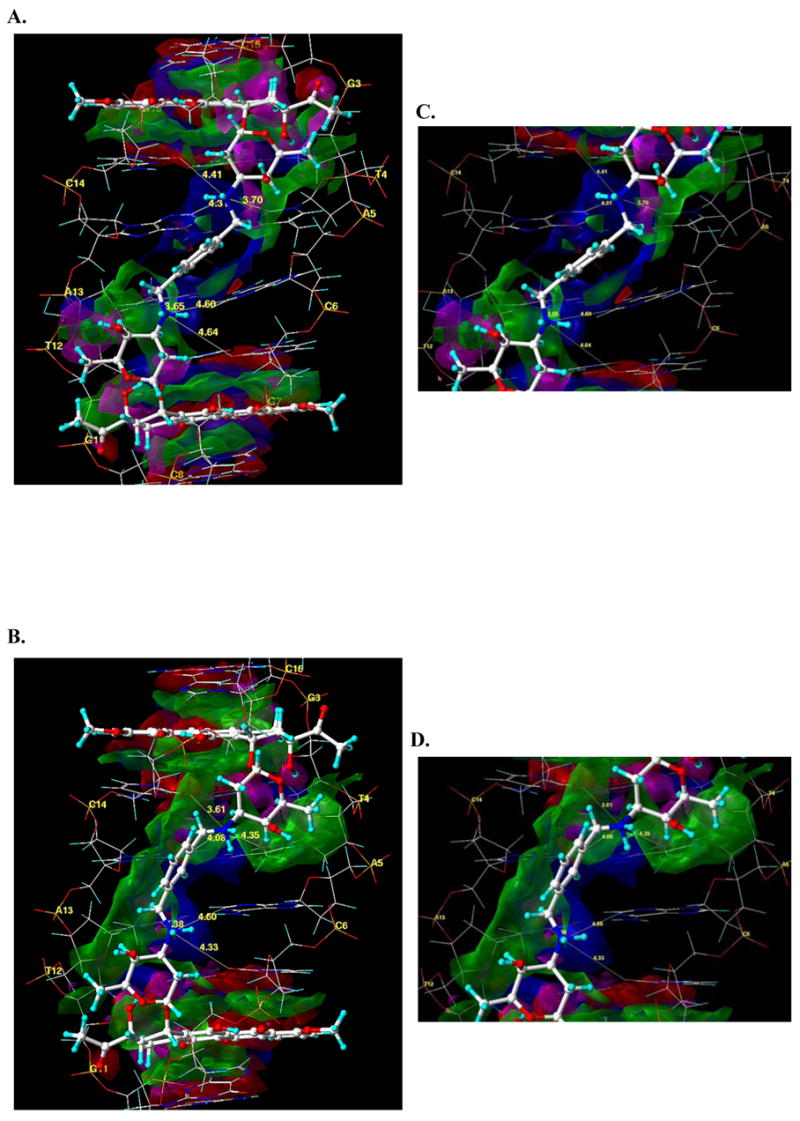
HINT interaction maps for the interaction of WP631 (A) and WP762 (B) with the 5′-GC|GTAC|GC-3′ base pair sequence of DNA, which displays, visually, the quality and magnitude of the various binding contacts involved in the interaction. The contour surfaces are color-coded by interaction type at a constant map density value of ±150. Blue surfaces represent favorable polar interactions, red surfaces represent unfavorable polar interactions; Green surfaces represent favorable hydrophobic interactions, magenta surfaces represent unfavorable hydrophobic interactions. The interatomic distance between the N3′ ammonium groups on the WP631 and WP762 sugar rings and select atoms of the surrounding base pairs of DNA is also indicated. At right are close-up views of the xylenyl linker region in the average structures of WP631 (C) and WP762 (D), indicating six hydrogen bonds (yellow lines) between the quaternary ammonium and various base pairs in the DNA.
There also appears to be a shorter distance between the cationic ammonium ions and several electronegative atoms of the base pairs in the DNA, resulting in stronger electrostatic interactions in WP762 than in WP631. These distances can be seen in the close-up figure of the linker regions of WP631 and WP762 (Figure 6: C & D). For example, the distance between the first quaternary ammonium (pictured on the top in figure 6C & D) and the C14 O2 has decreased from 4.41 Å to 3.61 Å, which strengthens the electrostatic interaction between the two functional groups. Likewise, the distance between this ammonium ion and the aromatic carbon of the A13 base also decreased from 4.31 Å to 4.08 Å. However, the distance between the ammonium and the T4 O2 increased slightly from 3.70 Å to 4.35 Å.
The second quaternary ammonium ion (pictured on the bottom in figure 6C & D) is also notably closer to both the T12 O2 (3.65 Å in WP762 compared to 3.38 Å in WP631) and the C6 O2 carbonyl atoms (4.64 Å compared to 4.33 Å), yielding stronger electrostatic interactions in WP762 versus WP631. Furthermore, inspection of Figure 6 shows that the m-xylenyl linker is closer to one side of the minor groove than the p-xylenyl linker. Additional evidence of this shift in position is seen with the increased favorable hydrophobic interactions between the linker region and the DNA backbone (Figure 6D). While we might expect to see additional movement along the molecular dynamics trajectory in regards to the xylenyl linker atoms in the wider minor groove for WP631, an analysis of the RMSD vs. time for both the WP631 and WP762 simulations indicates very little movement of these atoms in comparison to the DNA backbone atoms (Figure S1: Supporting information).
The calculated HINT interaction scores18–22 for each complex were 5384 for WP631 and 5701 for WP762, which gives a difference of 317. For comparison, a similar 1 ns molecular dynamics simulation was performed on daunorubicin, and the HINT score for the average structure of daunorubicin versus DNA was calculated at 3476. If we normalize the ΔGcalc (Table 1) to the ΔGt for daunorubicin, we get a ratio of -451 HINT score units equal to 1 kcal mol−1 free energy, which is very close to the estimated ratio of -515 HINT units equal to 1 kcal mol−1 free energy.18–22 If we use the normalized value of -451 for the HINT scores of WP631 and WP762, we obtain a calculated ΔGt of −11.9 kcal mol−1 for the binding of WP631 to DNA (experimental ΔGt = −11.4 kcal mol−1), and −12.6 kcal mol−1 for the binding of WP762 to DNA (experimental ΔGt = −13.6 kcal mol−1). While these values are notably below the experimentally-determined free energies (ΔGobs) in Table 1, it is important to consider here that these scores were calculated on the DNA/drug complex only, with no contribution of the solvent or counterions being included in this calculation, therefore ΔGt is the appropriate comparison term.
An initial evaluation of the biological activity of WP762 was undertaken. Table 2 shows the results of an in vitro cytotoxicity assay of the drug effects against Jurkat T lymphocytes, which are compared with those obtained for WP631 and daunorubicin. The effect of WP762 on the viability of Jurkat T cells was similar to that of WP631, and both bisanthracyclines were more cytotoxic than the monomeric daunorubicin. Figure 7 shows a cell cycle distribution analysis in Jurkat T cells treated with the IC75 concentration for WP762 (54 nM). Untreated cells maintained an almost uniform phase distribution during the experiments (Figure 7). In the presence of WP762, we observed that, within 24 hours, the G2/M fraction increased from 16.73% to 30.19%, with a decline in the number of cells in G1 phase. After 24 hour treatment, the cell cycle distribution appeared clearly altered, and after 48 hours the drug induced a cell distribution that could hardly be assigned to the different phases of the cell cycle, yet the sub-G1 cell population rose to about 18%. After longer periods of treatment, the cell distribution histograms become unsolvable (Figure 7), reflecting a clear dysfunction in the cell population, with a reduction in cell number due to cell death, as confirmed under the microscope by the presence of staining by Trypan blue dye in about 80% of the cells. In the presence of the IC75 for WP631 (60 nM), we have shown that Jurkat T cells suffered similar unpaired cell cycle distribution, but the altered cell cycle was observed much earlier with WP762, within 24 hours, while the cytotoxic effects of WP631 required a longer incubation with the drug.23 Comparison of the effects of WP631 and WP762 suggest an earlier (faster) effect of WP762 on cell viability. In this respect, WP762 effects on cell cycle and viability parallel those found with daunorubicin,2,24 although a much lower WP762 concentration was required (Table 2). However, an ‘internucleosomal ladder’ was observed in the presence of daunorubicin (Figure 8), which is a known inductor of apoptosis24, but not in the presence of either WP631 or WP762. While daunorubicin induced apoptosis from G1, the treatment with WP762 did not produce the presence of internucleosomal DNA cleavage, a qualitative hallmark of apoptosis (Figure 8) and cells died from G2 after entering mitosis. The behavior of cells treated with nanomolar concentrations of WP762 (Figure 12) suggests the presence of mitotic catastrophe occurring after the damaged cells overtook their temporal halt at the G2/M checkpoint (Figure 7), as we have described for WP631.23
Table 2.
Evaluation of the cytotoxicity of WP762, WP631 and daunorubicin in Jurkat T lymphocytes.
| IC50 [nM] | IC75 [nM] | |
|---|---|---|
| Daunorubicin | 82.6 ± 8.9 | 182.2 ± 13.3 |
| WP631 | 17.7 ± 6.0 | 60.0 ± 19.5 |
| WP762 | 17.0 ± 6.5 | 53.5 ± 4.8 |
IC50 and IC75 are the drug concentrations required to induce cell quiescence by 50% and 75% respectively, measured by the MTT assay (means ± SEM, for 3–6 experiments).
Figure 7.
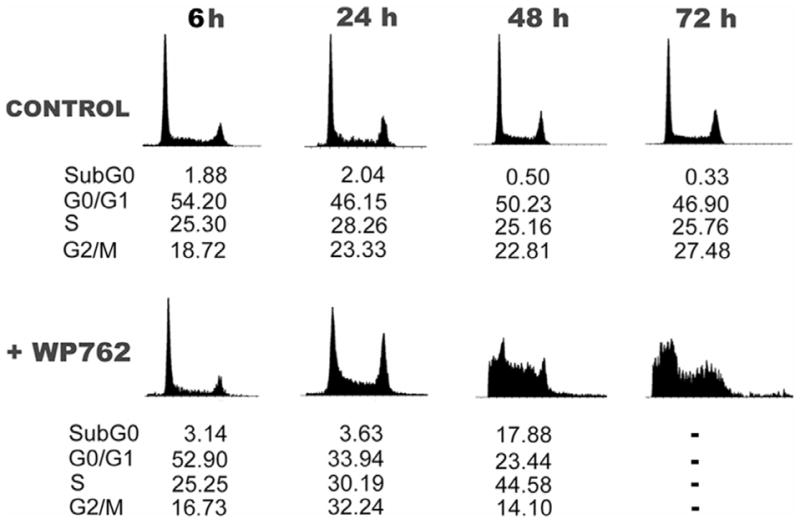
Flow cytometry analyses of Jurkat T cells treated with WP762. Cell cycle distribution was determined at the time intervals indicated. Data indicate the percentage of cells in each phase. (A) Cell cycle distribution of untreated Jurkat T cells. (B) Cell cycle distribution in the presence of 54 nM WP762 (its IC75). Transient changes in G2/M and sub-G1 peaks were evident after treatment.
Figure 8.
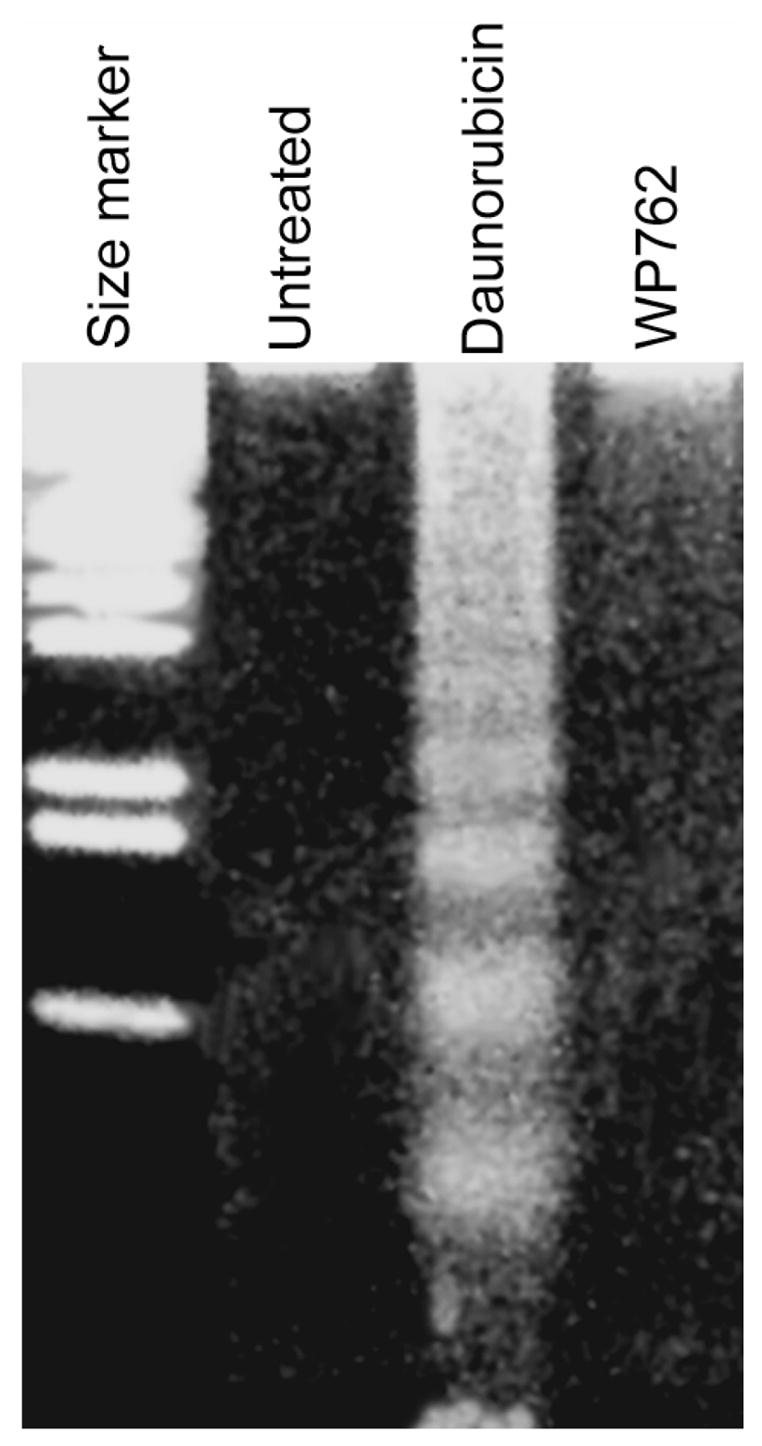
Agarose gel electrophoris analysis of the apoptosis-associated internucleosomal DNA fragmentation in Jurkat T cells treated with 54 nM WP762 (its IC75) for 48 h. For comparison, the effect of 180 nM daunorubicin —its IC75— (Table 2), which produced the typical internucleosomal cleavage in apoptotic cells, is shown as a positive control. Qualitatively, the gel suggested the absence of apoptosis in WP762-treated cells.
DISCUSSION
WP762 is a new bisanthracycline with picomolar affinity for DNA. Its affinity for DNA is among the highest yet observed for any small molecule, and is on par with the affinity of site-specific DNA binding proteins, even though WP762 has only a fraction of the molecular weight of protein molecules. The higher affinity of WP762, relative to its predecessor WP631, is the result of the increased favorable interaction of its xylenyl linker in the minor groove. WP762 retains the potent cytotoxicity of its predecessor. The bisanthracyclines hold great promise as potential anticancer agents,7 and serve as models for the modular design of new DNA binding agents.
Figure 9 shows a comparison of the thermodynamics of interaction between the intercalating compounds WP631, WP762, and daunorubicin with DNA. WP762 displays a larger favorable enthalpic contribution for the binding reaction compared to WP631. The binding enthalpy for both WP631 and WP762 is over twice the value of that observed for daunorubicin (c.f. Table 1 with Figure 9). This larger enthalpic contribution plausibly results from the intercalation of two chromophores of WP762 into DNA, with the formation of twice the number of stabilizing molecular interactions, along with additional interactions between the linker and the minor groove.
Figure 9.
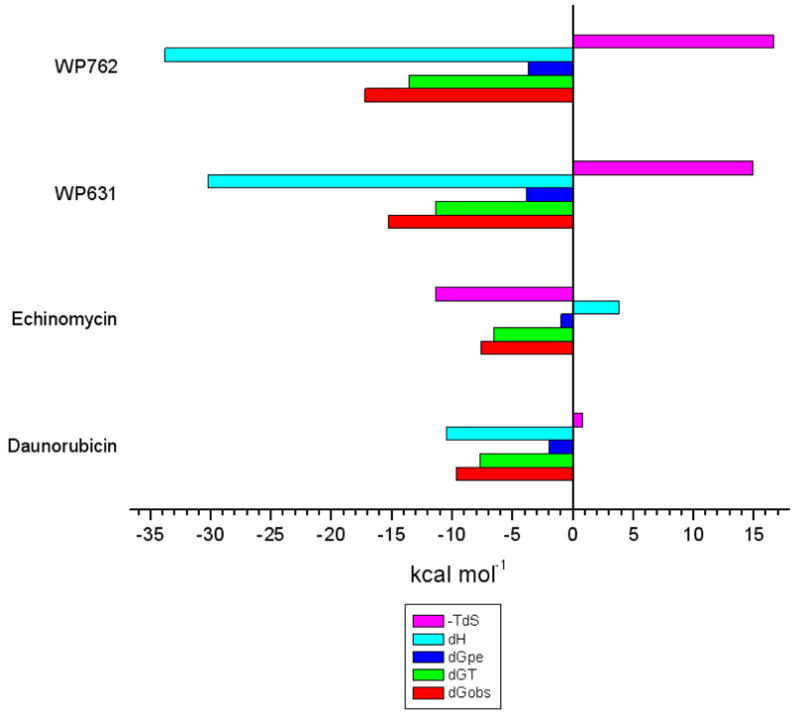
Comparison of the binding thermodynamics for several well characterized intercalating compounds: WP631, WP762, Daunorubicin, and Echinomycin.
Molecular modeling studies allow us to visualize the differences in the molecular interactions between WP631 and WP762 that rationalizes the higher affinity of WP762 for DNA. The HINT map shown in figure 6 shows that a large portion of the binding energy lies in the interaction with the intercalating chromophores of WP631 and WP762. We also can see favorable electrostatic interactions between the charged ammonium groups of both compounds and electron-rich functional groups present on the DNA base pairs. It is interesting to note the shorter distances between the cationic ammonium functional groups and electronegative functional groups on the DNA base pairs, resulting in increased favorable electrostatic interactions in WP762. One of the primary differences in the interactions between both compounds lies in the positioning of the m-xylenyl linker in WP762 near one side of the minor groove, providing greater van der Waal’s interactions with the DNA backbone (Figure 6: B & D). These interactions are absent in the p-xylenyl linker of WP631 (Figure 6: A & C). The more favorable positioning of the m-xylenyl linker shifts the positioning of the daunosamine sugar, which results in additional favorable van der Waal’s interactions between the daunosamine methyl groups of WP762 and the sugars of the DNA minor groove. The more favorable van der Waal’s interactions of the m-xylenyl linker of WP762 also help to compensate for its less favorable entropic component compared to WP631 (Table 1, Figure 9). While both WP631 and WP762 have a considerably less favorable entropic component than Daunorubicin, the entropic penalty for WP631 is less than for WP762.
The minor groove width of the DNA in the WP762 complex is slightly decreased by ~0.7 Å, compared to WP631, which points to less translational and rotational freedom in the WP762 complex. The lower minor groove width could be a consequence of the additional van der Waal’s interactions between the m-xylenyl linker and the sugar wall of the DNA minor groove.
It is interesting to compare the binding of WP762 to DNA with that of the hairpin polyamides. The hairpin polyamides are rationally designed groove binders that are capable of sequence-selective recognition of DNA.25–27. The polyamides can be designed to recognize sequences of different lengths. Polyamides that bind to specific 6 bp sequences vary in their affinity from 4 × 109 M−1 to 3 × 1011 M−1, depending on the exact sequence.28 The bisanthracyclines, which also bind selectively to a particular 6 base pair sequence, equal or slightly exceed the binding affinity of the polyamides. The bisanthracyclines possess both groove-binding and intercalating moieties, both of which contribute significantly to the binding affinity.
It is also of interest to compare the bisanthracylines with echinomycin, a natural product that was the first bisintercalator discovered.29 To provide a comparative modeling study with this compound, we used HINT to analyze the crystal structure of a DNA complex of echinomycin (pdb accession no.: 1pfe) (Figure S2: Supporting information).30,31 The HINT interaction score for the binding of echinomycin with DNA was found to be 5064, which correlates to an estimated ΔG of −9.8 kcal mol−1. While this is notably higher than the free energy of binding for daunorubicin and DNA, it is considerably lower than the free energy of binding for WP631 and WP762 (ΔΔGWP631 = −2.1 kcal mol−1/ΔΔGWP762 = −2.8 kcal mol−1). Unlike the binding of WP631 and WP762, the binding of echinomycin with DNA is primarily entropically driven, with an unfavorable enthalpic component.31 While echinomycin is a bis-intercalator like WP631 and WP762, its thermodynamic profile more closely resembles that of a groove binding compound such as Hoechst 33258, due to the bulky peptide moiety that fills the minor groove (Figure S2: Supporting information).31 Furthermore, the intercalating chromophore of echinomycin consists of two aromatic rings intercalated into the DNA in a parallel arrangement, rather than the perpendicular insertion typical of the anthracyclines. This form of intercalation, combined with the bulky peptide moiety, results in a significantly wider minor groove. There are also significant parts of the peptide moiety extending into the solvent and are not in contact with the DNA at all, which would also be entropically unfavorable.
WP762 also inhibits cell growth at nanomolar concentrations. Despite the potent effect of WP762 in the viability of Jurkat T cells, continuous treatment over 72 hours appears to produce only marginal apoptosis. The WP762 effects on cell growth extend our previous observations indicating that bisanthracyclines inhibit cell growth, killing cells by a mechanism that differs from classical apoptosis, as they appear to suffer mitotic catastrophe after transient stop at the G2/M checkpoint.23 Although we present an initial characterization of WP762, the similarities of its behavior with that of WP631 (which we previously analyzed in detail)12–14,32–34 suggest a common mechanism of action. Both drugs produced a transient stop of Jurkat T cells in G2, and it appears that cell death may have arisen as the result of overtaking the G2 checkpoint, resulting in mitotic catastrophe. At equitoxic concentrations (IC75), WP762 produced an unambiguous antiproliferative effect, which happened after shorter incubation times (cf. Figure 7 in ref.23), indicating that WP762 may easier penetrate cells to reach cytotoxic concentrations, or that the tighter DNA binding constant (Table 1) would result in a strong effect of this drug on gene transcription compared to WP631. Since we have shown that WP631 is a specific inhibitor of Sp1-activated transcription,33,34 it is worth conjecturing that WP762 may be a sturdy inhibitor of Sp1 binding to DNA. Taken together, these studies indicate a promising anticancer activity for bisanthracycline WP762, which also displays a cytotoxic activity higher than WP631 against a breast carcinoma cell line (details not reported). In any case, a further characterization of the antitumor capacities of WP762 will require the consideration of the determinant of tumor cell sensitivity to anthracyclines.3,34,35
Our results present several implications for drug design. A rational design process requires knowledge not only of the structures of drug-DNA complexes but also of the energetics of the binding reaction. We have used a thermodynamic approach to shed light on the binding process, which may be used to modify design strategies to improve binding affinity. The affinity of WP762 for DNA is ultratight, but its binding constant still falls short (by two orders of magnitude) of the expected 1014 M−1 value, based on the affinity of monomeric daunorubicin. We show that the lower than expected binding free energy results from a larger unfavorable entropic contribution for WP762 relative to that for daunorubicin. Our studies clarify that using a flexible linker that allows an induced fit of the ligand into the binding site can improve binding by bisintercalating ligands, but at the expense of obtaining the maximal possible binding free energy there is an energetic cost of restricting bond rotation. A modified design strategy in which a rigid, immobile linker was used could lead to even greater binding affinity (cf., the binding parameters for WP631 and WP762 in Table 1) and guide future rational design efforts. The challenge would be, however, to improve the link between monomeric units in such a way that they would be in exactly the proper orientation for bisintercalation. One caveat in this strategy is that bisintercalation may also carry an entropic cost for the lengthening and stiffening of the DNA upon binding. Little can be done to circumvent that necessary consequence of intercalation.
MATERIALS AND METHODS
Materials
The BPE buffer consisting of 6 mM Na2HPO4, 2 mM NaH2PO4, and 1 mM EDTA, at pH 7.0, was used in all of the binding experiments. A molar extinction coefficient of 13400 M−1 cm−1 at 480 nm was used to determine the free concentration of WP762, and a molar extinction coefficient of 17700 M−1 cm−1 at 508 nm was used to determine the WP762 bound to DNA.
Herring sperm DNA (Boerhinger Mannheim) was sonicated, phenol extracted, and purified as previously described. A molar extinction coefficient of 12858 M−1 (bp) cm−1 at 260 nm was used for DNA concentration determinations.
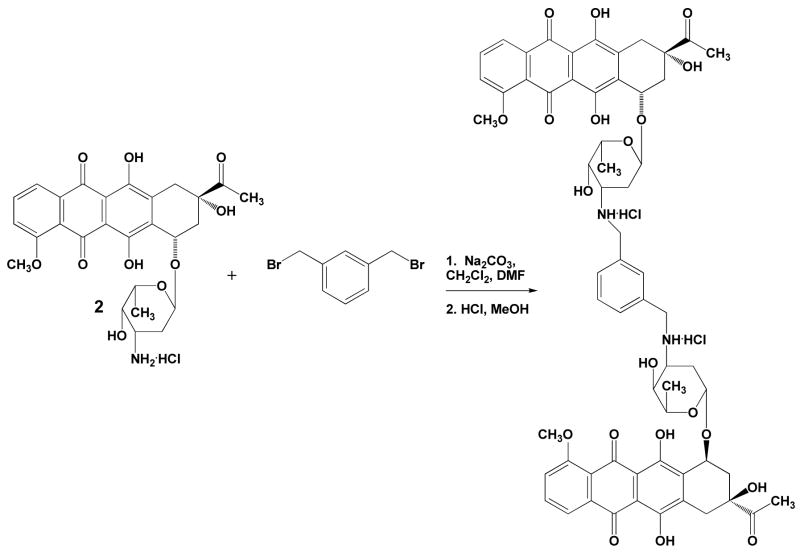
Synthesis of WP762
Daunorubicin hydrochloride (1.44 g, 2.4 mmol), and α,α′-dibromo-m-xylene (0.32 g, 1.2 mmol) were dissolved in the mixture of 12 mL dichloromethane and 6 mL dimethylformamide (Scheme 1). Sodium carbonate (1.5 g, 14.15 mmol) was added and the mixture was stirred at room temperature overnight. After the reaction was completed as determined by thin layer chromatography (TLC), the reaction mixture was diluted with 50 mL dichloromethane. Solid salts were filtered off, and filtrate was washed with water until neutral, then washed with brine, and was dried over anhydrous sodium sulfate. Drying agent was filtered off, and solvents were evaporated. Residue after evaporation was dissolved in 3 mL chloroform, and the mixture of products was precipitated with hexanes. The main product, WP762, was purified by column chromatography (Silicagel 60, Merck, Darmstadt) using chloroform, and then chloroform:methanol (98:2) as eluents, to give 0.833 g of red solid (yield 60%). Such obtained WP762 was characterized as a free base by 1H-NMR (data shown below):
1H-NMR (CDCl3, δ): 13.9, 13.3 (bs, 2H ea, 6-OH, 11-OH), 8.01 (dd, 2H, J = 8.4 Hz, J = 0.9 Hz, H-1), 7.77 (dd, 2H, J = J = 7.9 Hz, H-2), 7.38 (d, 2H, J = 8.7 Hz, H-3), 7.2 –7.1 (m, 4H, xylyl linker), 5.49 (d, 2H, J = 3.4 Hz, H-1′), 5.26 (s, 2H, H-7), 4.68 (bs, 2H, 9-OH), 4.07 (s, 6H, OMe), 4.04 (q, 2H, J = 6.5 Hz, H-5′), 3.75 (d, 2H, J = 12.7 Hz, CH2-xylyl linker), 3.64 (s, 2H, H-4′), 3.62 (d, 2H, J = 12.7 Hz, CH2-xylyl linker), 3.20 (dd, 2H, J = 18.8 Hz, J = 1.8 Hz, H-10), 2.94 (d, 2H, J = 18.8 Hz, H-10), 2.95-2.91 (m, 2H, H-3′), 2.4 (s, 6H, 14-CH3), 2.35 (d, 2H, J = 14.8 Hz, H-8), 2.08 (dd, 2H, J = 14.8 Hz, J = 4.1 Hz, H-8), 1.76 (ddd, 2H, J = J = 13.2 Hz, J = 3.8 Hz, H-2′a), 1.67 (dd, 2H, J = 13.2 Hz, J = 5 Hz, H-2′e), 1.35 (d, 6H, J = 6.5 Hz, H-6′).
Subsequently, WP762 was dissolved in a 1M solution of hydrochloride in methanol and then precipitated with ethyl ether as hydrochloride salt, which was washed with ethyl ether until neutral pH and dried under vacuum.
13C-NMR(DMSO-d6, δ): 212.21 (C-13), 186.73, 186.56 (C-5, C-12), 161.17 (C-4), 156.56, 154.81 (C-6, C-11), 136.59 (C-2, C arom. xylenyl linker), 135.79, 134,97, 134.91 (C-6a, C-10a, C-12a), 128.5 (C arom. xylenyl linker), 120.26 (C-1), 120.08 (C-4a), 119.34 (C-3), 110.98, 110.92 (C-11a, C-6a), 100.00 (C-1′), 75.57 (C-9), 70.74 (C-7), 66.65 (C-5′), 64.9 (C-4′), 57.01 (OMe), 53.29 (C-3′), 47.5 (CH2 xylenyl linker), 36.61 (C-8), 32.04 (C-10), 24.64 (C-14), 17.32 (C-6′). Anal. (C62H66Cl2N2O20·3H2O): C, H, Cl, N.
UV Melting Studies
Ultraviolet DNA melting curves were determined using a Cary 3E UV/Visible Spectrophotometer (Varian, Inc., Palo Alto, CA), equipped with a thermoelectric temperature controller. Solutions of herring sperm DNA in BPE buffer (final concentration of 20μM in bp) were prepared by direct mixing with aliquots from a WP762 stock solution, followed by incubation for 12 h to ensure equilibration. Samples were heated at a rate of 1°C min−1, while continuously monitoring the absorbance at 260 nm. Primary data were transferred to Origin software (Microcal, Inc., Northampton, MA) for plotting and analysis.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) experiments were undertaken in a Microcal MC2 instrument (Microcal, Inc.). Sonicated herring sperm DNA at a concentration of 1 mM bp in BPE buffer was used for all experiments. Primary data were corrected by subtraction of a buffer-buffer baseline and normalized to the concentration of DNA base pairs. Baseline-corrected normalized data were transferred to Origin software (Microcal, Inc.) for integration and plotting. Samples of DNA-WP762 complexes for DSC were prepared by weighing appropriate amounts of solid WP762, and dissolving it directly into 2 mL of a 1 mM DNA solution. Any undissolved drug was removed by low-speed centrifugation. The exact amount of WP762 bound to the DNA was then determined spectrophotometrically.
Determination of DNA Binding Constants
The DNA binding constant of WP762 was determined using the DSC results and ultraviolet-DNA melting studies. Assuming no interaction of ligand with single-stranded DNA, McGhee36 derived the equation:
| (1) |
where Tm0 is the melting temperature of the DNA alone, Tm is the melting temperature in the presence of saturating amounts of ligand, ΔHm is the enthalpy of DNA melting (per mol of bp), R is the gas constant, Ktm is the ligand binding constant at Tm, L is the free ligand concentration (approximated at Tm by the total ligand concentration), and n is the ligand site size.
The DNA binding constant of WP762 at lower temperatures was determined by use of the van’t Hoff equation:
| (2) |
where K is the DNA binding constant of WP762 at temperature T (Kelvin) and ΔHb is the enthalpy of binding of WP762 to DNA determined by DSC.
Salt Dependence of the Binding Constants
Differences between the DNA melting temperatures of DNA alone and in the presence of saturating amounts of WP762 at different NaCl concentrations were used to calculate DNA binding constants for WP762 and daunorubicin at the melting temperature. These DNA binding constants were corrected to 20°C by application of eq. 2, assuming that the binding enthalpy is independent of salt concentration. These data can be analyzed using the polyelectrolyte theory of Record et al.,15 considering:
| (3) |
where Z is the apparent charge of the ligands and Ψ is the fraction of counter ions associated with each DNA phosphate (Ψ= 0.88 for double-stranded B-form DNA).
Molecular Modeling
Molecular models of WP631 and WP762 in complex with the DNA octamer sequence (5′-GC|GTAC|GC-3′; the “|” symbol indicates the site of intercalation of each anthracycline ring system) were created using the NMR structure of DNA:WP631 complex as a starting structure (pdb accession code: 1AL9). One base pair was added to each end of the DNA hexamer to add stability to the double helix ends and ensure a stable realistic intercalation site. The xleap module in AMBER (Amber v. 8, Scripps Research Institute, La Jolla, CA 92037) was used to add Na+ ions to neutralize the systems, which were then solvated in an octahedral box of 3,573 TIP3P water molecules. Periodic boundary conditions and the particle mesh Ewald algorithm were used. A 2.0 fs time-step was used with bonds involving hydrogen frozen using SHAKE. Molecular dynamics (MD) calculations were carried out with the AMBER 7.0 mpi version of sander and general amber force field (gaff.dat) parameterization. Simulations were performed in the isothermic isobaric ensemble (P 1 atm; T 300 K). The systems were heated slowly and equilibrated for 125 ps with gradual removal of positional restraints on the DNA following this protocol:37 (i) minimize water; (ii) 20 ps MD (T 100 K) holding DNA fixed (100 kcal/mol Å-1); (iii) minimize water; (iv) minimize total system; (v) 20 ps MD (T 100 K) holding DNA fixed (100 kcal/mol Å-1); (vi) 20 ps MD (T 300 K) holding DNA fixed (100 kcal/mol Å-1); (vii) 20 ps MD (T 300 K) holding DNA fixed (50 kcal/mol Å-1); (viii) 20 ps MD (T 300 K) holding DNA fixed (10 kcal/mol Å-1); (ix) 20 ps MD (T 300 K) holding DNA fixed (1 kcal/mol Å-1). The production run consisted of unrestrained dynamics (T 300 K) for 1 ns after final equilibrium, which was used to obtain snapshots at 2 ps throughout the trajectory and average structures, (25 snapshots in the last 50 ps), which were subsequently minimized (5,000 cycles of steepest descent energy minimization). Analysis of the structures and trajectories utilized the ptraj module of AMBER, as well as the program Curves (Curves v. 5.1, Richard Lavery, Institut de Biologie Physico-Chimique, Paris, France), to analyze the DNA helicoidal parameters.17
The average structures were analyzed using the HINT molecular modeling software (SYBYL module, Tripos, Inc., St. Louis, MO 63144), using procedures previously described.18–22 The HINT model, based on the octanol-water partition coefficient (Log P), describes and scores specific hydrophobic (van der Waal’s) and polar (hydrogen bonding) interactions between two molecules.18–22 Previous HINT calculations have been shown to correlate with the free energy of drug – nucleic acid complexes in a ratio of approximately 515 HINT Score Units equal to –1 kcal mol−1.18,19
Cytotoxicity Assays
Jurkat T lymphocytes were maintained in RPMI 1640 medium (GibcoBRL) supplemented with 10% fetal calf serum (GibcoBRL), penicillin (100 μg/ml), streptomycin (100 μg/ml) and 2 mM L-glutamine (GibcoBRL), at 37°C in a humidified atmosphere with 5% CO2.
The effect of WP762 on Jurkat cells growth was determined by using the colorimetric MTT dye assay38 in 96-well microtiter plates with flat-bottomed wells (Corning Costar) in a total volume of 100 μL. Cells subcultured at a density of 5 × 104 cells/ml were incubated with various concentrations of daunorubicin, WP631 or WP762 at 37°C for 72 h. After incubation, MTT (3-(4,5-dymethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma) was added to each culture (50 μg/well). The dark-colored crystals produced by viable cells were solubilized with 30 mM HCl in 2-propanol. Absorbance was determined at 570 nm using a SPECTRAmax 250 microplate reader (Molecular Devices). For WP762, WP631 and daunorubicin, the IC50 — drug concentration required to inhibit cell growth by 50% — and the IC75 — drug concentration required to inhibit cell growth by 75% — were determined from dose-response curves.
Flow Cytometry
After treatment with 54 nM WP762 (its IC75 determined as described in the previous paragraph) for various periods of time, cells were harvested and stained with propidium iodide (Sigma) as described elsewhere.39 Nuclei were analyzed with a Coulter Epics-XL flow cytometer (Coulter Corporation) using the 488 nm line of an argon laser and standard optical emission filters. Percentages of cells at each phase of the cell cycle were estimated from their DNA content histograms after drug treatment by using the WinMDI 2.8 software (Purdue University Cytometry Laboratories, West Lafayette, IN).
Analysis of internucleosomal DNA damage
Qualitative analysis of DNA fragments resulting from internucleosomal cleavage in cells undergoing apoptosis was carried out as described elsewhere.32 Jurkat T cells treated with WP762, or with daunorubicin (Sigma), were analyzed by electrophoresis in 1.8% agarose gels, stained with ethidium bromide, and photographed under UV light.
Supplementary Material
Figure S1 (molecular dynamics trajectories for WP631 and WP762); Figure S2 (HINT map for the echinomycin-DNA complex); Elemental analysis of products of WP762 synthesis. This material is available free of charge via the Internet at http://pubs.acs.org.
Scheme 1.
Synthesis of WP762.
Acknowledgments
This work was supported in part by grant SAF2002-00371 (Spain) to J. Portugal and by a grant from The Welch Foundation (Houston, Texas) to W. Priebe. Research in the Chaires laboratory is supported by grant CA35635 from the National Cancer Institute. Jonathan B. Chaires holds the James Graham Brown Endowed Chair in Biophysics, and he is grateful to the James Graham Brown Foundation for their support.
References
- 1.Booser DJ, Hortobagyi GN. Anthracycline antibiotics in cancer therapy. Focus on drug resistance. Drugs. 1994;47:223–258. doi: 10.2165/00003495-199447020-00002. [DOI] [PubMed] [Google Scholar]
- 2.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacological Reviews. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 3.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochemical Pharmacology. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 4.Binaschi M, Bigioni M, Cipollone A, Rossi C, Goso C, Maggi CA, Capranico G, Animati F. Anthracyclines: selected new developments. Current Medicinal Chemistry - Anti-Cancer Agents. 2001;1:113–130. doi: 10.2174/1568011013354723. [DOI] [PubMed] [Google Scholar]
- 5.Chaires JB, Leng F, Przewloka T, Fokt I, Ling YH, Perez-Soler R, Priebe W. Structure-based design of a new bisintercalating anthracycline antibiotic. Journal of Medicinal Chemistry. 1997;40:261–266. doi: 10.1021/jm9607414. [DOI] [PubMed] [Google Scholar]
- 6.Priebe W. ACS Symposium Series. American Chemical Society; Washington, D.C: 1995. Anthracycline Antibiotics: New Analogues, Methods of Delivery and Mechanisms of Action. [Google Scholar]
- 7.Priebe W, Fokt I, Przewloka T, Chaires JB, Portugal J, Trent JO. Exploiting anthracycline scaffold for designing DNA-targeting agents. Methods in Enzymology. 2001;340:529–555. doi: 10.1016/s0076-6879(01)40441-1. [DOI] [PubMed] [Google Scholar]
- 8.Chaires JB. Advances in DNA sequence specific agents. JAI Press Inc; Greenwich, CT: 1996. Molecular recognition of DNA by daunorubicin; pp. 141–167. [Google Scholar]
- 9.Chaires JB, Herrera JE, Waring MJ. Preferential binding of daunomycin to 5′ATCG and 5′ATGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–6153. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 10.Frederick CA, Williams LD, Ughetto G, van der Marel GA, van Boom JH, Rich A, Wang AH. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 11.Rudnicki WR, Kurzepa M, Szczepanik T, Priebe W, Lesyng B. A simple model for predicting the free energy of binding between anthracycline antibiotics and DNA. Acta Biochimica Polonica. 2000;47:1–9. [PubMed] [Google Scholar]
- 12.Leng F, Priebe W, Chaires JB. Ultratight DNA binding of a new bisintercalating anthracycline antibiotic. Biochemistry. 1998;37:1743–1753. doi: 10.1021/bi9720742. [DOI] [PubMed] [Google Scholar]
- 13.Hu GG, Shui X, Leng F, Priebe W, Chaires JB, Williams LD. Structure of a DNA-bisdaunomycin complex. Biochemistry. 1997;36:5940–5946. doi: 10.1021/bi9705218. [DOI] [PubMed] [Google Scholar]
- 14.Robinson H, Priebe W, Chaires JB, Wang AH. Binding of two novel bisdaunorubicins to DNA studied by NMR spectroscopy. Biochemistry. 1997;36:8663–8670. doi: 10.1021/bi970842j. [DOI] [PubMed] [Google Scholar]
- 15.Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Quarterly Reviews of Biophysics. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 16.Chaires JB. Dissecting the free energy of drug binding to DNA. Anti-Cancer Drug Design. 1996;11:569–580. [PubMed] [Google Scholar]
- 17.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. Journal of Biomolecular Structure & Dynamics. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 18.Burnett JC, Kellogg GE, Abraham DJ. Computational methodology for estimating changes in free energies of biomolecular association upon mutation. The importance of bound water in dimer-tetramer assembly for beta 37 mutant hemoglobins. Biochemistry. 2000;39:1622–1633. doi: 10.1021/bi991724u. [DOI] [PubMed] [Google Scholar]
- 19.Fornabaio M, Cozzini P, Mozzarelli A, Abraham DJ, Kellogg GE. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 2. Computational titration and pH effects in molecular models of neuraminidase-inhibitor complexes. Journal of Medicinal Chemistry. 2003;46:4487–4500. doi: 10.1021/jm0302593. [DOI] [PubMed] [Google Scholar]
- 20.Cashman DJ, Kellogg GE. A computational model for anthracycline binding to DNA: tuning groove-binding intercalators for specific sequences. Journal of Medicinal Chemistry. 2004;47:1360–1374. doi: 10.1021/jm030529h. [DOI] [PubMed] [Google Scholar]
- 21.Cashman DJ, Scarsdale JN, Kellogg GE. Hydropathic analysis of the free energy differences in anthracycline antibiotic binding to DNA. Nucleic Acids Research. 2003;31:4410–4416. doi: 10.1093/nar/gkg645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellogg GE, Abraham DJ. Hydrophobicity: Is Log Po/w More than the Sum of its Parts? . European Journal of Medicinal Chemistry. 2000;35:651–661. doi: 10.1016/s0223-5234(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 23.Villamarin S, Mansilla S, Ferrer-Miralles N, Priebe W, Portugal J. A comparative analysis of the time-dependent antiproliferative effects of daunorubicin and WP631. European Journal of Biochemistry. 2003;270:764–770. doi: 10.1046/j.1432-1033.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 24.Mansilla S, Pina B, Portugal J. Daunorubicin-induced variations in gene transcription: commitment to proliferation arrest, senescence and apoptosis. Biochemical Journal. 2003;372:703–711. doi: 10.1042/BJ20021950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dervan PB, Burli RW. Sequence-specific DNA recognition by polyamides. Current Opinion in Chemical Biology. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 26.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Current Opinion in Structural Biology. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 27.Dervan PB. Molecular recognition of DNA by small molecules. Bioorganic & Medicinal Chemistry. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 28.Dervan PBDRM, Marques MA. Programmable DNA Binding Oligomers for Control of Transcription. Current Medicinal Chemistry. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- 29.Waring MJ, Wakelin LP. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974;252:653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- 30.Cuesta-Seijo JA, Sheldrick GM. Structures of Complexes between Echinomycin and Duplex DNA. Acta Crystallogr D Biol Crystallogr. 2005;61:442–448. doi: 10.1107/S090744490500137X. [DOI] [PubMed] [Google Scholar]
- 31.Leng F, Chaires JB, Waring MJ. Energetics of echinomycin binding to DNA. Nucleic Acids Research. 2003;31:6191–6197. doi: 10.1093/nar/gkg826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villamarin S, Ferrer-Miralles N, Mansilla S, Priebe W, Portugal J. Induction of G(2)/M arrest and inhibition of c-myc and p53 transcription by WP631 in Jurkat T lymphocytes. Biochemical Pharmacology. 2002;63:1251–1258. doi: 10.1016/s0006-2952(02)00865-1. [DOI] [PubMed] [Google Scholar]
- 33.Martin B, Vaquero A, Priebe W, Portugal J. Bisanthracycline WP631 inhibits basal and Sp1-activated transcription initiation in vitro. Nucleic Acids Research. 1999;27:3402–3409. doi: 10.1093/nar/27.17.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansilla S, Priebe W, Portugal J. Sp1-targeted inhibition of gene transcription by WP631 in transfected lymphocytes. Biochemistry. 2004;43:7584–7592. doi: 10.1021/bi036185e. [DOI] [PubMed] [Google Scholar]
- 35.Perego P, Corna E, De Cesare M, Gatti L, Polizzi D, Pratesi G, Supino R, Zunino F. Role of apoptosis and apoptosis-related genes in cellular response and antitumor efficacy of anthracyclines. Current Medicinal Chemistry. 2001;8:31–37. doi: 10.2174/0929867013373994. [DOI] [PubMed] [Google Scholar]
- 36.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 37.Trent JO. Molecular modeling of drug-DNA complexes: an update. Methods in Enzymology. 2001;340:290–326. doi: 10.1016/s0076-6879(01)40428-9. [DOI] [PubMed] [Google Scholar]
- 38.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 39.Doyle A, Griffiths JB, Newell DG. Cell & Tissue Culture: Laboratory Procedures. John Wiley & Sons; New York: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (molecular dynamics trajectories for WP631 and WP762); Figure S2 (HINT map for the echinomycin-DNA complex); Elemental analysis of products of WP762 synthesis. This material is available free of charge via the Internet at http://pubs.acs.org.