Abstract
Most secretory and membrane proteins are sorted by signal sequences to the endoplasmic reticulum (ER) membrane early during their synthesis. Targeting of the ribosome-nascent chain complex (RNC) involves the binding of the signal sequence to the signal recognition particle (SRP), followed by an interaction of ribosome-bound SRP with the SRP receptor. However, ribosomes can also independently bind to the ER translocation channel formed by the Sec61p complex. To explain the specificity of membrane targeting, it has therefore been proposed that nascent polypeptide-associated complex functions as a cytosolic inhibitor of signal sequence- and SRP-independent ribosome binding to the ER membrane. We report here that SRP-independent binding of RNCs to the ER membrane can occur in the presence of all cytosolic factors, including nascent polypeptide-associated complex. Nontranslating ribosomes competitively inhibit SRP-independent membrane binding of RNCs but have no effect when SRP is bound to the RNCs. The protective effect of SRP against ribosome competition depends on a functional signal sequence in the nascent chain and is also observed with reconstituted proteoliposomes containing only the Sec61p complex and the SRP receptor. We conclude that cytosolic factors do not prevent the membrane binding of ribosomes. Instead, specific ribosome targeting to the Sec61p complex is provided by the binding of SRP to RNCs, followed by an interaction with the SRP receptor, which gives RNC–SRP complexes a selective advantage in membrane targeting over nontranslating ribosomes.
INTRODUCTION
It has long been established that hydrophobic signal sequences direct most secretory and membrane proteins for cotranslational translocation across the endoplasmic reticulum (ER) membrane. However, the precise mechanism by which ribosome-nascent polypeptide chain complexes (RNCs) are targeted to the ER membrane remains unknown. It is clear that the signal recognition particle (SRP) plays an important role (for review, see Walter and Johnson, 1994; Rapoport et al., 1996). Initially, SRP binds weakly to the ribosome, but once a signal sequence in a nascent chain emerges from a translating ribosome and is recognized by the 54-kDa subunit of SRP (SRP54) (Krieg et al., 1986; Kurzchalia et al., 1986), the SRP–ribosome interaction becomes much stronger (Walter et al., 1981). Subsequently, the RNC–SRP complex is bound to the ER membrane by a GTP-dependent interaction between SRP and the SRP receptor (also known as docking protein) (Gilmore et al., 1982; Meyer et al., 1982; Connolly and Gilmore, 1989; Connolly et al., 1991). It is not clear whether this model suffices to explain specificity of targeting since ribosomes bind to the ER membrane irrespective of the presence or nature of a nascent chain (Borgese et al., 1974).
Recent experiments have shown that ribosomes bind to the Sec61p complex, a heterotrimeric protein complex in the ER membrane (Görlich et al., 1992; Kalies et al., 1994). The Sec61p complex is the major component of a protein-conducting channel (Görlich and Rapoport, 1993; Hanein et al., 1996), and the tight association between a translating ribosome and the Sec61p complex ensures that the elongating nascent chain is transferred directly from the channel in the ribosome into the membrane channel and then through it into the lumen of the ER (Crowley et al., 1994). The interaction between the translating ribosome and the Sec61p complex occurs in two consecutive steps (Jungnickel and Rapoport, 1995). When the nascent chain is just sufficiently long to allow binding of SRP, the subsequent interaction of the ribosome with the Sec61p complex is weak, as indicated by the fact that the RNCs can be removed from the membrane by extraction with high salt concentrations and that the nascent chain is sensitive to added proteases. Upon further chain elongation, a tighter interaction is achieved in which the RNCs become insensitive to proteases and can no longer be extracted by high salt. The tighter interaction can only occur in the presence of a functional signal sequence in the nascent chain. Signal sequence recognition requires the function of the Sec61p complex (Jungnickel and Rapoport, 1995) and, for most translocation substrates, the TRAM protein, another component of the translocation site (Voigt et al., 1996). Following signal sequence recognition in the membrane, the protein-conducting channel opens to the lumen of the ER (Crowley et al., 1994) and the nascent chain is inserted into the translocation channel (Jungnickel and Rapoport, 1995). At this stage, the nascent chain is committed to translocation.
If all ribosomes can bind to the Sec61p complex, is specificity of protein translocation exclusively determined in steps following the ribosome–membrane interaction? How does translocation occur efficiently if nontranslating ribosomes are able to compete with RNCs for common membrane-binding sites? What role does SRP play in targeting RNCs to the ER membrane? A model has recently been proposed to answer these questions. It suggests that an additional cytosolic factor, the nascent polypeptide-associated complex (NAC) (Wiedmann et al., 1994) binds initially to all ribosomes and nascent polypeptide chains emerging from translating ribosomes and serves as an inhibitor of ribosome–membrane interaction (Lauring et al., 1995a,b). When a signal sequence emerges from the ribosome, NAC would be replaced by SRP, thus removing the inhibitor of ribosome binding. In this model, the ribosome–membrane interaction would provide specificity of targeting and NAC and SRP would regulate the interaction. Because only RNCs with signal sequences would be bound to the ER membrane, the second signal sequence recognition step in the membrane, mediated by the Sec61p complex and TRAM (Jungnickel and Rapoport, 1995; Voigt et al., 1996), would exist merely to allow a double check.
The NAC model was derived from in vitro experiments with RNCs carrying truncated nascent chains that were generated either in the wheat germ or the reticulocyte translation system. When these RNCs were washed with high salt to remove bound NAC, they could be bound to canine microsomes in an SRP- and signal sequence-independent manner (Jungnickel and Rapoport, 1995; Lauring et al., 1995a,b). Readdition of NAC restored SRP- and signal sequence-dependent binding (Lauring et al., 1995a). However, experiments with salt-washed RNCs may not reflect physiological conditions. Indeed, high salt-washed ribosomes can bind more strongly than unwashed ribosomes to at least one other partner, the SRP, and the increased interaction may not be functional (Powers and Walter, 1996).
We report here that SRP-independent ribosome targeting to the ER membrane occurs in the absence or presence of wheat germ or reticulocyte lysate NAC, indicating that NAC is not an inhibitor of ribosome–membrane interaction. We found that in the absence of SRP, binding of RNCs to the ER membrane depends on the availability of sufficient Sec61p-binding sites in the membrane and is competitively inhibited by nontranslating ribosomes. In the presence of SRP, however, this competition is no longer observed. SRP also protects against ribosome competition when RNC binding is tested with reconstituted proteoliposomes containing only the Sec61p complex and the SRP receptor. Thus, our data are not consistent with inhibition of ribosome binding to the ER membrane by cytosolic factors. Rather, they support a model in which SRP plays a positive role, as originally proposed (Walter and Blobel, 1981b). Our results demonstrate that the interaction of SRP with its membrane receptor gives RNC-SRP complexes a significant advantage over nontranslating ribosomes in targeting to the Sec61p complex.
MATERIALS AND METHODS
Preparation of Ribosome-stripped Microsomes
Ribosome-stripped membranes (PK-RM) were prepared from rough microsomes (RM) by treatment with poromycin and potassium acetate, as follows. A suspension of RM containing 3.5 equivalents (Eq; for definition, see Walter et al., 1981) per μl was mixed with an equal volume of buffer B [100 mM HEPES/KOH, pH 7.6, 200 mM sucrose, 300 mM potassium acetate, 10 mM magnesium acetate, 3 mM dithiothreitol (DTT), 0.2 mM GTP, protease inhibitors (10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastatinal, 1 μg/ml pepstatin), 3 mM puromycin]. After homogenization, the mixture was incubated for 1 h at 0°C followed by 10 min at 37°C and 10 min at room temperature. The sample was centrifuged in a TLA 100.3 rotor for 30 min at 100,000 rpm at 2°C. The pellet was resuspended at 10 Eq/μl in buffer C (50 mM HEPES/KOH, pH 7.6, 500 mM sucrose, 800 mM CsCl, 15 mM magnesium acetate, 3 mM DTT, protease inhibitors) and mixed with an equal volume of a buffer identical to buffer C, except that it contained 1.95 M sucrose. The total volume was determined and additional CsCl was added to adjust to a final concentration of 700 mM. The sample was overlaid with 1 ml of buffer D (50 mM HEPES/KOH, pH 7.6, 800 mM sucrose, 700 mM CsCl, 15 mM magnesium acetate, 3 mM DTT, protease inhibitors) and 0.6 ml of buffer B in a 13 × 51-mm polycarbonate tube. After centrifugation in a TLA 100.3 rotor for 1 h at 100,000 rpm at 20°C, the top 0.2 ml was discarded and the next 2–2.5 ml, which contained the membranes, were collected. This fraction was diluted 1:4 in 50 mM HEPES/KOH (pH 7.6), 1 mM DTT, and protease inhibitors and centrifuged in a TLA 100.3 rotor for 30 min at 100,000 rpm at 2°C. The pellet was resuspended in buffer A (50 mM HEPES/KOH, pH 7.6, 250 mM sucrose, 150 mM potassium acetate, 2 mM magnesium acetate) containing 1 mM DTT and protease inhibitors. The membranes were washed two or three times by resuspension and centrifugation and finally taken up at 2–3 Eq/μl in buffer A containing 1 mM DTT.
Synthesis and Isolation of Ribosome–Nascent Chain Complexes
mRNAs coding for the N-terminal 86 amino acids of wild-type bovine preprolactin (pPL86), for the N-terminal 83 amino acids of a preprolactin mutant lacking three leucines in the hydrophobic core (pPLΔ13–15), or for N-terminal 77 amino acids of firefly luciferase (ffl77) were generated by in vitro transcription with SP6 polymerase, as described (Jungnickel and Rapoport, 1995). Truncated mRNAs were translated in a wheat germ or reticulocyte lysate system in the presence of [35S]methionine (Jungnickel and Rapoport, 1995). Translation in the wheat germ system was carried out for 13 min at 28°C followed by the addition of 2 μM edeine and further incubation for 3 min. Translation in the reticulocyte lysate was carried out for 25 min at 28°C.
To isolate RNCs, 100 μl of the translation mixture were diluted in 900 μl of 40 mM HEPES/KOH (pH 7.6), 2 mM magnesium acetate, and 2 mM DTT containing either 150 mM potassium acetate (for low salt-washed RNCs) or 500 mM potassium acetate (for high salt-washed RNCs). The samples were layered on top of a 1-ml cushion containing a low- or high-salt concentration (40 mM HEPES/KOH, pH 7.6, 0.5 M sucrose, 2 mM magnesium acetate, 2 mM DTT, and either 150 mM or 500 mM potassium acetate) in a 13 × 51-mm polycarbonate Beckman tube. After centrifugation for 1 h at 100,000 rpm in a TLA 100.3 rotor, the ribosome pellets were resuspended at a concentration of approximately 140 nM in buffer A (Lauring et al., 1995b).
Mock translation mixtures lacking mRNA and amino acids were prepared and incubated as described above. Salt-washed, nontranslating ribosomes were isolated from a mock translation mixture as described for the isolation of salt-washed RNCs. The mock translation was divided into a cytosolic and a ribosomal fraction by centrifugation for 1 h at 100,000rpm in a TLA 100 rotor. The ribosome pellet was resuspended in the original volume in buffer A.
RNCs produced in a reticulocyte lysate were isolated by centrifugation through a sucrose gradient containing 150 mM salt, as described for wheat germ RNCs, and incubated with 10 mM N-ethylmaleimide for 40 min on ice. The reaction was quenched with 50 mM DTT.
Ribosome Competition
A translation mixture containing approximately 200 fmol of RNCs (determined by measurement of the A260 nm absorption and assuming that a solution with 1 A260 nm absorption contains 16 nM ribosomes; Hanein et al., 1996) was mixed with different volumes of mock translation mixture in which the amounts of ribosomes were determined in the same manner. When subfractions of mock translations were used in competition experiments, the amounts added were equivalent to the original volume of the mock translation mixture. Where indicated, SRP (10 nM) was added after translation and before addition of the mock translation mixture. After addition of PK-RM (0.4 Eq per 5 μl final volume), incubation was carried out for 10 min on ice and for 5 min at 28°C. Experiments with isolated RNCs were performed in an analogous manner.
To assay membrane insertion of pPL86, the samples were incubated with 1 volume of 1.5 mg/ml proteinase K in buffer A containing 1 mM DTT and 8 mM magnesium acetate for 45 min on ice.
Translocation of membrane-targeted nascent chains was induced by incubation of the samples with 1.5 mM puromycin for 10 min on ice and 30 min at 37°C.
Targeting assays using flotation of membrane-targeted RNCs in a sucrose gradient were carried out as described (Jungnickel and Rapoport, 1993, 1995). The flotation of the membranes was confirmed by immunoblotting with antibodies against TRAPβ (previously called SSRβ; Görlich et al., 1990).
Photocrosslinking
One and a half pmoles of trifluoromethyl-diazirino-benzoic acid-lysyl tRNA were added to a 10-μl wheat germ translation mixture (Görlich et al., 1991). After membrane targeting of pPL86, as described above, or binding of NAC to ffl77 for 5 min on ice and 5 min at 28°C, the samples were irradiated for 10 min on ice. Cross-links of pPL86 to membrane proteins were analyzed by immunoprecipitation with Sec61α and TRAM antibodies as described (Görlich et al., 1992).
Reconstituted Proteoliposomes
The purification of the SRP receptor and the Sec61p complex as well as their reconstitution into proteoliposomes were carried out as described (Görlich and Rapoport, 1993; Jungnickel and Rapoport, 1995). The concentrations of the SRP receptor and Sec61p complex in the suspensions of proteoliposomes were 0.8–3 Eq/μl and 3–8 Eq/μl, respectively. Four-tenths microliters of the suspensions were used per 5 μl of final volume.
Sample Preparation
Following cross-linking and flotation experiments, the samples were precipitated with trichloroacetic acid and separated in 13.75% polyacrylamide gels. Samples from protease protection assays were trichloroacetic acid precipitated and separated in 12% Tris-Tricine gels. After drying, the gels were exposed to Fuji PhosphoImager screens and quantitated using a Fuji BAS1000.
RESULTS
SRP-independent Targeting Can Occur in the Presence of Cytosol
In previous experiments, we found that isolated RNCs, washed with high concentrations of salt, can be targeted to the ER membrane independently of the SRP/SRP receptor system (Jungnickel and Rapoport, 1995). On the basis of experiments by Lauring et al. (1995a,b), we assumed that this was due to the removal of cytosolic or ribosome-associated proteins, in particular removal of NAC. To test this assumption, we performed targeting reactions in the presence of a complete cytosol that includes NAC.
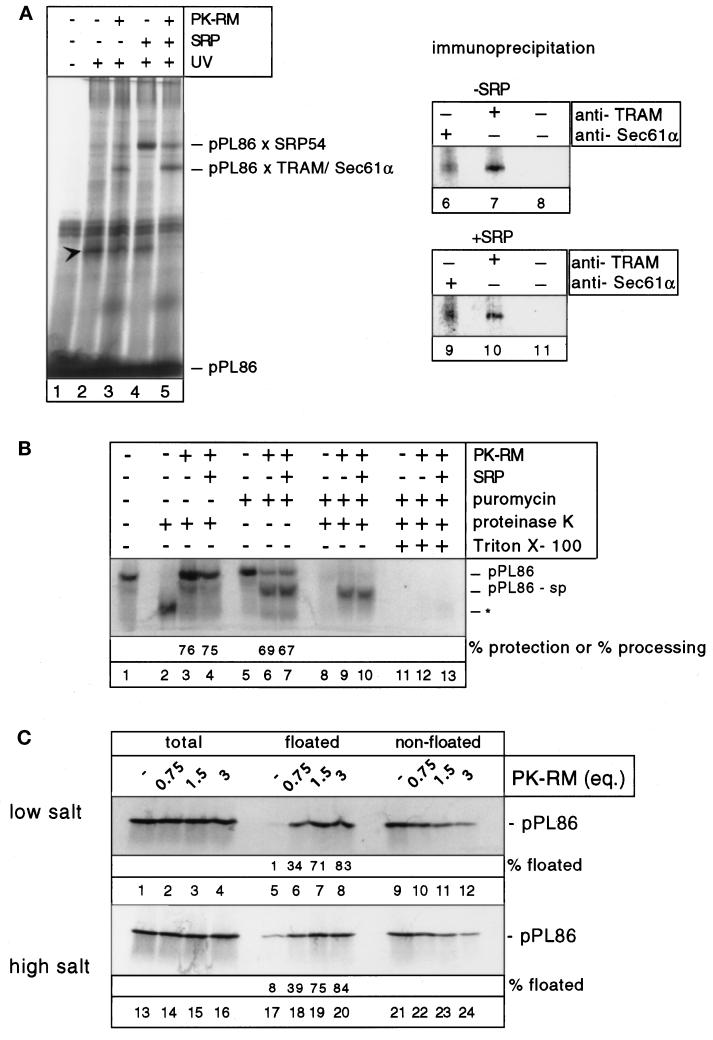
Truncated mRNA coding for the first 86 amino acids of preprolactin was translated in the wheat germ system, leading to stable RNCs with nascent chains that are long enough to be targeted to canine microsomes and to be firmly inserted into the translocation site (Jungnickel and Rapoport, 1995). The targeting of these RNCs mimics that in a normal cotranslational translocation reaction and this system has been used in previous studies (Lauring et al., 1995a,b). Uncoupling targeting from translation has several advantages: the nascent chains have a defined length; translational arrest by SRP (Walter and Blobel, 1981b) and nonspecific inhibition of translation by microsomes are irrelevant, and the time period given for membrane targeting of RNCs can be extended, allowing more efficient binding. Photocrosslinking was used to follow the path of a nascent chain from the cytosol into the membrane (Kurzchalia et al., 1986; Wiedmann et al., 1987; Görlich et al., 1992; Jungnickel and Rapoport, 1995). The 86-amino acid fragment of preprolactin (pPL86) was synthesized in the presence of a modified lysyl-tRNA, leading to the incorporation of photoreactive probes at positions of the nascent polypeptide chain where lysines would normally occur. Only the lysine derivatives at positions 4 and 9 of the signal sequence of pPL86 can give cross-links to cytosolic or membrane proteins; the other lysines are in the portion of the nascent chain that is buried inside the ribosome. In the absence of added SRP or microsomes, only cross-links to an unknown cytosolic protein in the wheat germ extract were observed (Figure 1A, lane 2 versus lane 1, arrowhead). When dog pancreatic microsomal membranes, stripped of ribosomes by treatment with puromycin at high-salt concentrations (PK-RM), were added, membrane protein cross-links appeared (Figure 1A, lane 3). Immunoprecipitation with specific antibodies demonstrated that they consisted of weak cross-links to Sec61α (α-subunit of the Sec61p complex; Figure 1A, lane 6) and stronger ones to TRAM (Figure 1B, lane 7). As reported previously, when dog pancreatic SRP was present, cross-links to the 54-kDa subunit of SRP (SRP54) were observed in the absence of microsomes (Figure 1A, lane 4) (see Krieg et al., 1986; Kurzchalia et al., 1986), and, upon addition of membranes, cross-links to SRP54 were reduced and those to Sec61α and TRAM appeared (Figure 1A, lane 5 and immunoprecipitations in lanes 9 and 10) (see Görlich et al., 1992; Jungnickel and Rapoport, 1995). Surprisingly, the membrane protein cross-links in the absence of SRP were not much weaker than in its presence (Figure 1A, lane 3 versus lane 5), indicating that SRP-independent transfer of the nascent chain into the translocation site of the ER membrane can occur efficiently even in the presence of all cytosolic factors in the wheat germ extract. It should be noted that there is very little endogenous SRP in the wheat germ extract used (note the absence of a strong band at the appropriate position in lane 2), and that wheat germ SRP is known to interact only poorly with the SRP receptor in canine microsomes (Prehn et al., 1987). We also consider it unlikely that residual SRP in the PK-RM is responsible for targeting since efficient targeting occurred in the absence of GTP (our unpublished results) and was also observed with isolated RNCs (see below, Figure 3A) which interact only poorly with SRP in a cross-linking assay.
Figure 1.
SRP-independent membrane binding of ribosome/nascent chain complexes. (A) Photocrosslinking of pPL86 to SRP and membrane proteins. pPL86 was synthesized in the wheat germ system in the presence of modified lysyl-tRNA. Dog pancreatic SRP and PK-RM were added as indicated. The samples were UV-irradiated, separated by SDS-PAGE, and analyzed with a PhosphoImager. pPL86 × SRP54 stands for the cross-linked product containing SRP54. To identify the cross-links to membrane proteins (pPL86 × TRAM/Sec61α), immunoprecipitations with antibodies to Sec61α or TRAM, as well as control precipitations without antibodies were carried out (lanes 6–11). Arrowhead denotes a cross-linked product containing an unknown cytosolic protein. (B) Membrane targeting and translocation of pPL86. pPL86 synthesized in the wheat germ system was incubated with PK-RM and SRP as indicated. Some samples (lanes 2–4) were treated with proteinase K, the others (lanes 5–13) with puromycin. To test for translocation after puromycin treatment, the samples were incubated with proteinase K in the absence or presence of Triton X-100. pPL86-sp indicates the nascent chain fragment after signal peptide cleavage. Asterisk indicates the 30-amino acid fragment protected from proteolysis by the ribosome. (C) Determination of membrane targeting by flotation. pPL86 was incubated with different amounts of PK-RM in the absence of SRP. The samples were then layered under a sucrose gradient under low- (150 mM) or high- (500 mM) salt conditions and subjected to centrifugation. The floated and nonfloated fractions were analyzed by SDS-PAGE.
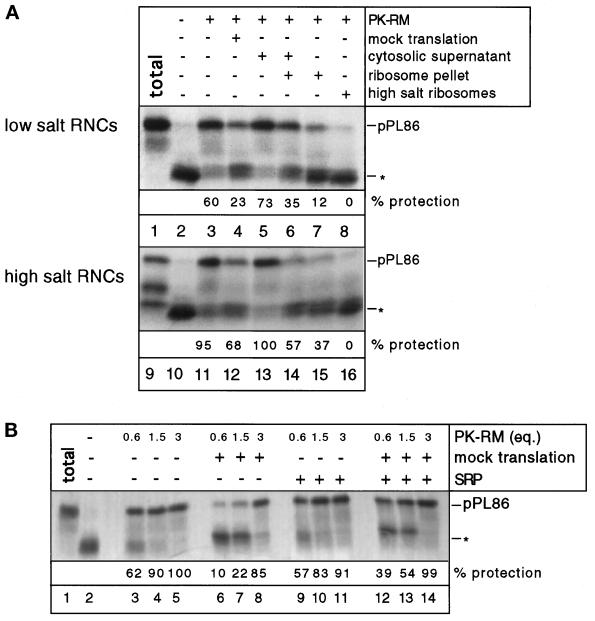
Figure 3.
Competition of RNCs with nontranslating ribosomes for membrane-binding sites. (A) Inhibition of SRP-independent targeting of isolated RNCs by ribosomes. pPL86 was synthesized in a wheat germ system and the RNCs were isolated under low- (150 mM) or high- (500 mM) salt conditions. As competitors, either a mock translation mixture was added or fractions containing the ribosome pellet or the cytosolic supernatant. In some experiments, ribosomes washed with high salt were used (high-salt ribosomes). Membrane binding to PK-RM was tested with a protease protection assay. Lanes 1 and 9 show the undigested pPL86 (total), all other samples were treated with proteinase K. Asterisk indicates the position of the ribosome-protected fragment of about 30 residues. (B) Dependence of targeting of RNCs on ribosomes and membrane-binding sites. pPL86 was synthesized in a wheat germ system and SRP was added where indicated. Some samples received a sixfold excess of a mock translation mixture. After addition of different amounts of PK-RM (given in Eq), membrane targeting was assayed by protease protection. Lane 1 shows the undigested pPL86 (total), all other samples were treated with proteinase K. Asterisk indicates the fragment of about 30 amino acids protected against the protease by the ribosome.
These results were confirmed using another targeting assay in which accessibility of a nascent chain to protease is determined; nascent chains that are inserted into the translocation site of the membrane become protected (Connolly et al., 1989; Jungnickel and Rapoport, 1995). When [35S]methionine-labeled pPL86 chains were treated with proteinase K in the absence of microsomes, a small fragment of about 30 amino acids was produced (Figure 1B, lane 2 versus lane 1, asterisk), representing the portion of the nascent chain that is buried inside the ribosome. Upon addition of PK-RM, the 30-amino acid fragment largely disappeared and instead a fragment of 86 amino acids appeared (Figure 1B, lane 3), indicating that the majority of nascent chains were targeted to the membrane. To obtain a quantitative estimate of the targeting efficiency, we calculated the percentage of radioactivity in the fragment of 86 amino acids compared with the total radioactivity in protease-protected material (the latter was found to be somewhat variable—compare, for example, Figure 1B, lanes 3 and 4—perhaps because of spontaneous hydrolysis of peptidyl-tRNA). The quantitation shows that in the absence or presence of SRP the targeting efficiency was identical (Figure 1B, lane 3 versus lane 4).
To prove that protease-protected nascent chains represent translocation intermediates, the polypeptides were released from the ribosome by puromycin; if nascent chains are properly inserted into the translocation channel, upon release they should move into the lumen of the ER and undergo signal sequence cleavage (Connolly and Gilmore, 1986). Puromycin-induced signal sequence cleavage was indeed observed (Figure 1B, lanes 6 and 7 versus lane 5); almost 70% of all preprolactin chains were processed. Again, addition of SRP did not have a significant effect (Figure 1B, lane 6 versus lane 7). The processed nascent chains were located in the lumen of the microsomal vesicles, since they were protected against protease in the absence but not presence of detergent (Figure 1B, lanes 9 and 10 versus lanes 12 and 13).
To test directly the ability of RNCs to bind to membranes in the absence of SRP and presence of cytosol, we incubated RNCs with increasing concentrations of PK-RM and subjected the membranes to flotation in a sucrose gradient (Figure 1C). At the highest membrane concentrations, more than 80% of all nascent chains floated with the PK-RM at physiological salt concentrations (150 mM) (Figure 1C, lane 8). Similar results were obtained under more stringent binding conditions (500 mM salt; Figure 1C, lane 20). Thus, all targeting assays indicate efficient SRP-independent targeting of RNCs to the ER membrane in a complete wheat germ system.
We considered the possibility that our wheat germ extract may contain particularly low levels of NAC, which would explain why we did not see inhibition of membrane targeting in the presence of cytosol. We therefore assayed the amount of NAC in five different wheat germ extracts by immunoblot using antibodies against mammalian NACα (Wiedmann et al., 1994; our unpublished results). The extracts differed by no more than a factor of three in their NAC concentration and the extract used in the experiments described here contained an intermediate concentration. The absolute concentration of NAC was estimated to be approximately 0.8 μM as judged from a comparison to recombinant mammalian NAC. NAC was present in roughly the same concentration in reticulocyte lysate.
Taken together, these data demonstrate that targeting and translocation of nascent chains can occur in the absence of added SRP even though all cytosolic factors are present. Thus, our results are in apparent contradiction with the hypothesis that cytosolic NAC inhibits SRP-independent membrane targeting (Jungnickel and Rapoport, 1995; Lauring et al., 1995a,b), as well as with the general view that SRP is essential for membrane targeting of RNCs (Walter and Blobel, 1981a).
Competition of Nontranslating Ribosomes with RNCs for Membrane-binding Sites
When comparing our targeting system with the previously described system in which SRP was essential for membrane targeting (Walter and Blobel, 1981a), there are two possible, major differences that could explain the discrepancy in results. First, the translation efficiency of our wheat germ extract may be higher than that in previous experiments which could result in a higher ratio of RNCs relative to nontranslating ribosomes. Second, the ribosome-stripped PK-RM used here have probably more binding sites for RNCs than previously used microsomes which contained bound ribosomes (rough microsomes or salt-washed microsomes) or ribosome remnants (salt- and EDTA-washed rough microsomes). The combined effect of these differences might be that RNCs encounter less competition from nontranslating ribosomes for common membrane-binding sites, allowing them to bind without SRP. The use of truncated nascent chains in our targeting reactions is not likely to account for the discrepancy, because SRP has previously been found to be required to target these chains (Connolly and Gilmore, 1986).
To test whether these explanations account for the differing results, we first investigated the effect of translation efficiency on SRP-independent targeting. To radiolabeled pPL86 in a wheat germ extract, we added increasing amounts of a translation mixture containing all components except mRNA and amino acids (mock translation). SRP-independent targeting of the nascent chains was indeed greatly reduced, regardless of whether the membranes were preincubated with the mock translation mixture (Figure 2A, lanes 3–7) or whether the mock- and mRNA-containing translation mixtures were added at the same time to the membranes (our unpublished results). When SRP was present, the targeting efficiency was much less affected by the addition of a mock translation mixture (Figure 2A, lanes 8–12), as indicated by the small percentage of change in the number of untargeted nascent chains that give rise to the 30-amino acid fragments. Addition of SRP alone did not lead to protease-protected fragments larger than 30 amino acids (our unpublished results). Thus, translation efficiency indeed appears to be a factor determining SRP-independent targeting, consistent with the idea that excess of nontranslating ribosomes may inhibit this process.
Figure 2.
Inhibition of membrane binding of pPL86 by ribosomes. (A) Microsomal membranes were preincubated with increasing amounts of mock translation mixture lacking mRNA and amino acids. In a separate tube, pPL86 was synthesized in a wheat germ system and SRP was added where indicated. The two mixtures were then combined (the numbers indicate the fold excess of the mock translation mixture over that containing mRNA). After incubation, a protease protection assay was used to determine membrane targeting of pPL86. Lane 1 shows the undigested pPL86 (total), all other samples were treated with proteinase K. Asterisk indicates the position of the ribosome-protected fragment of about 30 residues. (B) A competition experiment similar to that in A was carried out. The mock translation mixture was separated into a ribosome pellet and a cytosolic supernatant. The original mixture and both subfractions were used in a sixfold excess over mRNA-containing translation mixture in the competition experiments.
Next, we separated a mock translation wheat germ mixture into a ribosome pellet and a cytosolic supernatant. When the ribosome fraction was added to a complete translation system containing radiolabeled pPL86, inhibition of the SRP-independent targeting was as pronounced as with the complete mock translation mixture (Figure 2B, lanes 6 and 7 versus lane 4). Again, much less inhibition was seen in the presence of SRP (Figure 2B, lanes 9, 11, and 12). On the other hand, the cytosol fraction had no effect on targeting (Figure 2B, lanes 5 and 10). These data therefore support a model in which, in the absence of SRP, RNCs may be prevented from binding to the membrane by competing ribosomes; the interaction of RNCs with SRP may be required to overcome this competition. Cytosolic factors, including NAC, do not seem to inhibit ribosome binding to the membrane.
To approach conditions in which cytosolic factors, in particular NAC, were previously found to have a role in targeting, we performed experiments with RNCs isolated by sedimentation through a sucrose cushion. The cushion contained either a physiological salt concentration (150 mM; low salt RNCs), or a high-salt concentration (500 mM; high-salt RNCs) to deplete ribosome-associated proteins, such as NAC (Wiedmann et al., 1994). The presence of NAC in the low-salt washed RNCs and its absence from high-salt washed RNCs was confirmed by photocrosslinking experiments with a fragment of 77 amino acids of firefly luciferase (our unpublished results; see Wiedmann, et al., 1994).
Despite the fact that the RNCs washed with different salt concentrations differed in the amount of associated NAC, they behaved identically in the targeting assays (Figure 3A). Both types of RNCs bound in a SRP-independent manner to PK-RM (Figure 3A, lanes 3 and 11). When a mock translation mixture was added, membrane binding of both the low- and high-salt-washed RNCs was inhibited (Figure 3A, lanes 4 and 12). When the mock translation mixture was separated into a ribosome pellet and a supernatant fraction, the latter did not affect the targeting reaction (Figure 3A, lanes 5 and 13), even though it contained a large amount of NAC. On the other hand, the isolated ribosomes were even more inhibitory to targeting than the crude mock translation mixture (Figure 3A, lanes 7 and 15 versus lanes 4 and 12). Because the original level of inhibition was restored when ribosomes and supernatant were recombined (Figure 3A, lanes 6 and 14), it seems that the cytosol fraction has a moderate activity that either stimulates the targeting of RNCs or impairs the inhibition by nontranslating ribosomes. Ribosomes washed with high salt were more inhibitory than those kept at low salt (Figure 3A, lanes 8 and 16 versus lanes 7 and 15), suggesting that they may have a higher membrane affinity. Taken together, these data suggest that neither NAC nor any other cytosolic factor inhibits SRP-independent targeting of RNCs. Instead, nontranslating ribosomes have a pronounced inhibitory effect, supporting the idea that they compete with RNCs for common membrane-binding sites.
It should be noted that when targeting reactions with isolated RNCs were performed in the absence of SRP, they also lacked GTP. These conditions ensured that even if residual SRP was present in either the RNCs or PK-RM, it would not have been functional (Connolly and Gilmore, 1989; Rapiejko and Gilmore, 1997). We also found that the isolated RNCs do not interact well with SRP: whereas strong cross-links of the signal sequence of pPL86 to SRP54 were seen when SRP was added during synthesis in a crude translation mixture, addition of SRP to isolated RNCs resulted in only weak cross-links (our unpublished results). In addition, SRP had only a small effect in targeting assays with isolated RNCs, even if these were isolated under low-salt conditions and therefore contained NAC. Perhaps, upon removal of cytosolic chaperones, the nascent chain is folded into a conformation that does not allow an efficient, productive interaction with SRP.
If nontranslating ribosomes compete with RNCs for membrane-binding sites, one would expect the competition to be less pronounced if more membranes were present in the assay. Experiments in which targeting was assessed in a complete system with increasing amounts of membranes demonstrate that this is indeed the case (Figure 3B). In the absence of competing ribosomes, SRP-independent targeting of the RNCs to the membrane occurred to about the same extent at all membrane concentrations used (Figure 3B, lanes 3–5). When ribosomes were added, barely any targeting of the RNCs was observed at the lowest membrane concentration (Figure 3B, lane 6), whereas nearly complete targeting occurred at the highest concentration (Figure 3B, lane 8). In the presence of SRP, ribosome competition was much reduced at all membrane concentrations (Figure 3B, lanes 12–14). Thus, we conclude again that SRP binding allows RNCs to overcome the competition by nontranslating ribosomes which would otherwise inhibit their interaction with the microsomal membrane.
A Functional Signal Sequence Is Required to Prevent Ribosome Competition
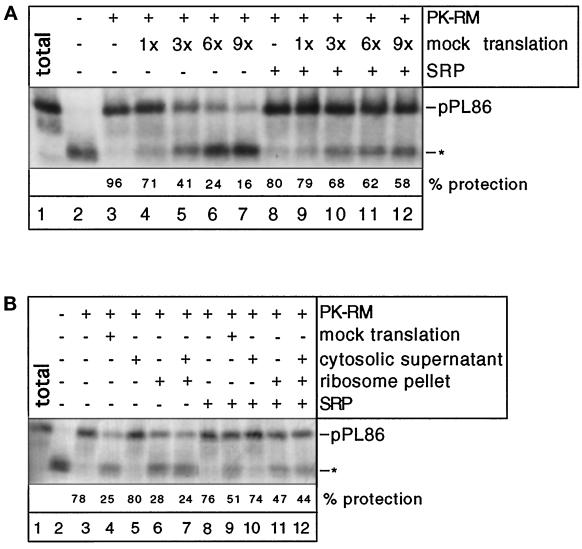
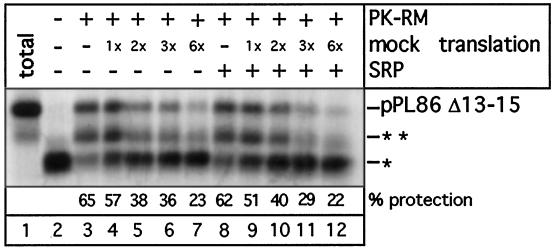
Next we tested whether the advantage that SRP-binding confers on RNCs is dependent on the nascent chain having a functional signal sequence. To this end, a mutant pPL86 chain that has three hydrophobic amino acids deleted from its signal sequence (pPL86 Δ13–15) was used in targeting assays. The full-length preprolactin bearing the deletion shows a much reduced level of translocation in vitro (about 0.5–2.5% of all molecules are translocated; Jungnickel and Rapoport, 1995). When tested in the absence of SRP in a protease protection targeting assay, a portion of the pPL86 Δ13–15 chains was completely inserted into the translocation site and became fully protected (Figure 4, lane 3 versus lane 2). Some chains were not targeted (30-amino acid fragments), but an additional membrane-protected band was also observed which corresponded to chains of about 50 amino acids (Figure 4, lane 3; indicated by two asterisks; see Jungnickel and Rapoport, 1995). When competing ribosomes were added, the intensity of the protected bands containing 86 or 50 amino acids was reduced and, correspondingly, the fragment of 30 amino acids became stronger (Figure 4, lanes 4–7 versus lane 3). Importantly, SRP did not prevent the competitive inhibition by ribosomes (Figure 4, lanes 8–12). Thus, a functional signal sequence and SRP are both required to overcome the competing effect of nontranslating ribosomes for membrane-binding sites.
Figure 4.
Ribosome inhibition of membrane targeting of RNCs with a mutated signal sequence in the absence and presence of SRP. A mutant form of pPL86 with a defective signal sequence (pPLΔ13–15) was synthesized in the wheat germ system. SRP and increasing amounts of a mock translation mixture (given as fold excess over the mRNA-containing mixture) were added as indicated. Membrane targeting was tested with a protease protection assay. Lane 1 shows the undigested pPL86 (total), all other samples were treated with proteinase K. The band designated with one asterisk is the ribosome-protected fragment of about 30 residues. The fragment indicated by two asterisks contains about 50 residues and is presumably an intermediate in the process of membrane insertion of pPL86.
SRP-independent Targeting in the Reticulocyte Lysate System
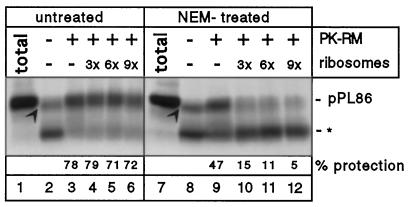
The experiments presented so far were performed with a translation system from wheat germ in combination with microsomes from dog pancreas, similar to many previous experiments (see, for example, Lauring et al., 1995a). To exclude that our results on SRP-independent targeting, in particular the lack of NAC inhibition, are restricted to this heterologous system, we performed targeting reactions with RNCs produced in the reticulocyte lysate system. This translation system contains endogenous SRP capable of interacting with canine microsomes (Meyer et al., 1982). RNCs were isolated by sedimentation through a sucrose cushion under physiological salt conditions which leaves SRP bound to them. In the absence of microsomes, the addition of protease led to the degradation of most of the nascent chains to fragments of 30 amino acids, as in the wheat germ system, but a portion was degraded to fragments slightly smaller than pPL86 (Figure 5, lane 2 versus lane 1, arrowhead), perhaps because of partial protection by a cytosolic protein. When PK-RM and GTP were present, a significant percentage of the nascent chains became fully protected from protease (Figure 5, lane 3 versus lane 2). The addition of nontranslating ribosomes had no effect on the membrane association, as seen before in the wheat germ system in the presence of SRP (Figure 5, lanes 4–6). To test for SRP-independent targeting, we inactivated SRP in isolated RNCs by treating them with N-ethylmaleimide (NEM). The membrane binding of ribosomes (Bacher et al., 1996) and their reaction with puromycin (our unpublished data) are not sensitive to this treatment, and the functions of NAC are also predicted to be insensitive because both subunits lack cysteines (Kanno et al., 1992; Yotov and St-Arnaud, 1996). The NEM-treated RNCs were still targeted to the membrane (Figure 5, lane 9 versus lane 8), but could now be competed off by nontranslating ribosomes (Figure 5, lanes 10–12). The isolated RNCs contained bound NAC, as demonstrated by photocrosslinking with a 77-amino acid fragment of firefly luciferase and pPL86 Δ13–15 (our unpublished results). Also, if the entire reticulocyte lysate was treated with NEM after synthesis of the RNCs, their membrane targeting was reduced only by about 30%. Thus, as with the wheat germ system, SRP-independent targeting was not inhibited by cytosolic factors, at least not by those that are NEM insensitive. Again, competitive inhibition of RNC targeting by nontranslating ribosomes was observed unless SRP was present.
Figure 5.
SRP-independent targeting in the reticulocyte lysate system. PPL86 was synthesized in a reticulocyte lysate system and RNCs were isolated by sedimentation through a sucrose cushion containing a low-salt concentration. One-half of the sample was treated with NEM, the other remained untreated. Before membrane targeting, low-salt-washed reticulocyte ribosomes, which were also treated with NEM, were added in increasing amounts as indicated (given as fold excess over RNCs). Membrane targeting of the RNCs was tested with a protease protection assay. Lanes 1 and 7 show the undigested pPL86 (total), all other samples were treated with proteinase K. Asterisk indicates the ribosome-protected fragment of about 30 amino acids. Arrowheads denote a fragment slightly smaller than pPL86 that is presumably protected from proteolysis by a cytosolic protein.
SRP Protection from Ribosome Competition in a Reconstituted System
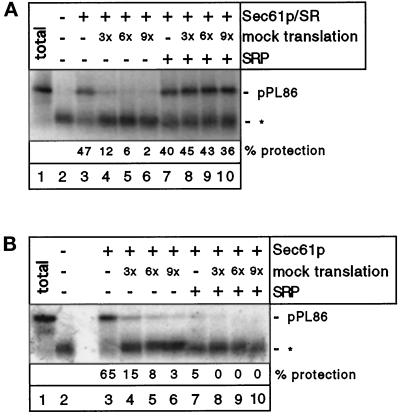
To address the mechanism by which SRP confers an advantage on RNCs in their competition with nontranslating ribosomes, we carried out targeting experiments with proteoliposomes containing only the Sec61p complex and the SRP receptor, both purified from canine pancreas microsomes (Görlich and Rapoport, 1993; Jungnickel and Rapoport, 1995). pPL86 synthesized in the wheat germ system was incubated with these proteoliposomes and its membrane targeting assessed by proteolysis (Figure 6A). As observed before with PK-RM, SRP-independent targeting was observed even though all cytosolic proteins were present (Figure 6A, lane 3). Nontranslating ribosomes competitively inhibited the targeting reaction (Figure 6A, lanes 4–6) unless SRP was added (Figure 6A, lanes 7–10). If proteoliposomes were used that contained only the Sec61p complex, SRP-independent targeting was still observed (Figure 6B, lane 3) and could be inhibited by competing ribosomes (Figure 6B, lanes 4–6). In the presence of SRP, little membrane targeting occurred (Figure 6B, lanes 7–10). This likely reflects the fact that, in the absence of the SRP receptor, SRP cannot be removed from the RNCs and thus the nascent chains cannot be transferred into the membrane. These data indicate that RNCs and nontranslating ribosomes compete for interaction with the Sec61p complex and that the binding of SRP to the RNCs, followed by an interaction with the SRP receptor, is necessary and sufficient to overcome the competition by nontranslating ribosomes.
Figure 6.
Membrane targeting of RNCs with reconstituted proteoliposomes. (A) pPL86 was synthesized in the wheat germ system. Where indicated, SRP and increasing volumes of a mock translation mixture (given in fold excess over the mixture containing the RNCs) were added before addition of reconstituted proteoliposomes containing the Sec61p complex (Sec61p) and the SRP receptor (SR). Membrane targeting was tested with a protease protection assay. Lane 1 shows the undigested pPL86 (total), all other samples were treated with proteinase K. The band designated with an asterisk is the ribosome-protected fragment of about 30 residues. (B) An experiment similar to that in A was carried out with proteoliposomes containing only the Sec61p complex.
DISCUSSION
We report here that targeting of RNCs to the ER membrane can occur without SRP, even when all cytosolic proteins, including NAC, are present. SRP-independent binding of RNCs to translocation sites in the ER membrane was competitively inhibited by nontranslating ribosomes. In the presence of SRP, however, efficient competition was no longer observed. A functional signal sequence in the nascent chain was required for the prevention of ribosome competition by SRP. Experiments with reconstituted proteoliposomes demonstrated that the interaction of SRP with its membrane receptor is sufficient to overcome the effect of competing ribosomes on the targeting of RNC–SRP complexes to the Sec61p complex. Taken together, our results indicate that the ribosome–Sec61p complex interaction is not inhibited by cytosolic factors such as NAC. Instead, as originally proposed (Walter and Blobel, 1981b), SRP appears to act as a positive effector that enables specific targeting of signal sequence-bearing RNCs, in spite of nonselective binding of ribosomes to the ER membrane.
The precise molecular mechanism by which SRP serves as a positive effector in the targeting process remains to be elucidated. In one model, SRP binding would simply increase the affinity of RNCs for the membrane by allowing RNC–SRP complexes to interact with two receptors, the SRP receptor and the Sec61p complex, rather than with only the latter. A functional signal sequence would be required to trigger tight binding of SRP to the RNCs (Walter et al., 1981). Our experiments with a signal sequence mutant have indeed shown that RNC binding to the membrane in the presence of competing ribosomes was no longer stimulated by SRP. However, it is not clear whether this effect is entirely due to a reduction of the RNC–SRP interaction since the signal sequence mutant can be cross-linked with 70% efficiency to SRP (Jungnickel and Rapoport, 1995). Although it is still possible that the binding constant of the RNC–SRP interaction is actually reduced to a greater extent than indicated by the cross-linking results, the experiments raise the possibility that SRP acts as a positive effector in a more complicated manner. For example, several rounds of cycling of the RNC between a free, SRP-bound, and membrane-bound state may be needed for a successful targeting event, and a slight reduction in the efficiency of each cycle might therefore have a strong overall effect. Alternatively, the displacement of competing ribosomes could in some way be signal sequence and SRP dependent.
Our results do not support a model in which the specificity of ribosome targeting is achieved by a cytosolic inhibitor of ribosome–membrane interaction. Although previous studies showed that NAC inhibited membrane binding of isolated RNCs unless SRP was present (Lauring et al., 1995a,b), we find that cytosol containing NAC does not inhibit the membrane interaction of isolated RNCs. Similarly, using complete translation systems and a variety of targeting assays, we have found no evidence for cytosolic inhibition of ribosome–membrane interaction. Although we cannot explain the differing results with isolated RNCs, experiments with complete translation mixture, such as those described here, may be more meaningful as they should more closely approximate physiological conditions than experiments using salt-washed RNCs.
Our results demonstrate that SRP-independent targeting can occur in both the complete wheat germ and reticulocyte lysate systems with a secretory protein that is typically regarded as SRP dependent. Why was SRP-independent targeting not seen before if NAC is not an inhibitor of ribosome binding? Two factors probably resulted in a higher efficiency of direct binding of RNCs to the Sec61p complex in our experiments as compared with previous studies (Walter and Blobel, 1981a). Both factors are expected to lead to a lower ratio of RNCs to membrane-binding sites in our experiments. One factor is the use of ribosome-stripped microsomes (PK-RM) which likely increases the concentration of unoccupied Sec61p complex molecules capable of binding newly targeted RNCs; all previous experiments were done with microsomes that contained either bound ribosomes or ribosome remnants and thus probably had fewer available Sec61p-binding sites. A second difference is the higher efficiency of current translation systems which leads to a higher ratio of RNCs to nontranslating ribosomes. As a result, there is less competitive inhibition of RNC binding to the Sec61p complex by nontranslating ribosomes. Another, probably less important, factor contributing to efficient SRP-independent targeting is the low level of SRP in the wheat germ extract used by us. Wheat germ SRP does not interact well with the mammalian SRP receptor (Prehn et al., 1987) and thus would not target RNCs. However, wheat germ SRP might sequester RNCs and prevent their membrane binding. Ironically, it now appears that the experimental conditions in the older experiments were quite fortunate since they did not favor SRP-independent translocation and therefore allowed the effect of SRP on translocation to be assayed (Walter and Blobel, 1981a,b; Walter et al., 1981). On the other hand, such conditions did not permit a test of the inhibitory effect of cytosolic factors on the targeting process.
The significance of SRP-independent targeting of RNCs in a system in which translocation and translation occur simultaneously remains unclear. In our experiments with truncated nascent chains of optimal length for membrane interaction, there is a long time period for productive membrane interaction. In a system in which translation and translocation are coupled, on the other hand, SRP-independent targeting would be reduced due to the small time window before the nascent chain becomes too long and folds into a conformation in which the signal sequence is no longer accessible. It is indeed one of the functions of SRP to delay elongation of the nascent polypeptide chain when the signal sequence has just emerged from the translating ribosome, thus extending the time period for membrane interaction (Walter and Blobel, 1981b).
Although SRP-independent targeting may not be significant for RNCs bearing a signal sequence, the fact that we have found no evidence for a cytosolic inhibitor of ribosome–membrane interaction suggests that other ribosomes can bind to the ER membrane in vivo. This assumption is consistent with the ability of high salt to strip a sizable percentage of ribosomes from rough microsomes (Adelman et al., 1973; Hanein, et al., 1996). Occasionally bound nontranslating ribosomes are probably simply displaced by SRP-containing RNCs, as suggested by our in vitro competition experiments. Whether ribosomes that synthesize nascent chains without a signal sequence can also be displaced is not yet known, but even if they could not be competed off while translating, they would presumably complete their translation at the membrane and then be released upon subunit dissociation or competition with newly arriving RNCs. A model in which membrane-bound ribosomes can synthesize polypeptide domains that ultimately reside in the cytosol is consistent with recent results on the synthesis of membrane proteins (Mothes et al., 1997). The membrane binding of a RNC would not automatically result in the productive insertion of the nascent chain into the translocation site, since a further signal sequence recognition step has to be passed inside the membrane (Jungnickel and Rapoport, 1995). Thus, the function of this second recognition step in cotranslational translocation may be mainly to prevent transport of cytosolic or other mistargeted proteins rather than to recheck a signal sequence that has passed the SRP step. The existence of a second signal sequence recognition step and the positive effect of SRP on the preceding targeting of RNCs to the membrane together may be sufficient to provide specificity in protein transport across the ER membrane.
ACKNOWLEDGMENTS
We are very grateful to Dr. Martin Wiedmann for sharing with us unpublished results, for providing materials, in particular antibodies and purified recombinant NAC, and for stimulating discussions. We thank C. Shamu and W. Prinz for critical reading of the manuscript. M.M.R. is a Howard Hughes Predoctoral Fellow. This work was supported by a grant from the National Institutes of Health to T.A.R. (GM-52586). T.A.R. is a Howard Hughes Medical Institute Investigator.
REFERENCES
- Adelman MR, Sabatini DD, Blobel G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membraneous components. J Cell Biol. 1973;56:206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher G, Lütcke H, Jungnickel B, Rapoport TA, Dobberstein B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- Borgese N, Mok W, Kreibich G, Sabatini DD. Ribosomal-membrane interaction: in vitro binding of ribosomes to microsomal membranes. J Mol Biol. 1974;88:559–580. doi: 10.1016/0022-2836(74)90408-2. [DOI] [PubMed] [Google Scholar]
- Connolly T, Collins P, Gilmore R. Access of proteinase K to partially translocated nascent polypeptides in intact and detergent-solubilized membranes. J Cell Biol. 1989;108:299–307. doi: 10.1083/jcb.108.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986;103:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Liao SR, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kurzchalia TV, Wiedmann M, Rapoport TA. Probing the molecular environment of translocating polypeptide chains by cross-linking. Methods Cell Biol. 1991;34:241–261. doi: 10.1016/s0091-679x(08)61684-2. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Herz J, Otto A, Kraft R, Wiedmann M, Knespel S, Dobberstein B, Rapoport TA. The signal sequence receptor has a second subunit and is part of a translocation complex in the endoplasmic reticulum as probed by bifunctional reagents. J Cell Biol. 1990;111:2283–2294. doi: 10.1083/jcb.111.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel B, Plath K, Kalies K-U, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. DIDS inhibits an early step of protein translocation across the mammalian ER membrane. FEBS Lett. 1993;329:268–272. doi: 10.1016/0014-5793(93)80235-m. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Görlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno M, Chalut C, Egly JM. Genomic structure of the putative BTF3 transcription factor. Gene. 1992;117:219–228. doi: 10.1016/0378-1119(92)90732-5. [DOI] [PubMed] [Google Scholar]
- Krieg UC, Walter P, Johnson AE. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Lauring B, Kreibich G, Wiedmann M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc Natl Acad Sci USA. 1995a;92:9435–9439. doi: 10.1073/pnas.92.21.9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Sakai H, Kreibich G, Wiedmann M. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995b;92:5411–5415. doi: 10.1073/pnas.92.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DI, Krause E, Dobberstein B. Secretory protein translocation across membranes- the role of the docking protein. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport TA. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between signal recognition particle and the ribosome. Curr Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- Prehn S, Wiedmann M, Rapoport T, Zwieb C. Protein translocation across wheat germ microsomal membranes requires an SRP-like component. EMBO J. 1987;6:2093–2097. doi: 10.1002/j.1460-2075.1987.tb02475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko PJ, Gilmore R. Empty site forms of SRP54 and SRα GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 1997;89:703–714. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of protein across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981a;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence and site specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981b;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Bielka H, Rapoport TA. Direct probing of the interaction between the signal sequence of nascent preprolactin and the signal recognition particle by specific cross-linking. J Cell Biol. 1987;104:201–208. doi: 10.1083/jcb.104.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotov WV, St-Arnaud R. Mapping of the human gene for the alpha-NAC transcriptional coactivator to chromosome 12q23–24.1. Mamm Genome. 1996;7:163–164. doi: 10.1007/BF03035343. [DOI] [PubMed] [Google Scholar]