Abstract
An efficient synthesis of a terminal alkyne derived from d-biotin was developed to provide an alternative for carboxylbased biotinylation. This approach was illustrated by the preparation of alkyne- and alkene-linked phenylalanine derivatives using palladium-catalyzed Sonogashira and Oh methodology. (Strept)avidin binding was observed using soluble and immobilized receptors. These results demonstrate the applicability of carbon-linked biotin derivatives for use in affinity-based purification and bioanalytical applications.
The interaction between biotin (Vitamin H) 1 and the glycoprotein avidin is one of the strongest noncovalent associations known (Ka ∼2.5 X 1015 M-1). Avidin is tetrameric and each subunit is capable of binding a biotin ligand. The high binding affinity and exceptionally slow dissociation rates of biotin results from a network of hydrogen bonds between receptor and the heterocyclic 7-oxo-3-thia-6,8-diazabicyclo[3.3.0]oct-4-yl ligand. The ureido nitrogens of biotin form hydrogen bonds with Thr35 and Asn118, the oxygen contacts Ser16 and Tyr33. Additional hydrogen bonding occurs between the biotin carboxylic acid and avidin residues Ala39, Thr40 and Ser75. The hydrophobic tetrahydrothiophene interacts with Phe79, Trp97, and Trp110.1
Avidin and the related tetrameric protein streptavidin share ∼33% conserved amino acids and strong biotin binding affinity (Ka ∼ 1.0 X 1012 M-1).2 Specific interactions include hydrogen bonding between biotin urea nitrogens with Ser45 and Asp128 and oxygen contacts with Asn23, Ser27 and Tyr43. Interactions also exist between Asn49 and Ser88 and the biotin carboxylic acid group. This network of hydrogen bonding in conjunction with hydrophobic interactions with four Trp residues (Trp 79, 92, 108, 120) and the tetrahydrothiophene result in high binding affinity.3
Biotin-(strept)avidin systems have been used for a variety of applications such as affinity isolation and purification, immunoassay, diagnostics, and localization.4,5,6 Nearly any biological entity can be labeled with biotin including peptides, proteins, oligonucleotides and antibodies. Antibody-(strept)avidin conjugates are used in the pretargeting approach to deliver radiolabeled biotin therapeutics and imaging agents.7 Other applications include drug delivery8 and material science.9
A variety of biotin-labeling agents are commercially available.10 Activated esters (N-hydroxysuccinimdyl ester and p-nitrophenyl esters), maleimide, and iodoacetyl derivatives are frequently used for coupling to substrates possessing amine or thiol groups.11 In vivo applications of amide-linked biotin derivatives can be problematic due to cleavage by the endogenous enzyme biotinidase.12,13
Several structural modifications have been described to prevent biotinidase cleavage however, these derivatives exhibit increased dissociation rates from (strept)avidin. Biotin amide derivatives containing a small α-substituent have provided the most effective balance of biotinidase stability and (strept)avidin affinity.14 Both N-methylation of the biotinamide linkage15 and homologation of the valeric acid chain provided increased resistance to cleavage16 as did replacement of the biotinamide connection with thiourea. Unfortunately, increased dissociation rates were observed with all of these linkagemodified derivatives.17
We have been interested in developing biotin conjugates of hydrophobic receptor ligands for affinity purification. Available biotin-coupling reagents are not suitable for derivatizing compounds that lack reactive functional groups such as amine, carboxylate or thiol. The installation of polar substituents on a hydrophobic receptor substrate is possible but may decrease receptor-binding affinity due to unfavorable electrostatic interactions, modified solvation characteristics, different lipophilicity (as measured by log P octanol/water), and increased steric interactions.18 As an alternative, we considered using non-polar linkages synthesized via catalytic C-C bond coupling methodology. Replacement of the biotin carboxamide with an alkyne or alkene connection would eliminate chemical or enzymatic hydrolysis. The success of this approach depends on the resulting C-C linked biotin substrates maintaining (strept)avidin affinity.
We elected to replace the carboxylic acid group with an alkyne that would allow entry into a wide variety of metal-catalyzed coupling procedures.19 The synthesis was initiated by acid catalyzed esterification of biotin. Selective reduction of the ester 2 was accomplished using diisobutylaluminum hydride (DIBAL) at —78 °C, affording alcohol 3 in 96% yield. Reaction of 3 with toluenesulfonyl chloride in pyridine at 0 °C provided the sulfonate ester 4 in 94% yield.20 The iodide 5a and bromide 5b were prepared by halide substitution of 4.
Low yields of the desired alkyne 6 were obtained from direct lithium acetylide substitution reactions of tosylate 4 or iodide 5a. Fortunately, the alkyl bromide 5b underwent efficient displacement with lithium acetylideethylenediamine in DMSO with careful temperature control ≤ 15 °C and produced the desired alkyne 6 in high yield. This synthesis provided alkyne-biotin derivative 6 with a combined yield of 66% over five steps from biotin.
We evaluated avidin binding of 6 in solution. Competitive displacement of 2-(4′-hydroxyphenylazo)benzoic acid (HABA) with biotin derivatives provides a convenient spectroscopic method for assessing avidin binding. In solution, HABA forms a complex with the biotin-binding site of avidin that is characterized by an absorbance band at 500 nm. Displacement of the HABA substrate by biotin results in decreased absorbance at 500 nm. This method has been widely used as a qualitative assay for evaluation of biotinylated substrates.21
We constructed a standardized biotin response curve for the HABA-avidin complex in 0.1 M phosphate buffer solution. The decrease in absorbance accompanying additions of 5 μL aliquots of d-biotin reference solution (5.0 × 10-4 M in 0.10 M NaH2PO4) were measured in triplicate and plotted against the concentration of added biotin. A response curve was generated analogously for alkyne 6 by addition to the standardized HABA-avidin solution. A decrease in absorbance was observed with the addition of 6 demonstrating HABA displacement. From the results of this assay it can be concluded that the association constant of 6 remained high since HABA (Ka = 6 × 106 M-1) is displaced.21 While the HABA displacement was attenuated relative to biotin, effective binding of 6 was retained despite replacement of the carboxyl group with alkyne.
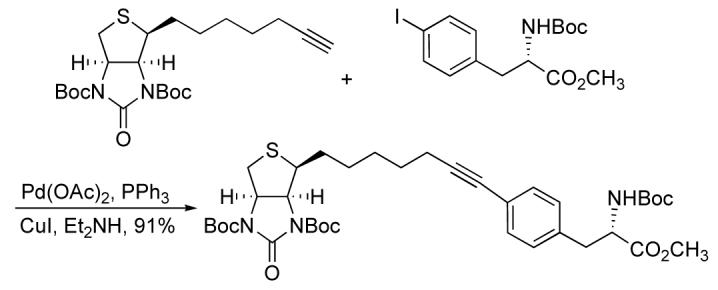
Encouraged by the affinity exhibited by 6, we proceeded to synthesize a phenylalanine biotin derivative. Amino acids labeled with biotin are important in peptide synthesis.8,22,23 Sonogashira cross coupling reactions have been used to produce alkynyl phenylalanine derivatives.24,25 In order to increase the solubility of 6 in organic solvents, we protected the urea as the di-tert-butyl carbamate 7. The 4-iodo-phenylalanine derivative 8 was coupled with alkyne 7 under standard conditions using catalytic Pd(OAc)2, PPh3, and CuI in diethylamine to give conjugate 9 in 91% (Scheme 2).
Scheme 2.
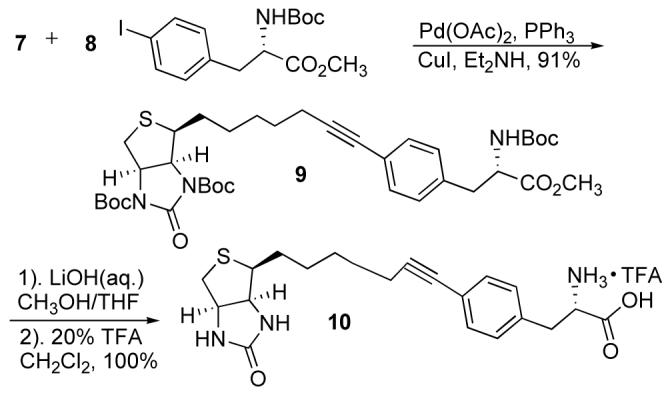
Synthesis of alkyne derivatives.
Hydrolysis of the ester with LiOH(aq.) in MeOH/THF at 0 °C, followed by deprotection of the tBoc-groups with trifluoroacetic acid (TFA) in CH2Cl2 gave the alkynelinked phenylalanine conjugate 10. Spectroscopic characterization by NMR and HPLC-MS confirmed product identity.
We wanted to investigate the affect of shortening the linkage between the biotin heterocycle and the phenylalanine on avidin affinity. Recently, Oh et al. reported a palladium-catalyzed insertion of arylboronic acids into terminal alkynes.26 Following this procedure, alkyne 7 was coupled with protected 4-boronic acid derivative 11 to afford alkene 12 in 73% yield (Scheme 3). The alkene protons appeared as doublets at δ 5.21 and 5.01 (J = 1.3 Hz) in the 1H-NMR spectrum, consistent with the expected alkene. No other alkene isomers were observed by 1H-NMR.
Scheme 3.
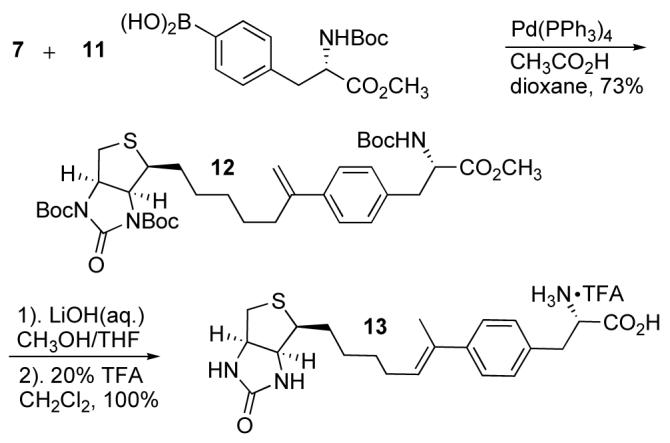
Synthesis of Alkene Derivatives.
The ester 12 was hydrolyzed with LiOH, followed by tBoc-deprotection with TFA/CH2Cl2. The product was characterized spectroscopically and by HPLC-MS. The exocyclic alkene isomerized to the more substituted (E)-alkene 13 during deprotection. The vinyl proton signal was observed as a triplet at δ 5.78 (J = 7.1 Hz) and the allylic methyl group appeared as a singlet at δ 1.96.
The avidin affinities of 10 and 13 were evaluated using the HABA assay as described for 6. Both derivatives displaced HABA from avidin with comparable efficiency. The sp2 hybridized carbon in alkene 13 corresponds to the position of the carboxyl in biotinamide derivatives and can be considered to be isosteric although not capable of hydrogen bonding. The linear alkyne connection in 10 is comparable to the overall length of homologated biotinamide conjugates. These minor differences in length and geometry did not significantly alter the observed binding affinity of 10 and 13. These derivatives exhibited decreased affinity for avidin relative to biotin as expected from the deletion of the carboxyl group.
Solid-supported streptavidin sorbents have been widely used for batch affinity isolation and purification of biotinylated substrates. Non-binding components are easily removed by elution and isolation of biotinylated substrates is accomplished by dissociation from the supported-streptavidin.27 The strong biotin-streptavidin interaction presents a problem in these applications because harsh conditions are required to dissociate the biotin from the support-matrix. Biotin derivatives with reduced binding affinity such as desthiobiotin and 2-imunobiotin are advantageous because elution occurs with milder conditions. We prepared the fluorescent alkynelinked phenylalanine 15 to evaluate the affinity for immobilized streptavidin (Scheme 4).
Scheme 4.
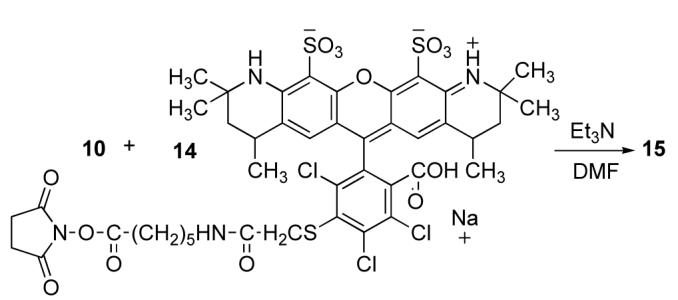
Synthesis of Biotin Alexafluor® conjugate.
The α-amino group of 10 was coupled with Alexafluor 546® succinimidyl ester 14 to give 15. Compound 15 was dissolved in 0.1M phosphate buffer pH 7.2 at a concentration of 3.27 × 10-6 M. A 3 mL aliquot (50% of the manufacture’s recommended loading) was added to centrifuge tube containing strepavidin (5 mL) on agarose support. The tube was agitated for 15 min, centrifuged for 5 min, and the supernate containing unbound 15 was isolated. The tube was washed with 10 bed volumes of water to collect unbound substrate 15. The concentration of free 15 was determined spectroscopically. The immobilized streptavidin retained 84% of 15, confirming the potential for C-linked derivatives of 6 in batch isolation methods.
The avidin binding of 15 was studied by fluorescence titration. It was observed that 15 displayed higher emission intensity in the presence of avidin. Lo et al. have attributed the observed fluorescence enhancement to greater hydrophobicity within the binding pocket. A standardized solution of solubilized avidin was prepared and titrated with 15. Fluorescence emission enhancement was monitored to determine the concentration required to saturate the receptor.28 From this titration experiment, a dissociation constant (Kd) of ca. 3.8 ×10-9 M was determined for avidin bound 15 using the method described by Srivastava et al.29 This dissociation constant lies between the values of desthiobiotin (Kd = 5.0 ×10-13 M)30 and 2-imunobiotin (Kd = 8.0 ×10-6 M)31 and is also comparable to other recently reported biotin-fluorophore conjugates.28,32
This study evaluated the potential for carbon-linked biotin conjugates in affinity-based methods. Alkyne 6 was efficiently synthesized from d-biotin and exhibited strong binding in the HABA assay despite the deletion of the carboxylic acid. The tBoc-protected alkyne 7 was effectively coupled using palladium-catalyzed reactions. Synthetic phenylalanine derivatives 10, 13, and fluorescent probe 15 exhibited affinity for soluble and immobilized (strept)avidin, as expected for binding to the preserved 7-oxo-3-thia-6,8-diazabicyclo[3.3.0]oct-4-yl core. This approach enables conjugation to substrates that cannot be labeled with activated carboxylate derivatives of biotin. The covalent carbon linkage that replaces the carboxamide group of traditional biotin conjugates decreases relative (strept)avidin binding but retains sufficient affinity for bioanalytical applications and provides complete protection from chemical or enzymatic hydrolysis. This approach may be extended to other alkyne coupling and cycloaddition procedures.
Supplementary Material
Scheme 1.

Synthesis of Biotin-Alkyne
Figure 1.

Emission Spectrum of 15.
Acknowledgment
This research was supported by NIH/SCORE GM08136. C. C. was supported by NIH/RISE GM61222. Instrumentation facility was funded by NIH RR16480 from the NCRR-INBRE Program.
Footnotes
Supporting Information Available: General methods, experimental details, copies of 1H-NMR and 13C-NMR spectra of compounds 1-3, 5-7, 9, 10, 12, 13, and HABA titration curves.
References
- (1).Puglise L, Coda A, Malcovati M, Bolognesi M. J. Mol. Biol. 1993;231:698. doi: 10.1006/jmbi.1993.1321. [DOI] [PubMed] [Google Scholar]
- (2).Pahler A, Hendrickson WA, Kolks MAG, Argarana CE, Cantor CR. J. Biol. Chem. 1987;29:13933. [PubMed] [Google Scholar]
- (3)(a).Chilkoti A, Stayton PS. J. Am. Chem. Soc. 1995;117:10622. [Google Scholar]; (b) Klumb LA, Chu V, Stayton PS. Biochemistry. 1998;37:7657. doi: 10.1021/bi9803123. [DOI] [PubMed] [Google Scholar]
- (4).Wilchek M, Bayer EA. Anal. Biochem. 1988;171:1. doi: 10.1016/0003-2697(88)90120-0. [DOI] [PubMed] [Google Scholar]
- (5).Diammandis EP, Christopoulos TK. Clin. Chem. 1991;35:625. [PubMed] [Google Scholar]
- (6).Diamandis EP, Christopoulos TK, editors. Immunoassay. Acedemic Press; San Diego, CA: 1996. [Google Scholar]
- (7).Boerman OC, van Schaijk FG, Oyen WJG, Corstens FHM. J. Nuc. Med. 2003;44:400. [PubMed] [Google Scholar]
- (8).Bhunilya S, Park SM, Kim BH. Org. Lett. 2005;7:1741. doi: 10.1021/ol050300r. [DOI] [PubMed] [Google Scholar]
- (9).Niemeyer CM. Angew. Chem. Int. Ed. 2001;40:4128. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- (10).Haugland RP. Chapter 4. In: Spence MTZ, editor. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 10th Invitrogen; Eugene, OR: 2005. p. 141. [Google Scholar]
- (11).Foulon CF, Alston KL, Zalutsky MR. Bioconjugate Chem. 1997;8:179. doi: 10.1021/bc970006m. [DOI] [PubMed] [Google Scholar]
- (12).Singh R, Maloney EK. Anal. Biochem. 2002;304:147. doi: 10.1006/abio.2002.5624. [DOI] [PubMed] [Google Scholar]
- (13).Bogusiewicz A, Mock NI, Mock DM. Anal. Biochem. 2004;331:260. doi: 10.1016/j.ab.2004.05.020. [DOI] [PubMed] [Google Scholar]
- (14).Wilbur DS, Hamlin DK, Chyan M, Kegley BB, Pathre PM. Bioconjugate Chem. 2001;12:616. doi: 10.1021/bc0100096. [DOI] [PubMed] [Google Scholar]
- (15).Pazy Y, Kulik T, Bayer EA, Wilchek M, Livnah O. J. Bio. Chem. 2002;277:30892. doi: 10.1074/jbc.M202874200. [DOI] [PubMed] [Google Scholar]
- (16).Wilbur DS, Hamlin DK, Pathare PM. Bioconjugate Chem. 1997;8:572. doi: 10.1021/bc9700852. [DOI] [PubMed] [Google Scholar]
- (17).Wilbur DS, Chyan M, Pathare PM, Hamlin DK. Bioconjugate Chem. 2000;11:569. doi: 10.1021/bc000024v. [DOI] [PubMed] [Google Scholar]
- (18).Schneider C, Scholer HF, Schneider RJ. Steroids. 2004;69:245. doi: 10.1016/j.steroids.2004.01.003. [DOI] [PubMed] [Google Scholar]
- (19).Negishi E, Anastasia L. Chem. Rev. 2003;103:1979. doi: 10.1021/cr020377i. [DOI] [PubMed] [Google Scholar]
- (20).DeLaLuz PJ, Golinski M, Watt DS, Vanaman TC. Bioconjugate Chem. 1995;6:558. doi: 10.1021/bc00035a009. [DOI] [PubMed] [Google Scholar]
- (21).Hermanson GT. Bioconjugate Techniques. Academic Press; San Diego, CA: 1996. p. 591. [Google Scholar]
- (22).Gallivan JP, Lester HA, Dougherty DA. Chem. Biol. 1997;4:739. doi: 10.1016/s1074-5521(97)90312-4. [DOI] [PubMed] [Google Scholar]
- (23).Kumar V, Aldrich JV. Org. Lett. 2003;5:613. doi: 10.1021/ol027044s. [DOI] [PubMed] [Google Scholar]
- (24).Sonogashira K, Tohda Y, Hagihara N. Tetrahedron Lett. 1975;50:4467. [Google Scholar]
- (25).Hoffmanns U, Metzler-Nolte N. Bioconjugate Chem. 2006;17:204. doi: 10.1021/bc050259c. [DOI] [PubMed] [Google Scholar]
- (26).Oh CH, Jung HH, Kim KS, Kim N. Angew. Chem. Int. Ed. 2003;42:805. doi: 10.1002/anie.200390214. [DOI] [PubMed] [Google Scholar]
- (27).Streptavidin Agarose: For Affinity Chromatography and Immunopecipitation of Biotinylated Proteins. Invitrogen; Eugene, OR: 2003. Techincal Document #25-0524. [Google Scholar]
- (28)(a).Lo KK, Hui W, Ng DC. J. Am. Chem. Soc. 2002;124:9344. doi: 10.1021/ja026598n. [DOI] [PubMed] [Google Scholar]; (b) Lo KK, Hui W. Inorg. Chem. 2005;44:1992. doi: 10.1021/ic049059n. [DOI] [PubMed] [Google Scholar]
- (29).Wang Z, Kumar NR, Srivastava DK. Anal. Biohem. 1992;206:376. doi: 10.1016/0003-2697(92)90381-g. [DOI] [PubMed] [Google Scholar]
- (30).Muller W, Ringsdorf H, Rump E, Wildberg G, Zhang X, Angermaier L, Knoll W, Liley M, Spinke J. Science. 1993;262:1706. doi: 10.1126/science.8259513. [DOI] [PubMed] [Google Scholar]
- (31).Reznik GO, Vajda S, Sano T, Cantor CR. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13525. doi: 10.1073/pnas.95.23.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Marek M, Kaiser K, Gruber HJ. Bioconjugate Chem. 1997;8:560. doi: 10.1021/bc970088e. [DOI] [PubMed] [Google Scholar]; (b) Lo KK, Lee TK. Inorg. Chem. 2004;43:5275. doi: 10.1021/ic049750q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


