Abstract
We have used DNA double crossover (DX) molecules to produce a translation system that generates unique molecular products. The particular species of DX molecule used contains an even number of half-turns between crossover points, so there is a continuous strand on both sides of the molecule. One of these strands acts as the input strand containing the message, and a second strand acts as the product of translation. The crossover strands carry the ‘code’ that connects the two sides of the molecule. This system is more robust, more extendable, and simpler than previous DNA-based translation systems that have been reported. It is designed to be useful in a variety of applications that utilize the concept of translating from one code to another.
Keywords: Molecular Translation, Unusual DNA Motifs, DNA Double Crossovers, DNA Self-Assembly, Molecular Encryption
Recently, we reported combining two sequence-dependent robust DNA-based 2-state nanomechanical devices with DNA parallelogram motifs to produce a translation machine.1 A picture of the device is shown in Figure 1a. The important parts of the machine are the gaps at the top, flanked by numbers that represent sticky ends. A series of DAE-type DNA double crossover molecules2 are used in this system to emulate aminoacyl-tRNA molecules. Their top strand corresponds to the amino acid, and the bottom domain of the DX molecules contains sticky ends complementary to the sticky ends in the gaps. The independently-addressable 2-state devices switch the components flanking the gaps, so that four different translation products can be produced, depending on the states of the two devices. One weakness of this device are that it is an extremely complex DNA construct for the current state of the art; another weakness is that it is a rotationally-based linear system, so that the size of the machine must be similar to the size of the product. In a different vein, Endo et al. have reported a system that entails more complex chemistry, but is conceptually much simpler.3 They have used two different strands of DNA coupled via their phosphates in an unusual linkage as the basis of a translation system; these strands link a ‘message’ strand and a ‘product’ strand. The system does not enforce the directionality of the product, because of possible swiveling around the unusual linkage.
Figure 1.
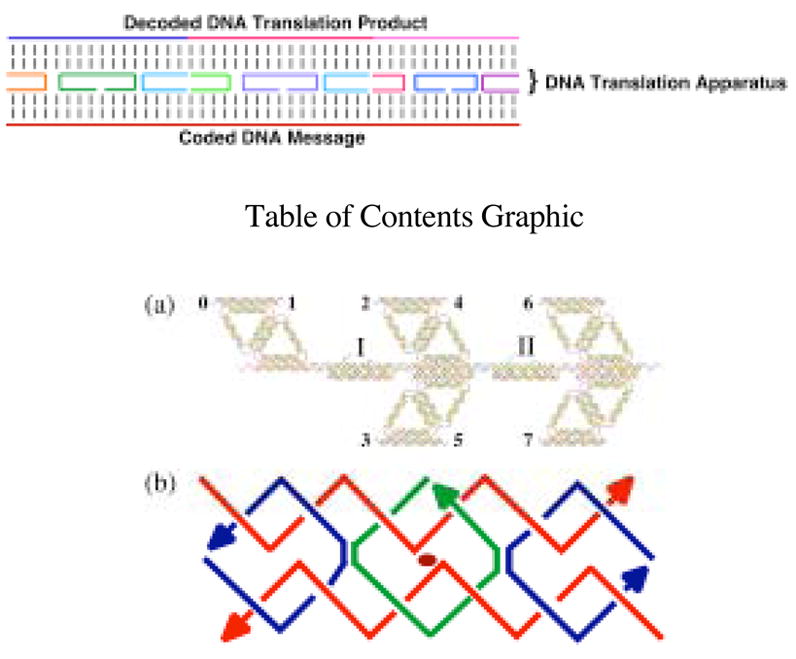
Schematic Drawings of DNA Molecules (a) A Nanomechanical Translation Apparatus. The Arabic numbers refer to sticky ends, and the Roman numerals indicate two independently addressable DNA-based nanomechanical devices. Double crossover (DX) molecules (analogous to aminoacyl tRNA molecules in protein synthesis) bind to the upper sticky ends, depending on the states of the devices. The setting of the devices shown would bind DX molecules flanked by sticky ends complementary to 1 and 2 and to 4 and 6. Switching the state of Device II, for example, would bind DX molecules complementary to 1 and 2 and to 4 and 7. Four states are available. (b) A DAE-type DX molecule. 3′ ends are indicated by arrowheads; the elliptical symbol at the center indicates the backbone dyad symmetry. The two red strands can function as a coded message and as the decoded translation product. The three crossover strands (two blue and one green) act as the translation apparatus, which decodes the message.
Here, we combine the simplicity of the Endo et al. system with the simplicity of DNA synthesis to produce a translation system that is unidirectional and easy to access: It is not necessary to do anything more complex than ordering oligonucleotides from a DNA synthesis facility or from a commercial vendor. The basis of the system we have tested is the DAE isomer of the DNA double crossover (DX) molecule. DNA double crossover molecules contain two DNA double helical domains that are linked to each other by two Holliday-like4 crossover points.2 In addition to their use in the rotary translation system described above, DX molecules have been used to build 2D periodic arrays,5 as components of nanomechanical devices,6 in algorithmic assemblies,7 and in assembling DNA nanotubes.8 There are a variety of DX molecules, but those in which the crossovers occur between strands of opposite polarity are the most robust when the separations between the crossovers are short, say two turns of DNA or less.2 These crossover points can be separated by an even or odd number of half-turns of the DNA double helix. Those separated by an even number of turns lead to molecules that contain continuous strands in both helical domains.
The schematic diagram in Figure 1b shows a DAE molecule whose crossovers are separated by a single turn of DNA. The two red strands are the continuous strands. One can serve as the message and the other as the translation product. The translation apparatus consists of the three strands that connect them, the two blue strands and the green strand they flank. If one designates, say, the bottom red strand as the message strand and the top red strand as the product strand, one can establish a code or correspondence relating the two by selecting the sequences of the crossover strands that connect them: The parts of the crossover strands that bind to the lower strand select a particular message strand, and the parts that bind to the upper strand select a particular product strand for which it codes. There is no need to limit the message strand to a strand from a single DAE unit. Indeed, the naturally-occurring messages that are used in protein synthesis (mRNA molecules) are long continuous strands that code for the entire length of a protein polymer. In the work reported here, we have used message strands that are two or three DAE units long. Thus, we are directing a specific product strand with several components from a particular message encrypted for all of those components.
The experimental systems we have used are shown in Figure 2. Panel 2a shows the first system that we used: Two DAE units connected laterally by the message strand on the bottom, labeled DAB09. To its right is a biotin-containing hairpin loop that terminates the assembly. We believed initially that we would need to use biotin-based magnetic streptavidin bead purification in this system to eliminate incomplete assemblies, as we had done in the previous system.1 However, that step proved not to be necessary; for convenience, we used the same biotinylated strand throughout this work, but never used a biotin-based purification. We refined our hybridization procedure based on the results obtained with this system: In the first attempts, the tiles were annealed separately, i.e., Tile A consisting of strands DA01, DA02, DA03 and DA04 and Tile B consisting of strands DB05, DB06, DB07 and DB08, following a fast annealing protocol, were then mixed together with strands Dbiotin and DAB09, heated to 40°C and cooled to 16 °C followed by ligation of strands DA04 and DB08. Strand DB08 contains a phosphate group on its 5′ end. Phosphorylation of the targeted end strand (DA04 in this case) did not lead to significantly different results (data not shown); this is also true of the (more successful) two-component and three-component systems described below. This protocol gave undesired products together with the expected one. So as to minimize undesired ligation products, all the strands were annealed together thereafter from 90 °C to 16 °C. The sequencing of the AB product did not give good results; presumably this was due to the difficulty in completely denaturing the hairpins on both strands DA04 and DB08, a necessary step during the PCR amplification done before sequencing. Even though the signal is very low starting from around the 40th base, the sequence is correct for the most part (see supplementary data).
Figure 2.
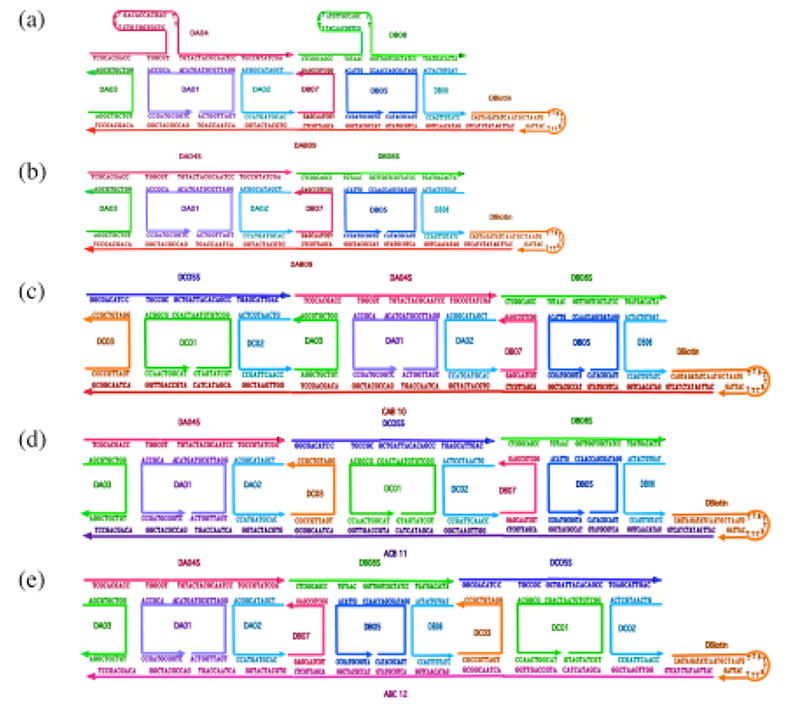
Schematic Drawings of the Systems Used Here. (a) An Unsuccessful System Containing Hairpins in the Product Structures. The hairpins interfered with PCR, and were abandoned. (b) A Simpler and Successful Two-Component System. There is a single 84-mer strand (DAB09 -- red) at the bottom acting as the message, and two product 42-mer strands (DA04S -- pink and DB08S -- green) shown before ligation. The translation strands with the crossovers in them are shown as well. The double DX nature of the molecules (fused by strand DAB09) is evident from the drawing. The space between the two helical domains of the DX molecules is exaggerated for clarity; there is only a single nucleotide backbone linkage between them. The biotin groups in the hairpin on the right are not used. (c-e) Three-component systems analogous to the 2-component system shown in (b).
These problems led us to design a modified system, illustrated in Figure 2b; it is related closely to the first system, but it lacks the extra loops in the product strands. The complex forms well, as demonstrated by non-denaturing gel electrophoresis, and the target band containing 84 nucleotides is the primary product, although some higher bands are visible on the denaturing gel that characterizes the products of the ligation reaction (Figure 3a). Most importantly, the sequence that we get is correct and is easy to read (see supplementary information).
Figure 3.
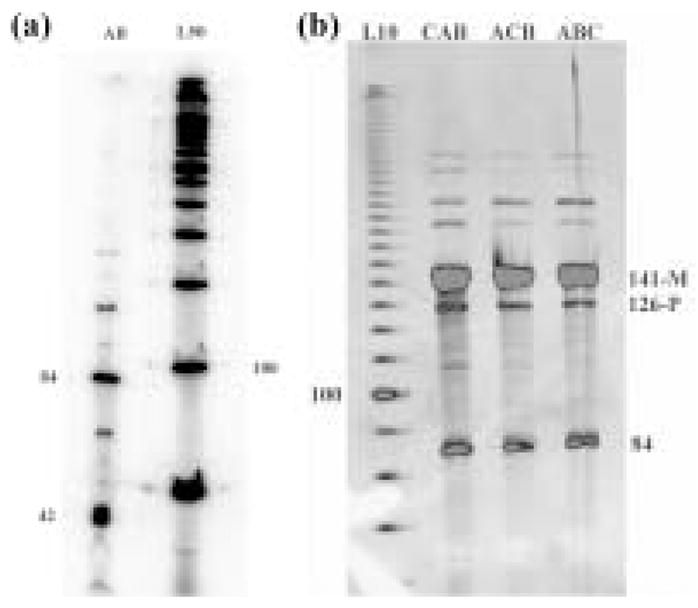
Denaturing Gels Showing the Products of Ligation. (a) Autoradiogram of the Ligation Products Corresponding to the Molecule shown in Figure 2b. A 50-mer linear marker lane (L50) is shown at the right. The target 84-mer is the major ligation product visible in the lane (AB) containing ligation products. (b) Scan of an Ethidium-Stained Gel Showing the Ligation Products Corresponding to the Molecules in Figures 2c-2e. A 10-mer linear marker lane (L10) is shown at the left. The products of systems CAB (Figure 2c), ACB (Figure 2d) and ABC (Figure 2e) are shown at the right. Dimer 84-mer molecules are visible. The ratio of 84-mers to target 126-mer products (126-P) are roughly 55:45 (CAB) and 63:37 (ACB and ABC). The message strand (141-M) is indicated as well.
These preliminary results encouraged us to move onward to three-component systems, illustrated in Figures 2c, 2d and 2e. These three molecules represent permutations of the same three sequences in the message strand, which should lead to corresponding permutations of the product strands in the ligated material. The first question that we address is whether the translation complexes form cleanly. Figure 4 contains non-denaturing gels illustrating that the hybridization products are concentrated into a single band of approximately the expected molecular weight. In general, we take this to be an indication that the complex has formed well.9 Smaller contaminants are just barely visible in each of the triple complexes, but these are not significant contributors to the overall population of molecules; this is why the biotin-based purification noted above was unnecessary. In each of the three cases shown in Figures 2c-2e, we obtain the target translation product as the major band on the denaturing gel analyzing the products of ligation. This gel is shown in Figure 3b. As is characteristic of these systems,1 there are some bands that represent failures of ligation. However, the key issue is whether the target product molecules have been assembled in the order prescribed by the message strand. In each case, the sequence and the length of the product is the target dictated by the message (see supplementary material). In addition, there are some bands longer than the targets, corresponding to unexpected products in both panels of Figure 3. For example, in Figure 3b, there are band pairs in all three lanes of lengths ~170 and ~190, and of lengths ~220 and ~240. It is possible to devise scenarios for their origins, e.g., template plus one monomer for a band in the first group. However, if the target length is known, as it is here, one can simply ignore them and analyze the target band; this is the procedure followed by Adleman in his original solution to the Hamiltonian path problem.10
Figure 4.
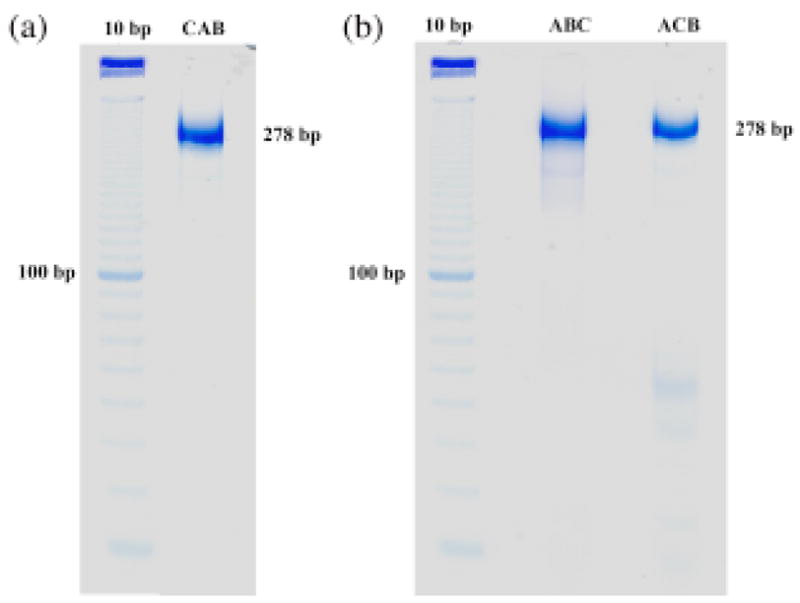
Non-Denaturing Gels of the Triple Combinations of Messages and Pre-Ligation Products. Both panels contain linear markers separated by 10 nucleotide pairs. The products have a mobility in the vicinity of their total mass, which is 278 nucleotide pairs. The single band seen is a clear indicator of the stability of the triple-DX complex.
We have described a simplified translation system combining the chemical simplicity of using conventional DNA with the simple translator method of Endo et al.3 There is no ambiguity about the products at the level of a three-unit message. It is unclear how large a message one could use in this system, but it seems likely that targets significantly longer than those attempted here would to be tractable. Although not explored here, messages that contain repeated elements ought to be amenable to this system. Recently, Tabor et al.11 have described another translation system based on ligation within the context of deoxyribozymes. This system is closely related to the work reported here, although its extension to systems larger than one connection between two components is not described by the authors. It is important to realize that our system does not require a continuous message, but could be expected to work with disjoint message segments that spanned two different DX units; the advantage of disjoint messages is that the entire message need not be determined at once, and the product can be used to describe the history of an evolving or oscillating system. Such an application would require a non-coding tail to connect the disjoint messages together on the translation apparatus, which would need to contain a complementary non-coding segment. In addition to making new DNA sequences, it is reasonable to expect systems that use pendent polymers12 could be introduced as product strands to produce more diverse products. In principle, this system is not limited to DAE molecules, but could employ other DX molecules containing two continuous strands that do not contain reverses, such as DPE, DPON or DPOW,2 as well as three-domain molecules (e.g., ref. 13), PX molecules14 or even complex bundles.15
We have shown that simple, commercially-available DNA sequences can be used to produce an encryption whose key is itself DNA. In an era when information on the internet can be used to produce a virus with unsophisticated DNA chemistry,16 we have shown here that a DNA message can be produced and translated (decoded) using the same basic approach. Without the key (the central strands of the DAE molecule), there is no simple way to break the code; with those strands, it is trivial to do so. It is worth noting that the system described cannot be used to evade bio-terrorism detection algorithms to produce sequences from bio-hazardous molecules, because both the message and product strands must be present in the translation solution. Although translation systems related to this one are not likely to be involved in nucleic acid metabolism, it is worth pointing out that meiotic intermediates are DPE-type2 DX molecules17 that also have continuous strands analogous to the message and product strands discussed here. PX molecules, which have been suggested as being involved in the search for homology14 in cellular systems have similar features.
Supplementary Material
The experimental methods and the sequencing results. This material is available free of charge via the Internet at http://pubs.acs.org/.
Acknowledgments
This research has been supported by grants GM-29554 from NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CCF-0523290 and CTS-0548774, CTS-0608889 from the NSF, 48681-EL from ARO, NTI-001 from Nanoscience Technologies, Inc., and a grant from the W.M. Keck Foundation, to N.C.S.
References
- 1.Liao S, Seeman NC. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu TJ, Seeman NC. Biochem. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 3.Endo M, Uegaki S, Majima T. Chem Commun. 2005:3153–3155. doi: 10.1039/b503247d. [DOI] [PubMed] [Google Scholar]
- 4.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 5.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Sun W, Shen Z, Seeman NC. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PWK, Papadakis N, Winfree E. PLOS Biology. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. J Am Chem Soc. 2004;126:16344–16352. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 9.Seeman NC. Protocols in Nucleic Acid Chemistry. John Wiley & Sons; New York: 2002. Unit 12.1. [Google Scholar]
- 10.Adleman L. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 11.Tabor JJ, Levy M, Ellington AD. Nucl Acids Res. 2006;34:2166–2172. doi: 10.1093/nar/gkl176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu L, Lukeman PS, Canary J, Seeman NC. J Am Chem Soc. 2003;125:10178–10179. doi: 10.1021/ja035186r. [DOI] [PubMed] [Google Scholar]
- 13.Seeman NC. NanoLett. 2001;1:22–26. [Google Scholar]
- 14.Shen Z, Yan H, Zhang X, Seeman NC. J Am Chem Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathieu F, Liao S, Mao C, Kopatsch J, Wang T, Seeman NC. NanoLett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cello J, Paul AV, Wimmer E. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 17.Schwacha A, Kleckner N. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The experimental methods and the sequencing results. This material is available free of charge via the Internet at http://pubs.acs.org/.


