Abstract
We have studied components of the endoplasmic reticulum (ER) proofreading and degradation system in the yeast Saccharomyces cerevisiae. Using a der3–1 mutant defective in the degradation of a mutated lumenal protein, carboxypeptidase yscY (CPY*), a gene was cloned which encodes a 64-kDa protein of the ER membrane. Der3p was found to be identical with Hrd1p, a protein identified to be necessary for degradation of HMG-CoA reductase. Der3p contains five putative transmembrane domains and a long hydrophilic C-terminal tail containing a RING-H2 finger domain which is oriented to the ER lumen. Deletion of DER3 leads to an accumulation of CPY* inside the ER due to a complete block of its degradation. In addition, a DER3 null mutant allele suppresses the temperature-dependent growth phenotype of a mutant carrying the sec61–2 allele. This is accompanied by the stabilization of the Sec61–2 mutant protein. In contrast, overproduction of Der3p is lethal in a sec61–2 strain at the permissive temperature of 25°C. A mutant Der3p lacking 114 amino acids of the lumenal tail including the RING-H2 finger domain is unable to mediate degradation of CPY* and Sec61–2p. We propose that Der3p acts prior to retrograde transport of ER membrane and lumenal proteins to the cytoplasm where they are subject to degradation via the ubiquitin-proteasome system. Interestingly, in ubc6-ubc7 double mutants, CPY* accumulates in the ER, indicating the necessity of an intact cytoplasmic proteolysis machinery for retrograde transport of CPY*. Der3p might serve as a component programming the translocon for retrograde transport of ER proteins, or it might be involved in recognition through its lumenal RING-H2 motif of proteins of the ER that are destined for degradation.
INTRODUCTION
The endoplasmic reticulum (ER) is the site of entry of proteins into the secretory pathway. The organelle has to guarantee proper folding of the newly synthesized membrane, secretory and lysosomal proteins before they are delivered to their site of action (Pelham, 1989; Pryer et al., 1992; Gaut and Hendershot, 1993). This requires a quality control system involved in proofreading and recognition of unassembled and malfolded proteins with subsequent delivery to a degradative machinery (Lippincott-Schwartz et al., 1988; Klausner and Sitia, 1990; Bonifacino and Lippincott-Schwartz, 1991; Bonifacino and Klausner, 1994). This process called ER degradation or ER-associated degradation had been thought to occur within the ER. An increasing number of substrates of this process had been found in yeast and mammalian cells; however, the proofreading and degradation mechanism remained essentially unknown (reviewed by Fra and Sitia, 1993). Most of the substrates found in mammalian cells are mutated forms of proteins such as the cystic fibrosis transmembrane conductance regulator (CFTR), α1-antitrypsin, or unassembled subunits of the T-cell receptor (Cheng et al., 1990; Wileman et al., 1991; Le et al., 1992; Amitay et al., 1992). Thus, it has been suggested that ER degradation could play an important role in the etiology of severe human diseases such as cystic fibrosis or certain forms of pulmonary emphysema. The ER degradation pathway is also involved in the regulated degradation of enzymes as 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase in response to changes in intracellular levels of certain metabolites (Hampton and Rine, 1994; McGee et al., 1996). Finally, it has been postulated that certain viruses such as the human cytomegalovirus and the human immunodeficiency virus type 1 could use this pathway to interfere with the normal immune response to increase the virulence of the infection (Wiertz et al., 1996a; Kerkau et al., 1997).
A set of unidentified ER resident proteinases had been proposed to be responsible for the degradation of unfolded and unassembled proteins retained in the ER. The finding that proteolytic destruction of CFTR (Jensen et al., 1995; Ward et al., 1995), the mutated translocon component Sec61–2 of yeast (Biederer et al., 1996), and HMG-CoA reductase (Hampton et al., 1996; McGee et al., 1996) were degraded via the cytoplasmic proteasome changed the view of specific ER resident proteases responsible for degradation of ER located membrane proteins. This view was further changed when the major histocompatibility class I complex was found to be degraded via the proteasome upon expression of cytomegalovirus proteins (Wiertz et al., 1996a,b). A radical change of the idea that specific ER-intrinsic proteinases were involved in degradation of malfolded proteins was initiated when degradation of a mutated and malfolded yeast CPY* encoded by the prc1–1 allele (Wolf and Fink, 1975; Finger et al., 1993; Knop et al., 1996a,b; Hiller et al., 1996) and mutant forms of pre-pro-α-factor and human α1 proteinase inhibitor (Werner et al., 1996), all located in the ER lumen, were found to be degraded via the cytoplasmic proteasome.
Degradation of these lumenal proteins by the cytoplasmic proteasome could only be explained by a retrograde transport of the soluble substrates from the ER lumen to the cytoplasmic side of the membrane. The finding of glycosylated and ubiquitinated CPY* being attached to the cytosolic face of membrane vesicles carrying the ER lumenal marker Kar2p demonstrated the existence of such a retro-translocation event (Hiller et al., 1996). A first hint to the nature of the postulated retrograde transporter came from the finding that in the presence of the human cytomegalovirus protein US2, major histocompatibility complex (MHC) class I heavy chain breakdown intermediates could be coimmunoprecipitated with Sec61p (Wiertz et al., 1996b). However, it remained unclear whether this was a finding specifically due to the action of the virally programmed protein. Using lumenal CPY* as substrate, studies on mutants defective in components of the translocon uncovered Sec61p, Sec63p, and Kar2p acting as a subcomplex in retrograde transport of the mutant protein in vivo (Plemper et al., 1997). In vitro studies on sec61 mutants using a mutated pro-α-factor as substrate corroborated the function of Se61p in retrograde transport (Pilon et al., 1997).
In a search for additional components involved in proofreading, recognition, and reprogramming the translocon for export and degradation of CPY*, we had isolated a variety of der mutants defective in the ER degradation process (Knop et al., 1996). Cloning of the genes by complementation of the respective mutations uncovered DER1 encoding a 211-amino acid ER membrane protein and DER2 coding for the ubiquitin-conjugating enzyme Ubc7, by this linking ER degradation of CPY* to modification with ubiquitin prior to proteolysis via the proteasome (Hiller et al., 1996). Here we report on the cloning of the DER3 gene, the characteristics and possible functions of the encoded protein.
MATERIALS AND METHODS
Yeast Strains and Growth Media
The DER3 wild-type strain used for all experiments was W303–1C (MATα prc-1-1 ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can-100) (Knop et al., 1996). Strain YAF29 carrying the der3–1 mutation was derived from EMS mutagenesis of the strain YAF6 (MATa pra1ΔSSprc1-1 leu2-3,112) (Finger et al., 1993). Strains W303–1CD, W303-CQ, and W303-CPQ contained a deletion in DER1, UBC7 and both UBC6 and UBC7, respectively (Hiller et al., 1996; Knop et al., 1996). Strain YRP086 (MATα) (Plemper et al., 1997) was derived from crossing strain W303–1C with YFP338 (MATα) kindly provided by M. Rose. To obtain strains W303–1CΔ3, W303–1CDΔ3, and YRP105 which carry a deletion in DER3, strains W303–1C, W303–1CD, and YRP066, respectively, were transformed with EcoRI–PvuII-digested DNA of the plasmid pUC/del3 followed by selection for His+ transformants. In all of the cases, the correct integration of the der3 allele harboring the HIS3 gene was checked by Southern blotting.
Yeast media were prepared according to Guthrie and Fink (1991).
Cloning of DER3
DER3 was isolated by transforming the YAF29 strain with the yeast genomic DNA library constructed by Cvrková and Nasmyth (1993) in the centromeric plasmid YCplac111. Leu+ transformants were replica-plated onto fresh plates of selective media covered by nitrocellulose membranes and incubated for 5 d to induce the accumulation of CPY*, essentially as described by Knop et al. (1996). Colonies were lysed according to Knop et al. (1996) and CPY* was detected with polyclonal CPY antisera and goat anti-rabbit horseradish peroxidase-conjugated antibodies. Blots were developed with 4-chloronaphthol. Nonstaining colonies were picked, retested by Western blotting, and the plasmids were rescued.
Molecular Biological Techniques and Plasmid Construction
Standard techniques of molecular biology were performed as described in Ausubel et al. (1988) and Sambrook et al. (1989).
Fragments of the complementing plasmid YCpDER3 isolated from the gene library were subcloned into YCplac111. A BamHI–EcoRI fragment of 2.8-kb contained the DER3 gene was cloned into the 2 μ plasmid YEp366 (Myers et al., 1986) to overexpress the DER3-encoded protein in yeast. To construct the DER3 disruption allele, an EcoRI–HindIII fragment of 3.8 kb of YCpDER3 was cloned into pUC19, yielding pUCDER3, and then a SalI–EcoRV fragment containing the complete open reading frame (ORF) of DER3 was replaced by the HIS3 gene, yielding the pUC/del3 plasmid. To construct a GST-Der3 fusion protein, plasmid pFUS1 was generated by cloning an EcoRI–EcoRV fragment including the C-terminal 139 codons of the ORF into the EcoRI–SmaI sites of pGEX-3X (Smith et al., 1988). Plasmid pDEG1 (Chen et al., 1993), containing a Deg1-β-galactosidase fusion, was kindly provided by M. Hochstrasser and T. Sommer. To overexpress DER3 in yeast cells under the control of the GAL1 promoter, plasmid pBM/DER3 was constructed by subcloning the whole ORF of DER3 into the vector pBM150 (Johnston and Davis, 1984) digested with BamHI and SalI. To create both restrictions sites flanking the coding region of DER3, PCR techniques were employed using as primers oligonucleotides GAL1 (5′-TAGATGTCGACTAATTTCCGC-3′) and GAL2 (5′-AGACAGGATCCTAATATGGTGCC-3′). Expression of Der3p encoded by pMB/DER3 was checked by Western blot and its ability to complement the der3–1 mutation. To construct the plasmid YCpDER3ΔR, pUCDER3 was digested with EcoRV for 5 min and religated, yielding pUCDER3ΔR lacking an EcoRV fragment of 342 bp of the DER3 ORF. The 3.5-kb EcoRI–HindIII fragment of pUCDER3ΔR was cloned into YCplac111 to yield YCpDER3ΔR.
Western Blotting
For Western blotting and immunodetection of CPY* and Der3p, yeast cells were grown on CM medium (Guthrie and Fink, 1991) until an OD600 of 3.1-ml aliquots of culture were lysed according to Yaffe and Schatz (1984). After trichloroacetic acid precipitation, pellets were resuspended in 100 μl of urea buffer [5% SDS, 8 M urea, 200 mM Tris-HCl (pH 6.8), 0.1 mM EDTA, bromophenol blue] and shaken for 10 min at 60°C. After a short centrifugation, 15–20 μl of the supernatants were loaded on a 8% SDS-polyacrylamide gel with subsequent electrophoresis (Laemmli et al., 1970) followed by Western blotting.
Cell extracts enriched in membranes were prepared as described by Serrano (1988).
Protease-protection experiments were basically performed as described by Hiller et al. (1996) with the following modification. The trichloroacetic acid precipitates were resuspended in 100 μl of urea buffer and samples of 20 μl were separated on a 8% SDS-polyacrylamide gel, blotted, and the specific immunoreactive material was detected with the respective antibodies.
In all cases, a suitable horseradish peroxidase-conjugated secondary antibody was used and the blots were developed with the ECL detection kit (Amersham, Arlington Heights, IL).
Subcellular Fractionation and Enzymatic Assays
Subcellular fractionation was performed as described by Antebi and Fink (1992), modified by Knop et al. (1996).
Guanosine diphosphatase (GDPase) activity was measured as described by Abeijon et al. (1989).
The kinetics of degradation of the Deg1-β-galactosidase fusion protein was performed as described in Plemper et al. (1997). Briefly, exponentially growing yeast cells on CM medium were harvested by centrifugation and resuspended in fresh CM medium adjusted to 3 units of OD600. Cycloheximide was added up to a final concentration of 0.5 mg/ml. Samples of 90 μl of culture were taken after 0, 30, 60, and 90 min, and activity of β-galactosidase was measured. β-Galactosidase activity was calculated as described by Fürst et al. (1988).
Cell Labeling and Immunoprecipitation
For CPY* pulse-chase experiments, yeast cells were grown on CM medium until an OD600 of 2 was reached. Cells from 5 ml of culture concentrated in 1 ml of labeling medium were labeled with [35S] methionine for 20 min at 30°C. Thereafter, 1 ml of chase medium was added and aliquots of 450 μl were removed for each time point. Growth, labeling, and chase medium and further treatment of cells, immunoprecipitation, and SDS-PAGE were done as described by Finger et al. (1993).
For pulse-chase experiments performed for detection of the Sec61p, exponentially growing cells in SD medium were shifted to the restrictive temperature of 38°C and labeled with [35S] methionine for 10 min. Conditions of growth and labeling, disruption of cells, immunoprecipitation, and SDS-PAGE were performed as described previously (Biederer et al., 1996). Labeled Sec61p was visualized by autoradiography and quantitated with a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Antibodies
For detection of Der3p, polyclonal antibodies recognizing the 139-amino acid containing C-terminal region of the protein were generated. Using the plasmid pFUS1, a GST-Der3 fusion protein was expressed in Escherichia coli and purified by affinity chromatography (Ausubel et al., 1989). One milligram of the purified protein was used as antigenic material for immunization of rabbits.
Polyclonal anti-Der3p was diluted 1:2500 for Western blotting and 1:250 for immunofluorescence. Polyclonal anti-CPY (Finger et al., 1993), monoclonal anti-CPY (Molecular Probes), polyclonal anti-Kar2p (a gift from R. Schekman and H.K. Rudolph), and monoclonal antibodies recognizing the 100-kDa subunit of the vacuolar membrane ATPase (Molecular Probes) were diluted 1:10,000 for immunoblotting. Polyclonal antibodies specific for Sec61p were kindly supplied by T. Sommer.
Immunofluorescence
Immunofluorescence for intracellular localization of Der3p and Der3ΔRp was performed as previously described for Der1p (Knop et al., 1996). Cy3-fluorescence was monitored with a Zeiss confocal laser scanning microscope.
RESULTS
Cloning of the DER3 Gene
The der3–1 mutation had been introduced into yeast strain YAF6 by EMS mutagenesis (Knop et al., 1996). This allele leads to high accumulation of CPY*. The DER3 gene was cloned by functional complementation of the der3–1 mutation. A yeast genomic DNA library (Cvrková and Nasmyth, 1993) in the centromeric LEU2-based plasmid YCplac111 was transformed into the der3–1 mutant strain. Leucine prototrophic colonies were replica-plated onto plates containing selective medium and covered with nitrocellulose membranes. Thirty thousand transformants were tested for the absence of CPY antigenic material on the nitrocellulose filters. Positive clones were picked and steady-state levels of CPY* were tested by Western blotting. One clone contained a plasmid which was clearly able to complement the mutant phenotype associated with the der3–1 mutation and therefore restored degradation of CPY* to wild-type levels. Plasmid YCpDER3 with a genomic DNA insert of 10 kb was rescued from the complementing clone. After subcloning, a 2.8-kb BamHI–EcoRI fragment was found to be the smallest piece of DNA able to complement the der3–1 phenotype. Complete sequencing of this region and subsequent search in the data bank revealed the presence of one ORF. It corresponded to the ORF YOL013c (embl/774755/SCYOL013) located on chromosome XV and was named DER3 (Figure 1). Recently, a publication appeared showing that the same gene, which had been called HRD1, encodes a gene product involved in the regulation of HMG-CoA reductase (Hampton et al., 1996).
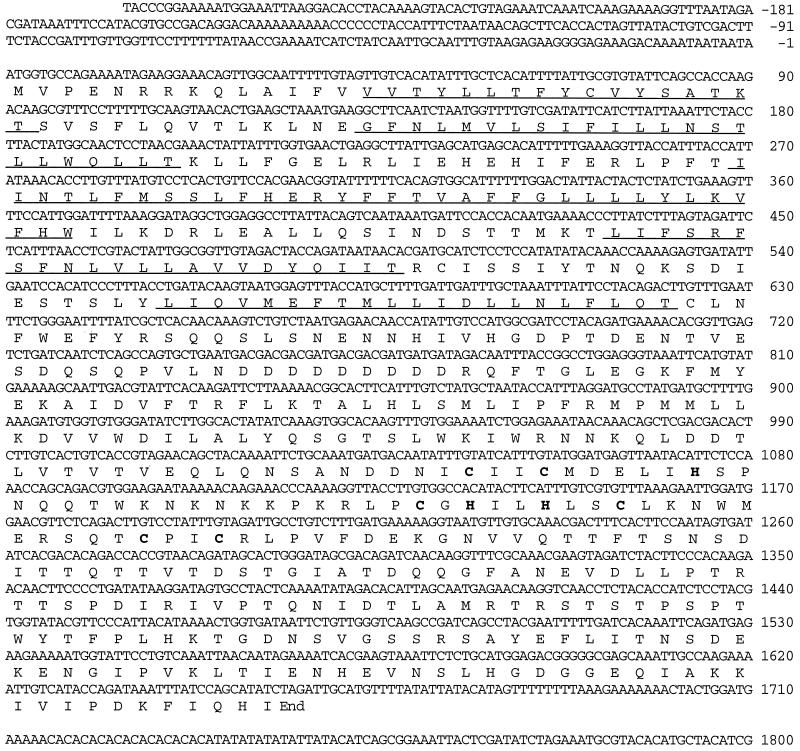
Figure 1.
Structure of the DER3 gene. Nucleotide sequence of DER3 and the predicted amino acid sequence of Der3p are shown. The regions with the predicted transmembrane domains are underlined; the H2-RING finger motif is marked with bold letters.
DER3 encodes a protein of 551 amino acids with an estimated molecular mass of 63,494 Da. The predicted structure for Der3p shows a hydrophobic N-terminal region with five putative transmembrane domains and a long hydrophilic carboxyl-terminal tail containing a RING-H2 finger motif (Freemont, 1993).
No significant homology with proteins in the data bank was found. The Der3 protein sequence does not contain any known ER retention signal such as HDEL (Pelham et al. 1988) or KKXX (Townsley and Pelham, 1994). Two putative N-glycosylation sites are located in positions 58 and 137. A specific DER3 mRNA of 1.8 kb was identified by Northern blot analysis (our unpublished observations).
Chromosomal Loss of DER3 Causes a Defect in ER Degradation of CPY*
To investigate the function of the Der3p in the ER-associated degradation, we determined the phenotype of a yeast strain entirely lacking the DER3 gene. We constructed a strain carrying a DER3 disruption allele by replacing the entire ORF of DER3 by the HIS3 gene. As found for mutants carrying the der3–1 allele, der3 null mutants were viable and showed highly increased levels of CPY* on immunoblots when compared with an isogeneic wild-type strain (Figure 2A). Pulse-chase analysis showed that accumulation of CPY* is due to a block in CPY* degradation (Figure 2B). Deletion of DER3 led to a complete stabilization of CPY* even after a 3-h chase, whereas CPY* cannot be detected any more after 60 min of chase in a strain harboring the DER3 wild-type allele. We had to confirm that the der3–1 mutation was in fact a lesion in the DER3 gene. We therefore crossed strain YAF29 carrying the der3–1 allele with strain W303–1CΔ3 harboring the DER3 deletion. The diploid exhibited high levels of CPY*, suggesting that both alleles resided in identical genes. As expected, diploids constructed from the DER3 wild-type strain W303–1C and the der3–1 mutant strain YAF29 exhibited wild-type behavior with respect to degradation of CPY*.
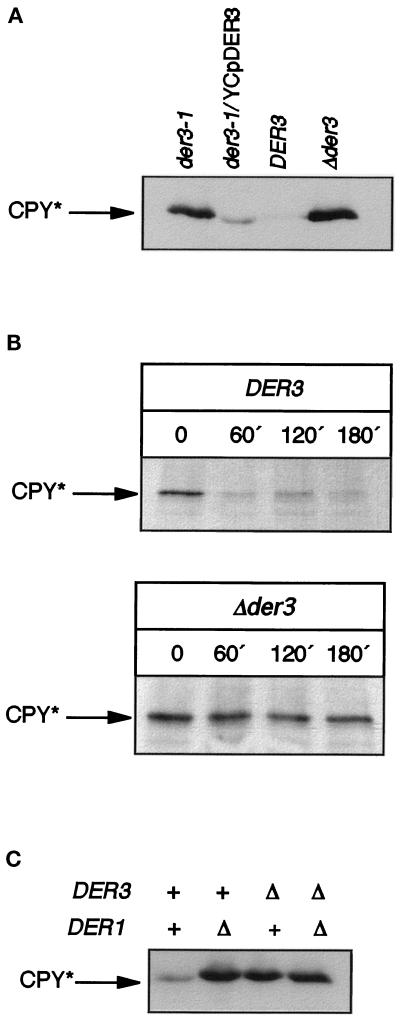
Figure 2.
(A) Steady-state levels of CPY* in DER3 wild-type and mutant strains. Western blotting was performed with crude extracts prepared from early stationary phase yeast cells grown on CM medium of the strains YAF29 (der3–1), YAF29 harboring the centromeric complementing plasmid YCpDER3, wild-type strain W303–1C (DER3), and DER3 deleted strain W303–1CΔ3 (Δder3). (B) Kinetics of CPY* degradation in DER3 wild-type and mutant strains. A pulse-chase experiment performed with the wild-type strain W303–1C (DER3) and W303–1CΔ3 (Δder3) is shown. Cells were labeled for 20 min with [35S]methionine, and samples were taken at the indicated chase periods. (C) Steady-state levels of CPY* in DER1 and DER3 mutant and wild-type strains. Crude extracts from early stationary phase cells of the strains W303–1C (wild-type), W303–1CD (Δder1), W303–1CΔ3 (Δder3), and W303–1CDΔ3 (Δder1 Δder3) were prepared. CPY-immunoreactive material was detected after SDS-PAGE and Western blotting with monoclonal anti-CPY antibodies.
Strains carrying a deletion in DER3 did not show any additional phenotype when growth of cells at different temperatures was measured or when cells were treated with dithiothreitol and β-mercaptoethanol, substances which induce misfolding of proteins. Also on inositol-free media Δder3, cells did not show any difference as compared with the DER3 wild-type strain.
A similar phenotype as found for der3 mutants, a defect in CPY* degradation, had previously been described for mutant strains defective in the DER1 gene encoding an ER membrane protein (Knop et al., 1996). We investigated a possible interaction between Der1p and Der3p by overexpressing either DER3 in a strain carrying the der1–2 mutation or by overexpressing DER1 in the der3–1 background. In no case could a restoration of CPY* degradation be observed (our unpublished results). A Δder1Δder3 double mutant strain did not show any further increase of CPY* steady-state levels as compared with the respective single mutants (Figure 2C).
Der3p Is a Protein of the ER Membrane
We studied the intracellular localization of Der3p. For this purpose, specific polyclonal antibodies able to detect this protein in yeast cells by Western blot, immunofluorescence, or immunoprecipitation were obtained. Briefly, a Der3p-GST fusion protein containing the last 139 amino acids of the C-terminal hydrophilic tail of the protein was expressed in E. coli, purified, and used as antigen to generate an immune response in rabbits. The obtained antisera exclusively recognized a protein of around 64 kDa in wild-type crude extracts after SDS-PAGE on a Western blot (Figure 3 A). This protein band was considerably increased upon overexpression of DER3 on the multicopy plasmid YEpDER3. No immunoreactive protein could be observed in der3–1 or Δder3 mutant cells (Figure 3A).
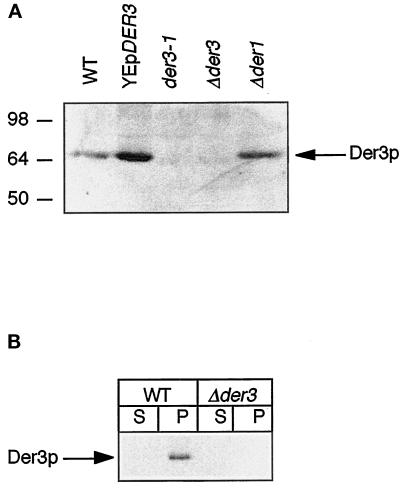
Figure 3.
(A) Identification of the Der3 protein. Crude extracts of exponentially growing cells on CM medium of strains W303–1C [wild type (WT)], W3031-C harboring the high-copy plasmid YEpDER3, YAF29 (der3–1), W303–1CΔ3 (Δder3), and W303–1CD (Δder1) were prepared, separated by SDS-PAGE, and immunoblotted. Levels of Der3p were detected with polyclonal Der3p antisera. (B) Der3p is an integral membrane protein. Crude extracts enriched in membranes were prepared from exponentially growing cells on CM medium of strains W303–1C (WT) and W303–1CΔ3 (Δder3). Aliquots of the soluble (S) and membrane (P) fractions were loaded onto a 8% SDS-polyacrylamide gel and separated by electrophoresis followed by Western blotting. Der3p antigenic material was visualized with anti-Der3p polyclonal antibodies.
The N-terminal hydrophobic region of Der3p with its five putative transmembrane domains pointed to a membrane localization of the protein. Der3p can indeed be detected in a membrane-enriched fraction of yeast cell extracts and not in the soluble fraction (Figure 3B). Although the Der3p sequence contains two recognition sites for N-glycosylation, the protein is not glycosylated. No shift of Der3p-specific protein bands could be observed after SDS-PAGE and immunoblotting upon endoglycosidase F treatment (our unpublished observations).
To investigate intracellular membrane localization of Der3p, we performed a subcellular fractionation in yeast cells harboring the high-copy plasmid YEpDER3. As can be seen in Figure 4A, Der3p cofractionates with the ER lumenal chaperone Kar2p. Der3p does not cofractionate with a protein of the vacuole, Vph1p, or the Golgi localized GDPase (Abeijon et al., 1989; Manolson et al., 1992). ER localization of Der3p was confirmed by immunofluorescence experiments performed in cells overexpressing Der3p. A typical pattern for the ER, perinuclear with stained regions along the plasma membrane (Preuss et al., 1991), was observed when Der3p was visualized (Figure 4B). When subcellular fractionation was repeated in yeast cells harboring only the chromosomal copy of DER3, faint, but clearly visible Der3p immunoreactive bands which cofractionate with the ER resident protein Kar2p were seen, confirming the ER localization of the protein (Figure 4C).
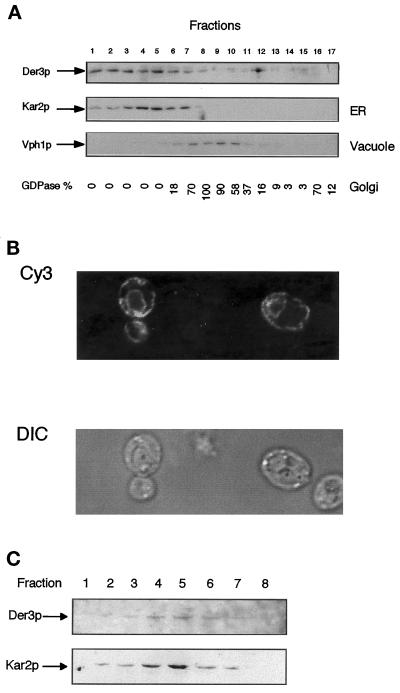
Figure 4.
Intracellular localization of Der3p. (A) Der3p cofractionates with the ER resident protein Kar2p. Spheroplasts of the W303–1C (wild-type) strain harboring the high-copy plasmid YEpDER3 were prepared, and, after a gentle lysis, the homogenate was fractionated on a 10-step sucrose gradient (18–54%, steps of 4% sucrose). Aliquots of each fraction were subjected to SDS-PAGE and Western blotting. Levels of Der3p, Kar2p (ER marker), and the 100-kDa subunit of the vacuolar membrane ATPase were detected with the respective specific antibodies. The activity of GDPase, a Golgi marker, is given as percentage of the highest activity value measured. (B) Der3p is localized in ER-like structures. Exponentially growing cells of the strain W303–1C transformed with the 2 μ plasmid YEpDER3 were fixed and stained with polyclonal anti-Der3p antibodies and goat anti-rabbit Cy3 antibody. Cy3 fluorescence was monitored with a laser confocal microscope (Cy3, Cy3 fluorescence; DIC, Nomarski optics). (C) Also when expressed from the chromosomal copy of the gene, Der3p cofractionates with Kar2p. A subcellular fractionation was performed as in A using strain W303–1C. For detection of Der3p, 500 μl of each fraction were diluted up to 10-fold with 10 mM HEPES (pH 7.5) and the 100,000-g membrane pellets (1 h) were resuspended in urea buffer and loaded onto a 8% SDS-polyacrylamide gel. After Western blotting Der3p and Kar2p were detected with suitable antibodies. Only fractions 1–8 are shown; no band could be detected in fractions 9–17.
The Hydrophilic Tail of Der3p Is Oriented to the ER Lumen
Studies on the mechanism of function of Der3p require the knowledge of the orientation of its large 351 amino acids containing hydrophilic C-terminal domain harboring a RING-H2 finger motif. The function of this class of motifs is unknown, but has been proposed to be involved in protein–protein interactions (Saurin et al., 1996).
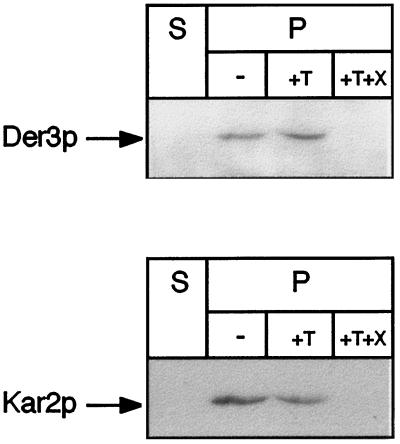
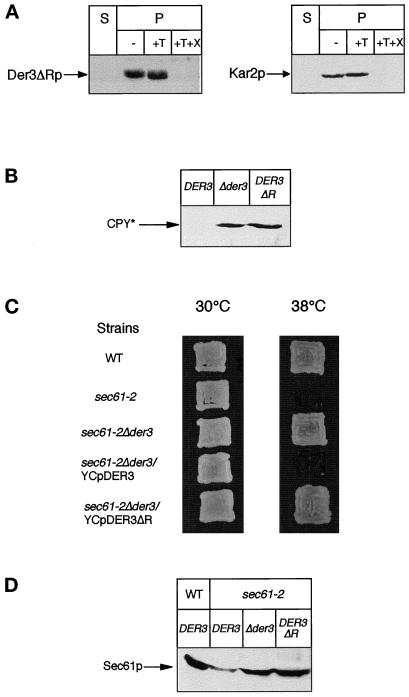
Microsomes of wild-type cells were isolated and treated with trypsin. If the C-terminal tail of Der3p was located to the cytoplasmic side of the ER, one would expect reduction of the molecular mass of Der3p by trypsin treatment by about 40 kDa after SDS-PAGE. As can be seen in Figure 5, no shift of the Der3p-reactive band can be observed after trypsin treatment of microsomes. Only when Triton X-100 is added, the C-terminal region of Der3p becomes accessible to the protease and is completely degraded. Since the antibody is only directed against the hydrophilic C-terminus of Der3p, this treatment leads to complete loss of the ability of the antibodies to detect any fragment of the protein (Figure 5). Furthermore, only after treatment of the microsomes with Triton X-100 is Kar2p accessible to trypsin digestion. This demonstrates that the hydrophilic C-terminal tail of Der3p is located in the lumen of the ER.
Figure 5.
The C-terminal tail of Der3p is oriented to the ER lumen. Yeast spheroplasts of the wild-type strain W303–1C were subjected to gentle lysis. The lysate was centrifuged to separate a soluble fraction (S) from the pellet fraction (P) containing intact yeast microsomes. Aliquots of microsomes were incubated for 30 min on ice in the absence (−) or in the presence of trypsin (+T) or trypsin and Triton X-100 (+T+X). Treatments were stopped by trichloroacetic acid precipitation (10% final concentration). Pellets were resuspended in urea buffer, immunoblotted after SDS-PAGE, and stained with anti-Der3p or anti-Kar2p polyclonal antibodies.
Loss of Der3p Causes an Accumulation of CPY* Inside the ER
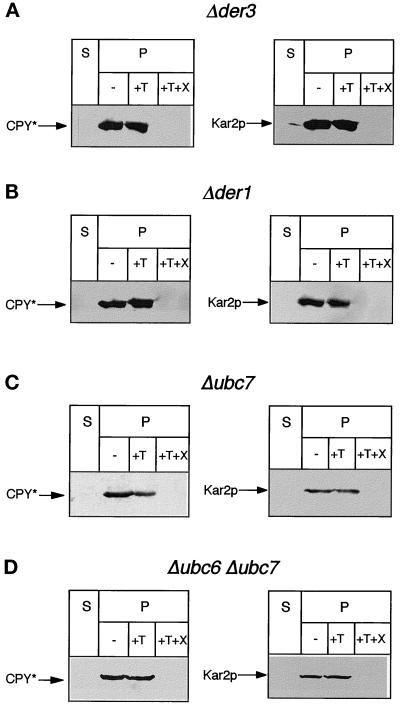
To shed some light on the function of Der3p in the ER-associated degradation pathway, we investigated the locus of the accumulation of CPY* in cells harboring the deleted DER3 gene. Microsomes were prepared and treated with trypsin in the absence or presence of Triton X-100 and CPY antigenic material was visualized with CPY monoclonal antibodies. As can be seen in Figure 6A, CPY* is protected by the ER membrane from proteolytic digestion by trypsin. Only upon lysis of membranes with Triton X-100 does CPY* become digested. The integrity of the vesicles was controlled with the ER lumenal chaperone Kar2p which behaves identical to CPY* in this experiment. Thus, CPY* accumulates inside the ER in yeast cells lacking the DER3 gene.
Figure 6.
Localization of CPY* in different mutants with a reduced ER degradation. Protease protection experiments were performed with intact yeast microsomes of the strains W303–1CΔ3 (Δder3), W303–1CD (Δder1), W303-CQ (Δubc7), and W303CPQ (Δubc6 Δubc7). Aliquots of the soluble fraction (S) and ER vesicles (P) not treated (−) or treated with trypsin (+T) or with trypsin and Triton X-100 (+T+X) were subjected to Western blotting and levels of CPY* and for control Kar2p were detected with suitable antibodies.
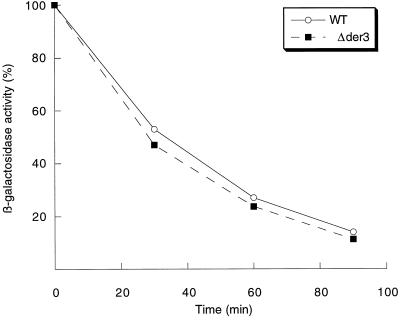
The integrity of the Ubc6-Ubc7 ubiquitination pathway is required for ER degradation of CPY* (Hiller et al., 1996). We therefore tested whether a defect in Der3 affected the function of this pathway. For this purpose, the kinetics of degradation of Deg1-β-galactosidase, a fusion protein of β-galactosidase with the Deg1 degradation domain of the Matα2 repressor protein, which had been proven to be a specific substrate for Ubc6-Ubc7-dependent ubiquitination (Chen et al., 1993), was studied in a wild-type and Δder3 background (Figure 7). No significant difference in degradation of this substrate could be observed in Δder3 cells as compared with wild type. This indicates that Der3p is not involved in any function connected with recruitment of the ubiquitin-conjugating enzymes Ubc6 and Ubc7 or the proteasome to the ER membrane for degradation of CPY*.
Figure 7.
The Ubc6-Ubc7 ubiquitination and proteasome pathway is not affected by the absence of Der3p. The kinetics of the degradation of a Deg1-β-galactosidase fusion protein in the strains W303–1C (WT) and W303–1CΔ3 (Δder3) was measured. Early stationary phase yeast cells grown on CM medium of both strains harboring the high-copy plasmid pDeg1 were harvested and resuspended in fresh CM medium containing cycloheximide (0.5 mg/ml) and incubated at 30°C. Samples of the culture were taken after 0, 30, 60, and 90 min, and the activity of β-galactosidase was measured. Enzymatic activity was set at 100% at time 0.
Also Loss of Der1p and Ubc6p/Ubc7p Leads to Accumulation of CPY* Inside the ER
Besides components of the translocon and Kar2p (Plemper et al., 1997), as well as the proteasome (Hiller et al., 1996), we had previously described additional gene products, Der1p, Ubc7p/Der2p, and Ubc6p to be required for a proper ER-associated degradation of CPY* (Hiller et al., 1996; Knop et al., 1996). Deletion of these genes also leads to accumulation of CPY* in cells. Using the same methods as applied for localization of CPY* in Δder3 mutants, we found CPY* also localized inside the ER in Δder1 mutants (Figure 6B). Interestingly, in an isogeneic strain harboring a UBC7 deletion allele, a partial accumulation of CPY* is visible in microsomes (Figure 6C). However, in a strain lacking both ubiquitinating enzymes Ubc6 and Ubc7, most of CPY* accumulates in the ER lumen (Figure 6D). Thus, components localized in the ER membrane (Der1p, Der3p) as well components functionally localized in the cytoplasm (Ubc6p, Ubc7p) are necessary for retrograde transport of CPY* from the ER lumen to the cytosol.
Deletion of DER3 Suppresses the Temperature-sensitive Growth of Cells Carrying the sec61–2 Mutation and Leads to a Stabilization of the Sec61–2 Mutant Protein In Vivo
Sec61p is a major component of the protein import machinery of the ER membrane (Deshaies et al., 1991; Görlich et al., 1992; Hartmann et al., 1993). A mutant form of this protein is encoded by the sec61–2 allele (Deshaies and Schekman, 1987). Like CPY* (Hiller et al., 1996), this mutant protein is selectively degraded by the ubiquitin-proteasome pathway at the restrictive temperature of 38°C (Biederer et al., 1996). Since the lumenal CPY* and the Sec61–2p are degraded via the same proteolytic pathway, it was interesting to elucidate whether Der3p was involved in some step necessary for degradation of the membrane-located Sec61–2p prior to ubiquitination and proteasomal proteolysis.
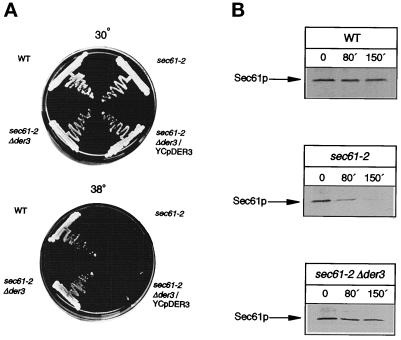
Strains carrying the sec61–2 allele show a clear temperature-sensitive growth, not surviving at the restrictive temperature of 38°C (Deshaies and Schekman, 1987). We therefore investigated whether the deletion of DER3 is a suppressor of the growth defect induced by the sec61–2 mutation. A strain carrying the sec61–2 allele and a deletion of DER3 was constructed. Although the strain carrying the sec61–2 mutation was unable to grow at 38°C due to degradation of the Sec61–2 protein and by this being completely devoid of the ability to import proteins into the ER, the sec61–2 Δder3 double mutant was able to grow at 38°C (Figure 8A). As expected sec61–2 mutant cells stopped growing again, when the DER3 gene was introduced into the Δder3 mutant on a plasmid. This indicated that lack of the Der3 protein stabilized the mutated translocon component Sec61–2p, allowing protein import into the ER. Thus, a defective degradation of Sec61–2p in the absence of Der3p has to be assumed.
Figure 8.
(A) Disruption of DER3 suppresses the temperature-sensitive growth phenotype of the sec61–2 mutant. Strains W303–1C (WT), YRP086 (sec61–2) and YRP105 (sec61–2 Δder3) harboring the LEU2 containing plasmid YCplac111, and YRP105 (sec61–2 Δder3/YCpDER3) containing the DER3 gene in a centromeric plasmid were tested for growth at 30 and 38°C on CM plates without leucine for 36 h. (B) Deletion of DER3 blocks the degradation of Sec61–2p at restrictive temperature. Pulse-chase experiments were performed in strains W303-1C (WT), YRP086 (sec61–2), and YRP105 (sec61–2 Δder3). Exponentially growing cells on SD medium were shifted to 38°C for 2 h and labeled with [35S]methionine. After the chase aliquots of cells were taken at the indicated chase points, lysed, and immunoprecipitated with specific anti-Sec61p antibodies.
We measured the half-life of the Sec61–2 protein in DER3 wild-type and Δder3 mutants in a pulse-chase experiment at the restrictive temperature of 38°C. After SDS-PAGE of the Sec61p antigenic material, labeled Sec61–2 protein was visualized by autoradiography and its half-life was measured using a Phosphoimager. As can be seen in Figure 8B, wild-type Sec61p is rather stable even after 150 min of chase at nonpermissive temperature. The sec61–2 allele encodes a rapidly degraded Sec61–2 protein at 38°C which is barely detectable after 150 min of chase. Deletion of DER3 leads to a considerable stabilization of the Sec61–2 protein. The half-life of the mutant protein is increased threefold in the Δder3 mutant strain as compared with DER3 wild type.
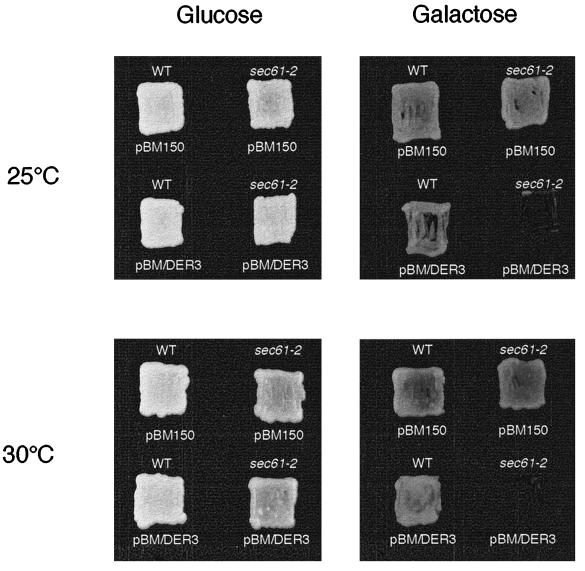
We expressed DER3 under the control of the GAL1 promoter in isogeneic wild-type and sec61–2 mutant strains. While on glucose-containing medium both strains were clearly able to grow at 25° and 30°C; induction of overexpression of DER3 on galactose-containing medium led to a dramatic slow down of growth of the strain harboring the sec61–2 allele at 25°C and 30°C (Figure 9). Again, this indicates that Der3p is involved in the degradation of the Sec61–2 mutant protein. As after deletion of UBC7, a sec61–2 mutant strain overexpressing DER3 becomes viable (our unpublished observations), the lethality of high-level expression of Der3p in sec61–2 strains at permissive temperature must be explained by a faster degradation of the mutated Sec61–2 protein.
Figure 9.
Overexpression of DER3 is lethal in a sec61–2 mutant at permissive temperature. Strains W303–1C (WT) and YRP086 (sec61–2) harboring the URA3 containing plasmid pBM150 or the DER3 gene under the control of the GAL1 promoter cloned into pBM150 plasmid (pBM/DER3) were grown on CM plates without uracil with glucose or galactose as carbon source at 25 and 30°C for 36 h.
Integrity of the Hydrophilic Tail of Der3p Is Essential for ER Degradation of CPY* and Sec61–2p
To investigate the role of the hydrophilic C-terminal tail of Der3p containing the RING-H2 finger motif, we constructed a mutant form of the protein lacking a fragment of 114 residues between position 307 and 419, including the RING-H2 domain (see Figure 1). After expression in yeast cells, this Der3ΔR truncated protein of about 50 kDa could be easily detected in crude extracts by Western blot analysis and located to the ER membrane by immunofluorescence techniques (our unpublished observations). Correct orientation of the C-terminal tail of Der3ΔR to the ER lumen was confirmed by a protease-protection experiment performed with intact yeast microsomes (Figure 10A). Mutated Der3p can be detected in the ER vesicle fraction after treatment with trypsin and only after addition of a detergent, the lumenal tail becomes accessible to the protease and is degraded.
Figure 10.
Integrity of the lumenal tail of Der3p is required for a proper degradation of CPY* and Sec61–2p. (A) The C-terminal hydrophilic tail of the truncated protein Der3ΔR is oriented to the ER lumen. A protease protection assay was performed with intact yeast microsomes from strain W303–1CΔ3 harboring the plasmid YCpDER3ΔR. Levels of Der3ΔRp and Kar2p were detected after Western blotting in the soluble fraction (S) and in ER vesicles (P) not treated (−) or after treatment with trypsin (+T) or with trypsin and Triton X-100 (+T+X). (B) Steady-state levels of CPY* in crude extracts prepared from yeast cells from strain W303–1C expressing wild-type Der3p (DER3), W303–1CΔ3 (Δder3), and W303–1CΔ3 harboring the plasmid YCpDER3ΔR (DER3ΔR) grown on CM medium were immunodetected with monoclonal anti-CPY antibodies after SDS-PAGE using Western blotting. (C) Expression of the truncated Der3ΔR protein is not able to revert the suppression of the temperature-sensitive growth phenotype caused by the disruption of the DER3 gene. Strains W303–1C (WT) and YRP086 (sec61–2) harboring the LEU2-based centromeric plasmid YCplac111 and strain YRP105 containing the plasmids YCplac111(sec61–2Δder3), YCpDER3 (sec61–2Δder3/YCpDER3), or YCpDER3ΔR (sec61–2Δder3/YCpDER3ΔR) were tested for growth on CM plates without leucine at 30and 38°C for 36 h. (D) Der3ΔR protein cannot support degradation of the mutated Sec61–2p at 38°C. Exponentially growing cells on CM medium at 25°C from strains W303–1C (WT/DER3), YRP086 (sec61–2/DER3), YRP105 (sec61–2/Δder3), and YRP105 harboring the plasmid YCpDER3ΔR (sec61–2/DER3ΔR) were shifted to 38°C for 1 h. Levels of Sec61p were detected after SDS-PAGE and Western blotting with specific antibodies.
We investigated the ability of Der3ΔRp to support ER degradation of CPY*. For this purpose we compared steady-state levels of CPY* in yeast cells expressing wild-type Der3p and mutated Der3ΔRp as well a strain carrying the Δder3 deletion. As can be seen in Figure 10B, while in DER3 wild-type cells CPY* can be hardly detected, Der3ΔRp-harboring cells show high accumulation of CPY* which is equivalent to a Δder3 null mutant strain. This indicates that the intact lumenal tail is necessary for the function of Der3p in ER degradation of CPY*.
We also studied the function of the truncated Der3ΔRp in ER degradation of the integral membrane Sec61–2 mutant protein at the restrictive temperature of 38°C. As can be seen in Figure 10C, yeast cells harboring the sec61–2 and der3ΔR alleles are able to grow at 38°C, suggesting a block in degradation of the mutated component of the ER translocon. This result was confirmed by measuring levels of Sec61p in cells after a 1-h shift to the restrictive temperature (Figure 10D). While in a sec61–2 DER3 strain the intensity of the Sec61–2p immunoreactive band was clearly reduced; in a isogeneic strain expressing Der3ΔRp, levels of Sec61–2p reached the level found in a strain harboring a deletion in DER3. From this we conclude that the lumenal tail of Der3p and most likely its RING-H2 domain plays an important role in the degradation pathway for both misfolded lumenal and integral membrane proteins of the ER.
DISCUSSION
We have cloned the DER3 gene by complementation of the der3–1 mutation which leads to a dramatically reduced degradation rate and thus to highly increased steady-state levels of CPY*, a lumenal substrate for ER-associated protein degradation in the yeast Saccharomyces cerevisiae (Finger et al., 1993; Knop et al., 1996). After sequencing and the subsequent search in the data bank, we found DER3 to be identical to the ORF YOL013 located on chromosome XV. Recently, a report on the identification and sequencing of three genes (called HRD) involved in the regulation of the ER membrane-bound HMG-CoA reductase via proteasome-triggered degradation appeared (Hampton et al., 1996). It turned out that DER3 is identical to HRD1. Deletion of DER3 resulted in mutant strains, which, like the der3–1 point mutant, exhibit an almost complete retardation of the proteolysis of CPY* and by this accumulated considerable amounts of the mutant protein (Figure 2, A and B). No other phenotype was observed for Δder3 cells.
The amino acid sequence of Der3p predicts a protein of about 64 kDa and a structure of an N-terminal hydrophobic region with five putative transmembrane domains followed by a 333-amino acid containing hydrophilic C terminus (Figure 1). A protein of similar size was identified after SDS-PAGE. Cell fractionation and immunofluorescence studies located Der3p to the endoplasmic reticulum, most likely to the ER membrane (Figures 3 and 4). Proteinase-protection experiments with isolated microsomes indicated the hydrophilic C-terminal tail of Der3p to be located in the lumen of the ER (Figure 5). This C-terminal region of Der3p contains a RING-H2 finger motif which is defined by the order and distance of cysteine and histidine residues and the flanking regions (Freemont et al., 1993).
Protease-protection experiments located CPY* in the lumen of the ER in Δder3 cells. Other components, which are known to be necessary for ER degradation of CPY* and respective mutants of these proteins thus accumulate CPY*, are components of the translocon and Kar2p (Plemper et al., 1997), Der1p (Knop et al., 1996), the ubiquitin-conjugating enzymes Ubc7p/Der2p and Ubc6p as well as the 26S proteasome (Hiller et al., 1996). Besides mutations in translocon components, Kar2p (Plemper et al., 1997) and, as shown here, Der3p, also disruption of the DER1 gene by depleting cells of the ER membrane-located Der1p (Knop et al., 1996) leads to accumulation of CPY* inside the ER (Figure 6B). Interestingly, disruption of the UBC7/DER2 gene that encodes a cytoplasmically localized ubiquitin-conjugating enzyme (Jungmann et al., 1993) only leads to partial accumulation of CPY* inside the ER (Figure 6C). Obviously only some CPY* is retrograde transported to the cytoplasmic surface of the ER in Δubc7/Δder2 cells, where it is ubiquitinated, most likely by Ubc6p (Hiller et al., 1996). Interestingly, when ubiquitination was completely blocked by disruption of UBC7/DER2 and UBC6, encoding a cytoplasmically oriented ubiquitin-conjugating activity in the ER membrane (Sommer and Jentsch, 1993), CPY* remained in the ER lumen (Figure 6D). This strongly indicates the involvement of ubiquitinating enzymes Ubc6p and Ubc7p, either directly or indirectly in the retrograde transport process of CPY* across the ER membrane.
The fact that DER3 had also been found to be a necessary gene for the degradation of HMG-CoA reductase, localized to the cytoplasmic face of the ER membrane, anticipated that Der3p might represent a protein generally involved in the ER degradation process of lumenal and membrane proteins. A well-known protein degraded via the same components as CPY*, the ubiquitin-conjugating enzymes Ubc6p and Ubc7p as well as the proteasome, is a mutant form of the ER translocon channel protein Sec61p (Biederer et al., 1996). Mutants carrying the sec61–2 allele are unable to grow at 38°C due to degradation of Sec61–2p. The Δder3 mutation turned out to be a suppressor of the sec61–2 mutation (Figure 8A). As expected from the suppressor phenotype, Δder3 mutants considerably stabilized the mutant Sec61–2 protein by allowing import of proteins into the ER and thus growth (Figure 8B). In addition, high-level expression of the Der3 protein from the GAL1 promoter was lethal for sec61–2 cells at permissive temperature (Figure 9), suggesting that an increased degradation of the mutated Sec61–2p takes place. These results indicate that Der3p has a central role in the ER degradation pathway and that it is necessary in general for removal of lumenal and membrane-bound proteins. This is in contrast to the ER membrane Der1 protein, which has only been found to be necessary for the degradation of the lumenal CPY* so far.
RING-H2 finger motifs are thought to be involved in protein–protein interaction (Saurin et al., 1996). We constructed a truncated version of Der3p lacking a region of 114 amino acids in the C-terminal tail which includes the RING-H2 finger domain and demonstrated its correct expression and localization in cells. Interestingly, this mutant Der3p is able to mediate proper degradation of neither CPY* nor Sec61–2p (Figure 10). This result suggests that the lumenal tail and here, most likely, the RING-H2 finger plays an important role in the function of Der3p. This function could reside in the delivery of proteins to be degraded into close proximity to the translocation machinery, of which Sec61p and Sec63p are important components and/or open the translocation channel for lateral gating and export of the proteins to the cytoplasm. Here, the ubiquitin-conjugating enzymes, Ubc6p and Ubc7p, and the proteasome with its ATPase subunits, with or without the help of additional chaperones, might comprise the machinery which provides the driving force for export and degradation.
ACKNOWLEDGMENTS
We thank M. Knop and M. Rose for providing strains, F. Cvrková and K. Nasmyth for the CEN-LEU2 library, M. Hochstrasser and T. Sommer for Deg1-β-galactosidase fusion plasmid, T. Sommer for providing anti-Sec61p antibodies, and R. Schekman and H.K. Rudolph for anti-Kar2p antibodies. We thank Monica Bredschneider for her assistance with the confocal microscope and S. Jäger, M. Laquai, and J. Strayle for helpful comments and discussions. This work was supported by the Bundesministerium für Forschung und Technologie (Bonn, Germany), Zentrales Schwerpunktprojekt Bioverfahrenstechnik (Stuttgart, Germany), and the Fonds des Chemischen Industrie (Frankfurt, Germany). J.B. is a fellow of the Spanish Ministerio de Educación y Cultura (Plan F.P.I. en el extranjero).
Note Added in Proof
Recently a gene, CUE1, was identified which encodes an ER membrane protein involved in the recruitment of the ubiquitin-conjugating enzyme Ubc7p to the ER membrane essential for Ubc7p-dependent protein degradation. A triple mutant deleted in Cue1p and the ubiquitinating enzymes Ubc6p and Ubc7p were shown to be defective in the export of CPY* from the ER, and the importance of ubiquitination for this process was suggested. (Biederer et al. Science 278, 1806–1809, 1997). Our studies demonstrate that deletion of the ubiquitin-conjugating enzymes Ubc6p and Ubc7p is sufficient for a block of retrograde transport of CPY* giving direct proof for the necessity of ubiquitin conjugation for the reverse translocation event.
REFERENCES
- Abeijon C, Orlean P, Robbins PW, Hirschberg CB. Topography of glycosylation in yeast: characterization of GDP-mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitay R, Shachar I, Rabinovich, Haimovich E, J, Bar-Nun S. Degradation of secretory immunoglobulin M in B lymphocytes occurs in a post endoplasmic compartment and is mediated by a cysteine protease. J Biol Chem. 1992;267:20964–20700. [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca2+-ATP-ase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley Interscience; 1992. [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Degradation of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-protesome pathway. EMBO J. 1996;15:2069–2077. [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Klausner RD. Cellular Proteolytic Systems. Vol. 15. A. Ciechanover and A.L. Schwarz, New York: Wiley-Liss; 1994. Degradation of proteins retained in the endoplasmic reticulum; pp. 137–160. [Google Scholar]
- Bonifacino JS, Lippincott-Schwartz J. Degradation of proteins within the endoplasmic reticulum. Curr Opin Cell Biol. 1991;4:592–560. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Cvrková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of the yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Fra AM, Sitia R. The endoplasmic reticulum as a site of protein degradation. Subcell Biochem. 1993;21:143–168. doi: 10.1007/978-1-4615-2912-5_7. [DOI] [PubMed] [Google Scholar]
- Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- Fürst P, Hu S, Hackett R, Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- Gaut JR, Hendershot LM. The modification and assembly of proteins in the endoplasmic reticulum. Curr Opin Cell Biol. 1993;5:589–595. doi: 10.1016/0955-0674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kallies K-U, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–21. [PubMed] [Google Scholar]
- Hampton R, Y, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Sommer T, Prehn G, örlich S, D, Jentsch S, Rapoport TA. Evolutionary conservation of the components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Reins H-A, Scobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;62:661–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Kerkau T, Bacik I, Bennink JR, Yewdell JW, Hünig T, Schimpl A, Schubert U. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J Exp Med. 1997;185:1295–1305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop M, Hauser N, Wolf DH. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 1996;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le A, Ferrell GA, Dishon DS, Le Q-Q, Sifers RN. Soluble aggregates of human PiZ α1-antitrypsin variant are degraded within the endoplasmic reticulum by a mechanism sensitive to inhibitors of protein synthesis. J Biol Chem. 1992;267:1072–1080. [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Bonifacino JS, Yuan L, Klausner RD. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988;54:209–229. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- McGee TP, Cheng HH, Kumagai H, Omura S, Simoni RD. Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J Biol Chem. 1996;271:25630–25639. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- Myers AL, Tzagoloff A, Kinney DM, Lusty CJ. Yeast shuttle and integrative vectors with multiple cloning sites for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Scheckman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saurin AJ, Borden KLB, Boddy NB, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena saliva roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Pelham HR. The KKXX signal mediates retrieval of membrane proteins from the Golgi to the ER in yeast. Eur J Cell Biol. 1994;64:211–216. [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:769–779. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky J, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996a;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wiertz EJHJ, Tortorella D, Bogyo Yu, M, J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996b;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Wileman T, Kane LP, Carson GR, Terhorst C. Depletion of cellular calcium accelerates protein degradation in the endoplasmic reticulum. J Biol Chem. 1991;266:4500–4507. [PubMed] [Google Scholar]
- Wolf DH, Fink GR. Proteinase C (carboxypeptidase Y) mutant of yeast. J Bacteriol. 1975;123:1150–1156. doi: 10.1128/jb.123.3.1150-1156.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]