Abstract
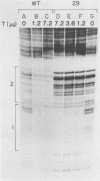
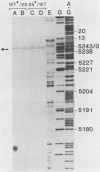
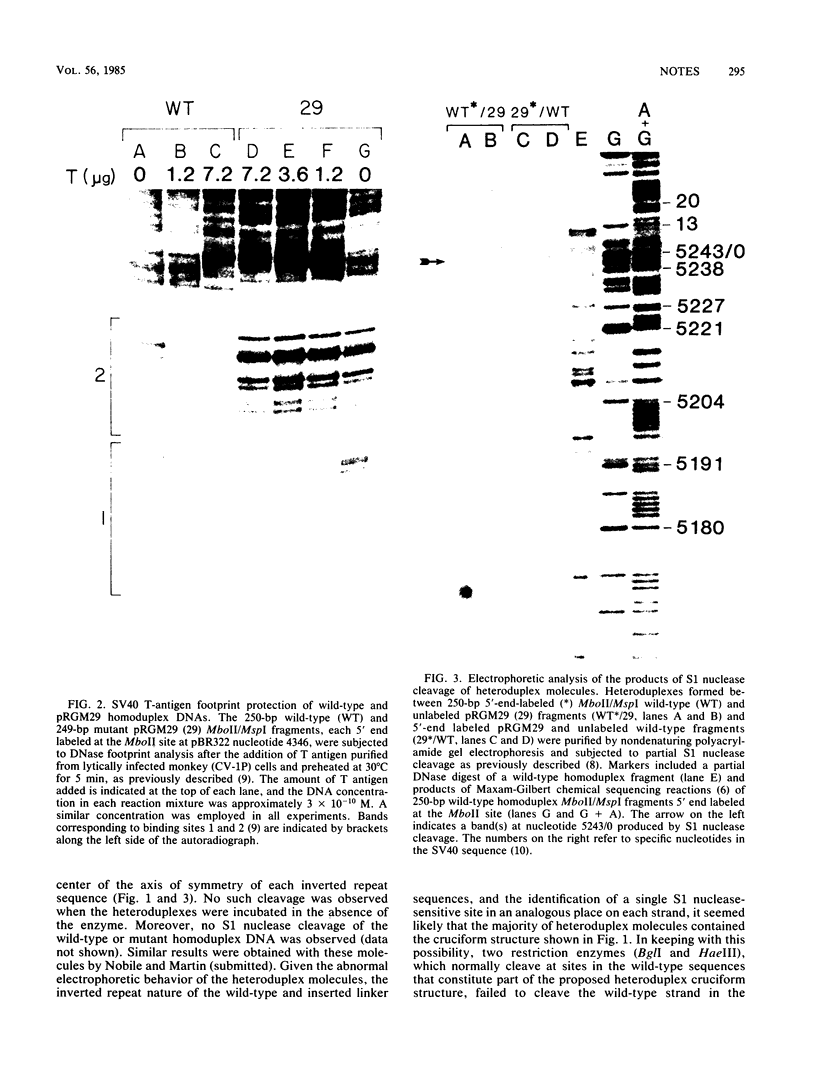
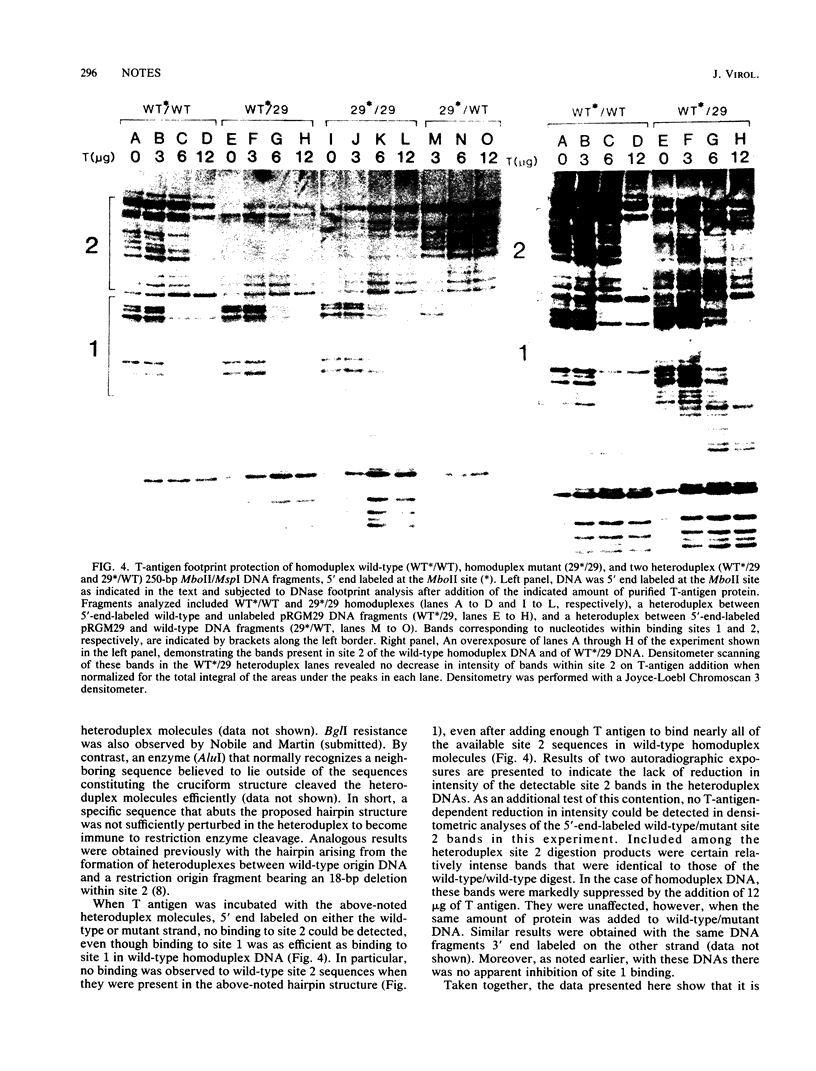
Heteroduplex DNA molecules were formed by annealing an intact simian virus replication origin-containing fragment to a mutant derivative lacking the indigenous wild-type 27-base-pair (bp) inverted repeat within this structure and containing a nonhomologous 26-bp inverted repeat sequence in its place. Results of restriction enzyme and S1 endonuclease cleavage analyses strongly suggested that a 13-bp stem-loop structure formed at the site of nonhomology between these two DNAs. This structure lies within the boundary of simian virus 40 T-antigen-binding site 2, and its presence inhibited T-antigen binding to that sequence but not to an adjacent higher-affinity binding site (site 1). Therefore, the conformation of sequences within an otherwise intact T-antigen-binding site can have major effects upon T-antigen binding there.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A. L., Lewton B. A., Tjian R., Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983 Apr;46(1):143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewton B. A., DeLucia A. L., Tegtmeyer P. Binding of simian virus 40 a protein to DNA with deletions at the origin of replication. J Virol. 1984 Jan;49(1):9–13. doi: 10.1128/jvi.49.1.9-13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Rio D. C., Robbins A. K., Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981 Aug;25(2):373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- Tenen D. G., Livingston D. M., Wang S. S., Martin R. G. Effect of a stem-loop structure within the SV40 replication origin upon SV40 T antigen binding to origin region sequences. Cell. 1983 Sep;34(2):629–639. doi: 10.1016/0092-8674(83)90395-1. [DOI] [PubMed] [Google Scholar]
- Tenen D. G., Taylor T. S., Haines L. L., Bradley M. K., Martin R. G., Livingston D. M. Binding of simian virus 40 large T antigen from virus-infected monkey cells to wild-type and mutant viral replication origins. J Mol Biol. 1983 Aug 25;168(4):791–808. doi: 10.1016/s0022-2836(83)80075-8. [DOI] [PubMed] [Google Scholar]