Abstract
Although the efficacy of pharmacotherapy for tobacco dependence has been previously demonstrated, there is substantial variability among individuals in treatment response. We performed a systems-based candidate gene study of 1295 single nucleotide polymorphisms (SNPs) in 58 genes within the neuronal nicotinic receptor and dopamine systems to investigate their role in smoking cessation in a bupropion placebo-controlled randomized clinical trial. Putative functional variants were supplemented with tagSNPs within each gene. We used global tests of main effects and treatment interactions, adjusting the P-values for multiple correlated tests. An SNP (rs2072661) in the 3′ UTR region of the β2 nicotinic acetylcholine receptor subunit (CHRNB2) has an impact on abstinence rates at the end of treatment (adjusted P = 0.01) and after a 6-month follow-up period (adjusted P = 0.0002). This latter P-value is also significant with adjustment for the number of genes tested. Independent of treatment at 6-month follow-up, individuals carrying the minor allele have substantially decreased the odds of quitting (OR = 0.31; 95% CI 0.18–0.55). Effect of estimates indicate that the treatment is more effective for individuals with the wild-type (OR = 2.14, 95% CI 1.20–3.81) compared with individuals carrying the minor allele (OR = 0.83, 95% CI 0.32–2.19), although this difference is only suggestive (P = 0.10). Furthermore, this SNP demonstrated a role in the time to relapse (P = 0.0002) and an impact on withdrawal symptoms at target quit date (TQD) (P = 0.0009). Overall, while our results indicate strong evidence for CHRNB2 in ability to quit smoking, these results require replication in an independent sample.
INTRODUCTION
Use of tobacco is the single greatest preventable cause of death in the USA, with a substantial impact on morbidity and mortality worldwide. One in five Americans is a current smoker, and available therapies are efficacious for only a small fraction of those who attempt to quit (1). Bupropion, an anti-depressant therapy, is among the only two FDA-approved non-nicotine pharmacotherapies available for the treatment of nicotine dependence. While its efficacy is well-documented, there is substantial inter-individual variability in therapeutic response (2–4), and most smokers do not maintain long-term abstinence (5,6). Pharmacogenetic studies may improve the outcomes of bupropion therapy for smoking cessation by identifying smokers most and least likely to benefit based on inherited differences in drug metabolism and CNS targets. Such studies may also identify genetic markers of relapse risk, thereby identifying smokers who may require more intensive treatment.
Genes coding for nicotinic acetylcholine receptor (nAChRs) and genes within the dopamine reward system are plausible candidates for pharmacogenetic investigations of bupropion. Nicotine activates α4β2 nAChRs to stimulate dopamine release, which plays a key role in signaling the reward system (7). Bupropion inhibits dopamine re-uptake, resulting in higher levels of dopamine, which is believed to be responsible for alleviating the cognitive and affective symptoms of nicotine withdrawal, as demonstrated in preclinical (8,9) and human studies (8,10,11). Consistent with these data, several studies have identified associations of single nucleotide polymorphisms (SNPs) in dopaminergic and nAChR subunit genes with nicotine dependence (12–16) and with smoking cessation during the treatment with bupropion (17–22). While the α4β2 nAChRs account for the majority of high affinity binding related to dopamine release, nAChRs on dopaminergic neurons may also contain α5, α6 and β3 subunits, which may contribute to receptor targeting, localization and receptor permeability (23).
The present analysis was undertaken to extend prior pharmacogenetics research on prospective smoking cessation beyond individual polymorphisms or single gene analyses. Using data from a placebo-controlled randomized clinical trial of bupropion for smoking cessation (20), we explored the associations of 1295 SNPs within 58 candidate gene regions in the dopamine system, including nAChRs and intracellular signaling proteins. A systems-based approach and analysis has many advantages over the evaluation of single functional polymorphisms and single candidate gene approaches. By taking a more expansive approach to SNP selection and gene selection, we hope to provide more complete coverage of the underlying genetic variation within a suspected etiologic system. This is accomplished in two ways. In addition to the inclusion of putative functional polymorphisms, we have included a set of SNPs to comprehensively capture the underlying genetic architecture within each candidate gene. Likewise, by broadening our selection of candidate genes to include multiple genes in a interconnected system, rather than simply focusing on single putatively interesting genes, we hope to provide a more complete picture of the impact of the entire system. This potentially allows for the identification of sets of markers in the presence of some system redundancy. Thus, we have leveraged biological knowledge in the selection of both SNPs and genes while also including broad SNP coverage within each gene and an expanded selection of genes across the entire dopamine system. Our analysis mimics this structure in which we first perform tests of single SNPs for main effects and SNP X treatment interactions, while adjusting the level of significance for all SNPs within each gene. Then, taking a more global perspective, we further adjust for the number of genes used to characterize the entire system. Then, based upon findings from this analysis, we use our biological knowledge to guide us in testing the haplotypic effects and interactions with other relevant candidate genes.
RESULTS
Descriptive data
The genetic association analyses were performed on persons with self-identified European ancestry only (Table 1). This included 222 individuals receiving bupropion and 195 individuals receiving placebo. There were slightly more females (54%) than males, and the average age was 44 (SD = 11) years. Similar distributions of the Fagerström Test for Nicotine Dependence (FTND) were observed between the treatment and placebo groups with a mean FTND of 5.1 (SD = 2.1) and 5.2 (SD = 2.2), respectively. At the end of treatment (EOT), there was a 32.0% abstinence rate in the bupropion group when compared with 21.5% in the placebo group. This corresponds to a treatment OR = 1.76 (95% CI 1.12–2.76). Abstinence rates were decreased at 6-month follow-up with 25.7% of those individuals receiving bupropion remaining abstinent and 17.4% of those in the placebo group abstinent (treatment OR = 1.66; 95% CI 1.03–2.69). These treatment effects are consistent with prior research (24). For individuals who relapsed during the 6-month follow-up period, the average for time to first cigarette was 45.2 days (SD = 50.5) in the bupropion group and 28.2 days (SD = 38.9) in the placebo group (P = 0.0016). At target quit data (TQD), the bupropion group had higher mean withdrawal symptoms (32.3, SD = 7.2) compared with the placebo group (30.7, SD = 7.3) (P = 0.04). Of our original 1295 candidate gene SNPs, 97 (7.5%) were excluded from further analysis after genotyping because they were monomorphic or had low (< 0.01) minor allele frequencies in our samples. The remaining 1198 SNPs had an r2 of 0.8 or greater with 85% of all the common (>0.05%) SNPs within the 58 candidate genes in the current version of HapMap (Genome Build 36, Table 2) (see Supplementary Material, Table S3 for information regarding all the SNPs).
Table 1.
Study characteristics
| Buproprion, n (%) | Placebo, n (%) | ||
|---|---|---|---|
| 217 | 195 | ||
| Age | 44.0 ± 11.8 | 44.6 ± 11.2 | |
| Gender | Female | 120 (55.3%) | 106 (54.4%) |
| Male | 97 (44.7%) | 89 (45.6%) | |
| FTNDa | 0 | 2 (0.9%) | 2 (1%) |
| 1 | 9 (4.1%) | 13 (6.7%) | |
| 2 | 10 (4.6%) | 10 (5.1%) | |
| 3 | 22 (10.1%) | 15 (7.7%) | |
| 4 | 42 (19.4%) | 31 (15.9%) | |
| 5 | 42 (19.4%) | 30 (15.4%) | |
| 6 | 33 (15.2%) | 32 (16.4%) | |
| 7 | 26 (12%) | 34 (17.4%) | |
| 8 | 19 (8.8%) | 21 (10.8%) | |
| 9 | 10 (4.6%) | 6 (3.1%) | |
| 10 | 2 (0.9%) | 1 (0.5%) | |
| Mean | 5.1 ± 2.1 | 5.2 ± 2.2 | |
| End of treatmentb | Abstinent | 70 (32.3%) | 42 (21.5%) |
| Relapsed | 147 (67.7%) | 153 (78.5%) | |
| 6-month follow-upb | Abstinent | 56 (25.8%) | 34 (17.4%) |
| Relapsed | 161 (74.2%) | 161 (82.6%) | |
| Average withdrawal symptoms at TQDc | 32.3 ± 7.2 | 30.7 ± 7.3 | |
| Average days to smoking relapsed | 45.2 ± 50.5 | 28.2 ± 38.9 |
All the subjects were of European ancestry.
aScores for the Fagerström test for nicotine dependence (FTND) were assessed at baseline.
bSmoking abstinence or relapse was biochemically assessed and verified at the end of treatment (8 weeks following target quit date [TQD]) and 6-months after the target quit date.
cWithdrawal symptoms at the TQD were assessed on 18 items (e.g. irritability, insomnia, nausea), each scored from 0 (no symptoms) to 3 (severe symptoms). Scores in this study ranged from 18 to 60.
dDays to smoking relapse up to the 6-month follow-up period was self-reported by subjects. The average days to relapse exclude subjects who self-reported abstinence throughout the entire study period.
Table 2.
Single nucleotide polymorphism (SNP) distribution for candidate genes
| Gene symbol | Gene name | Chromosome | Total HapMap SNPsb | tagSNPs |
Coveragee | |
|---|---|---|---|---|---|---|
| Selectedc | Analyzedd | |||||
| DRD1IP (Calcyon) | dopamine receptor D1 interacting protein | 10 | 18 | 6 | 6 | 0.82 |
| ADCYAP1 | adenylate cyclase activating polypeptide 1 | 18 | 74 | 24 | 23 | 0.83 |
| ANKK1_DRD2a | 11 | 194 | 55 | 52 | 0.97 | |
| ANKK1 | ankyrin repeat and kinase domain containing 1 | |||||
| DRD2 | dopamine receptor D2 | |||||
| BDNF | brain-derived neurotrophic factor | 11 | 87 | 28 | 26 | 0.96 |
| CALM1 | calmodulin 1 (phosphorylase kinase, delta) | 14 | 53 | 11 | 10 | 0.82 |
| CDK5 | cyclin-dependent kinase 5 | 7 | 26 | 11 | 10 | 0.87 |
| CHRNA2 | cholinergic receptor, nicotinic, alpha 2 | 8 | 97 | 27 | 26 | 0.88 |
| CHRNA5_CHRNA3_CHRNB4a | 15 | 108 | 30 | 30 | 0.93 | |
| CHRNA5 | cholinergic receptor, nicotinic, alpha 5 | |||||
| CHRNA3 | cholinergic receptor, nicotinic, alpha 3 | |||||
| CHRNB4 | cholinergic receptor, nicotinic, beta 4 | |||||
| CHRNB3_CHRNA6a | 8 | 79 | 8 | 7 | 0.50 | |
| CHRNA6 | cholinergic receptor, nicotinic, alpha 6 | |||||
| CHRNB3 | cholinergic receptor, nicotinic, beta 3 | |||||
| CHRNA4 | cholinergic receptor, nicotinic, alpha 4 | 20 | 61 | 12 | 12 | 0.62 |
| CHRNA7 | cholinergic receptor, nicotinic, alpha 7 | 15 | 168 | 33 | 33 | 0.91 |
| CHRNB2 | cholinergic receptor, nicotinic, beta 2 | 1 | 48 | 13 | 13 | 0.92 |
| CLIC6 | chloride intracellular channel 6 | 21 | 73 | 21 | 18 | 0.98 |
| CNR1 | cannabinoid receptor 1 (brain) | 6 | 91 | 21 | 21 | 0.74 |
| COMT | catechol-O-methyltransferase | 22 | 84 | 35 | 28 | 0.88 |
| CREB1 | cAMP responsive element binding protein 1 | 2 | 63 | 18 | 17 | 0.94 |
| CREBBP | CREB binding protein (Rubinstein-Taybi syndrome) | 16 | 229 | 30 | 27 | 0.80 |
| CRHR1 | corticotropin releasing hormone receptor 1 | 17 | 76 | 18 | 16 | 0.86 |
| DBH | dopamine beta-hydroxylase (dopamine beta-monooxygenase) | 9 | 129 | 42 | 36 | 0.85 |
| DDC | dopa decarboxylase (aromatic l-amino acid decarboxylase) | 7 | 334 | 50 | 47 | 0.87 |
| DRD1 | dopamine receptor D1 | 5 | 89 | 34 | 32 | 0.80 |
| DRD3 | dopamine receptor D3 | 3 | 107 | 26 | 24 | 0.95 |
| DRD4 | dopamine receptor D4 | 11 | 33 | 12 | 11 | 0.63 |
| DRD5 | dopamine receptor D5 | 4 | 41 | 12 | 8 | 0.91 |
| EPB41 | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | 1 | 202 | 48 | 44 | 0.99 |
| EPB41L1 | erythrocyte membrane protein band 4.1-like 1 | 20 | 230 | 25 | 16 | 0.97 |
| EPB41L2 | erythrocyte membrane protein band 4.1-like 2 | 6 | 445 | 51 | 49 | 0.81 |
| FLNA | filamin A, alpha (actin binding protein 280) | X | 35 | 11 | 5 | 1.00 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 19 | 27 | 7 | 7 | 0.58 |
| GNAS | GNAS complex locus | 20 | 127 | 32 | 31 | 0.85 |
| GNAZ | guanine nucleotide binding protein (G protein), alpha z polypeptide | 22 | 114 | 24 | 22 | 0.94 |
| GRB2 | growth factor receptor-bound protein 2 | 17 | 110 | 23 | 23 | 0.97 |
| HCRTR1 | hypocretin (orexin) receptor 1 | 1 | 37 | 11 | 11 | 1.00 |
| HCRTR2 | hypocretin (orexin) receptor 2 | 6 | 224 | 56 | 54 | 0.98 |
| HTR1A | 5-hydroxytryptamine (serotonin) receptor 1A | 5 | 26 | 6 | 6 | 0.88 |
| HTR1B | 5-hydroxytryptamine (serotonin) receptor 1B | 6 | 66 | 29 | 27 | 0.93 |
| KCNJ9 | potassium inwardly-rectifying channel, subfamily J, member 9 | 1 | 32 | 11 | 10 | 0.95 |
| KLF16 | Kruppel-like factor 16 | 19 | 24 | 6 | 6 | 0.68 |
| MAOA_MAOBa | X | 223 | 42 | 40 | 0.95 | |
| MAOA | monoamine oxidase A | |||||
| MAOB | monoamine oxidase B | |||||
| MAPK1 | mitogen-activated protein kinase 1 | 22 | 141 | 28 | 27 | 0.97 |
| NCK1 | NCK adaptor protein 1 | 3 | 86 | 25 | 25 | 0.94 |
| FREQ (NCS1) | frequenin homolog (Drosophila) | 9 | 114 | 42 | 40 | 0.80 |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 2 | 33 | 10 | 9 | 0.83 |
| NTRK1 | neurotrophic tyrosine kinase, receptor, type 1 | 1 | 120 | 14 | 10 | 0.22 |
| OPRM1 | opioid receptor, mu 1 | 6 | 421 | 46 | 44 | 0.47 |
| POMC | proopiomelanocortin | 2 | 35 | 16 | 12 | 0.85 |
| PPP1R1B | protein phosphatase 1, regulatory (inhibitor) subunit 1B (DARPP-32) | 17 | 26 | 9 | 9 | 1.00 |
| PRKAR2B | protein kinase, cAMP-dependent, regulatory, type II, beta | 7 | 153 | 36 | 32 | 0.95 |
| PICK1 (PRKCABP) | protein interacting with PRKCA 1 | 22 | 51 | 16 | 14 | 1.00 |
| SLC6A3 | solute carrier family 6, member 3 (dopamine transporter) | 5 | 145 | 27 | 26 | 0.78 |
| SNCA | synuclein, alpha (non A4 component of amyloid precursor) | 4 | 243 | 39 | 39 | 0.99 |
| TDO2 | tryptophan 2,3-dioxygenase | 4 | 56 | 12 | 12 | 0.82 |
| TH | tyrosine hydroxylase | 11 | 50 | 16 | 15 | 0.67 |
| 5958 | 1295 | 1198 | 0.88 | |||
aWhen selecting SNPs, overlapping genes were treated as a single gene region.
bThe total number of HapMap SNPs was obtained from Genome Build 36.
ctagSNPs were selected using pairwise r2 within gene regions.
dThose that had a call rate of zero or minor allele frequency lower than 0.01 were excluded from the analysis.
eCoverage is calculated as the proportion of HapMap SNP with r2 greater than 0.80 with the tagSNPs analyzed.
Associations with abstinence at end of treatment and 6-month follow-up
Results from the single SNP analyses are presented in Table 3 for EOT and 6-month follow-up, respectively. Only SNPs with an adjusted P-value <0.05 from a likelihood ratio test (LRT) and with stable effect estimates are presented in the tables. Results for all the SNPs are presented in Supplementary Material, Tables S1 and S2. Two SNPs within the 3′ UTR of the CHRNB2 (cholinergic receptor, nicotinic, beta 2) gene show a substantial impact on relapse at the EOT with an observed LRT P-value for the most significant SNP (rs2072661) of 9.97 × 10−4 and an adjusted P-value of 1.33 × 10−2. The two SNPs in CHRNB2 are in strong linkage disequilibrium (r2 = 0.96) with each other and their effect estimates following a similar trend with both SNPs demonstrating a dominant genetic model. For simplicity in presentation, we focus on reporting the result for rs2072661. Irrespective of treatment, having the minor allele for rs2072661, which was present in 23% of our subjects, decreases the odds of quitting substantially (OR = 0.40; 95% CI 0.25–0.67, P = 0.0004). The interaction effect with treatment is not significant at an α = 0.05 level (P = 0.97); treatment OR for wild-type (WT) genotype = 1.77 (95% CI 1.02–3.07) and for minor allele OR = 1.81 (95% CI 0.79–4.14). At 6-months follow-up, the genetic effect for rs2072661 becomes more pronounced (OR = 0.31; 95% CI 0.18–0.55; P = 0.00006) and there is a more noticeable difference in the treatment effects by genotype, with treatment being substantially more effective for quitting the WT genotype group (OR = 2.14; 95% CI 1.20–3.81) when compared with the treatment effect within the individuals carrying the minor allele (OR = 0.83; 95% CI 0.32–2.19). A test for SNP-treatment interaction yields a P-value = 0.10. In addition to the two original SNPs within CHRNB2 showing an effect at EOT, there are three additional SNPs in LD with rs2072661 (rs1127314, rs2131902, and rs3766927) 3′ of the CHRNB2 gene with substantial genetic effects and significantly adjusted P-values from the LRT at 6-month follow-up.
Table 3.
Single nucleotide polymorphism (SNP)-specific results for abstinence rates
| SNP | Gene | Chromosome | Position | Minor allele frequencya | Genetic modelb | SNP OR (95% CI) | Treatment OR | Test of interactionc | Likelihood ratio testd |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type (95% CI) | Variant (95% CI) | Observed P | Adjusted P | ||||||||
| End of treatment | |||||||||||
| rs2072661 | CHRNB2 | 1 | 152,815,504 | 0.23 | Dom | 0.40 (0.25–0.67) | 1.77 (1.02–3.07) | 1.81 (0.79–4.14) | 9.70 × 10−1 | 9.97 × 10−4 | 1.33 × 10−2 |
| rs2072660 | CHRNB2 | 1 | 152,815,345 | 0.23 | Add | 0.50 (0.32–0.76) | 1.51 (0.88–2.59) | 2.33 (1.11–4.88) | 3.28 × 10−1 | 2.86 × 10−3 | 2.34 × 10−2 |
| rs12961210 | ADCYAP1 | 18 | 890,523 | 0.32 | Add | 1.85 (1.33–2.57) | 2.15 (1.07–4.31) | 1.86 (1.14–3.03) | 6.74 × 10−1 | 1.15 × 10−3 | 2.59 × 10−2 |
| rs2565059 | CHRNA2 | 8 | 27,392,895 | 0.19 | Add | 2.32 (1.46–3.68) | 1.76 (0.98–3.16) | 2.06 (1.11–3.83) | 7.97 × 10−1 | 1.49 × 10−3 | 3.14 × 10−2 |
| rs1936158 | HTR1B | 6 | 78,241,094 | 0.41 | Dom | 0.53 (0.33–0.84) | 3.14 (1.52–6.49) | 1.14 (0.63–2.07) | 3.51 × 10−2 | 1.20 × 10−3 | 3.43 × 10−2 |
| rs10517626 | TDO2 | 4 | 157,050,894 | 0.10 | Add | 1.81 (1.10–2.99) | 1.39 (0.84–2.31) | 3.98 (1.6–9.91) | 4.26 × 10−2 | 4.03 × 10−3 | 4.16 × 10−2 |
| 6-month follow-up | |||||||||||
| rs2072661 | CHRNB2 | 1 | 152,815,504 | 0.23 | Dom | 0.31 (0.18–0.55) | 2.14 (1.20–3.81) | 0.83 (0.32–2.19) | 1.01 × 10−1 | 1.54 × 10−5 | 2.40 × 10−4e |
| rs2072660 | CHRNB2 | 1 | 152,815,345 | 0.23 | Dom | 0.33 (0.19–0.58) | 2.04 (1.15–3.63) | 0.86 (0.33–2.26) | 1.32 × 10−1 | 4.33 × 10−5 | 6.36 × 10−4e |
| rs2069454 | CDK5 | 7 | 150,383,915 | 0.14 | Dom | 2.07 (1.18–3.62) | 1.12 (0.61–2.05) | 5.03 (1.93–13.1) | 9.38 × 10−3 | 3.71 × 10−4 | 4.73 × 10−3 |
| rs1127314 | CHRNB2 | 1 | 152,822,890 | 0.30 | Add | 0.52 (0.35–0.78) | 1.71 (0.92–3.15) | 1.53 (0.79–2.96) | 7.95 × 10−1 | 3.67 × 10−3 | 2.77 × 10−2 |
| rs2238687 | FOSB | 19 | 50,665,006 | 0.13 | Add | 2.22 (1.4–3.52) | 1.74 (0.97–3.15) | 1.59 (0.74–3.43) | 8.48 × 10−1 | 3.68 × 10−3 | 3.05 × 10−2 |
| rs10517626 | TDO2 | 4 | 157,050,894 | 0.10 | Add | 1.96 (1.17–3.28) | 1.31 (0.76–2.28) | 3.46 (1.37–8.76) | 6.98 × 10−2 | 3.23 × 10−3 | 3.34 × 10−2 |
| rs2131902 | CHRNB2 | 1 | 152,826,222 | 0.31 | Dom | 0.47 (0.29–0.76) | 1.78 (0.94–3.37) | 1.43 (0.67–3.04) | 6.62 × 10−1 | 6.42 × 10−3 | 4.47 × 10−2 |
| rs3766927 | CHRNB2 | 1 | 152,830,765 | 0.30 | Dom | 0.47 (0.29–0.78) | 1.83 (0.97–3.46) | 1.35 (0.63–2.90) | 5.47 × 10−1 | 6.79 × 10−3 | 4.53 × 10−2 |
| rs13152449 | TDO2 | 4 | 157,055,252 | 0.10 | Add | 1.94 (1.15–3.25) | 1.34 (0.77–2.31) | 3.45 (1.35–8.82) | 7.74 × 10−2 | 4.61 × 10−3 | 4.55 × 10−2 |
aEstimate minor allele frequency in our study sample.
bFor each SNP, dominant (Dom) and additive (Add) genetic models were estimated and the model yielding the lower adjusted P-value was selected.
cThe test of interaction is the observed P-value from a test of heterogeneity for the treatment odds ratios.
dThe likelihood ratio test observed P-value was obtained using a joint 2-df test combining the genetic marginal effect and the gene–treatment interaction effect. Adjusted P-values were estimated by accounting for the correlation between observed SNPs within the respective gene regions. This adjustment was also used when selecting a genetic model.
eAfter adjustment, rs2072661 and rs2072660 reached pathway level significance (P < 0.0009) at 6-month follow-up. All models were adjusted for age, gender, and FTND.
SNP rs10517626 within the TDO2 (tryptophan 2,3,-dioxygenase) gene was also found to be noteworthy and has similar patterns of effects at EOT and at 6-month follow-up. Specifically, at EOT there is a genetic effect enhancing quit rates for each additional minor allele (OR = 1.81; 95% CI 1.10–2.99; P = 0.01) with the treatment effect being more effective within those individuals carrying the minor allele (OR = 3.98; 95% CI 1.60–9.91) when compared with the treatment effect within individuals with the WT genotype (OR = 1.39; 95% CI 0.84–2.31). The difference in treatment effects by genotype is suggestive of an observed P-value from the interaction test of 4.26 × 10−2. Additional polymorphisms at EOT are indicated in ADCYAP1 and HTR1B or at 6-month follow-up in CDK5 and FOSB, but none are consistent with the analyses at both time points.
To examine the LD patterns within the CHRNB2 gene, we investigated the block structure using Haploview (25) and the CEPH (Centre d'Etude du Polymorphisme Humain) from Utah (CEU) HapMap (version 23a) samples. Figure 1 presents the LD patterns for 24 SNPs spanning the CHRNB2 gene with HapMap genotype data. Of these, 22 SNPs have MAF >0.05 and two SNPs were forced in the selection process because of prior information (rs2072658 and rs2072661). Our 13 tagSNPs captured 20 of the 22 total common HapMap SNPs with SNPs rs9427092 and rs3766925 not tagged at an r2 of 0.8 or greater. The LD structure indicates two distinct block regions with significant and suggestive SNPs from the single SNP analyses located within the second block. Using the eight genotyped SNPs within this region, we estimated six common haplotypes explaining 98% of the total frequency in the sample (Table 4). A haplotype analysis using the haplotype carrying the most common alleles for all the SNPs as the referent indicates that there is a significant global LRT at EOT (P = 0.03) and at 6-month follow-up (P = 0.007). In the analysis at both time points, the haplotypes carrying the minor allele for rs2072661 and rs2072660 (haplotypes 11001011 and 11101011) have odd ratios indicating increased relapse rates. Any haplotype without these minor alleles shows no effect further indicating that either rs2072661 or rs2072660 are driving the effect in this region. We further confirmed the independent effect of either r2072661 or rs2072660 by performing a joint regression model on a subset of SNPs in this region (results not shown). However, because of the high amount of correlation between these two SNPs, we were unable to differentiate the independent effect of each SNP.
Figure 1.
Block structure for CHRNB2 from HapMap Genome Build 36. The linkage disequilibrium (LD) plot was obtained using Haploview (25) and HapMap Build 36. The scale at the top of the figure depicts the HapMap region for CHRNB2 (Chromosome 1: 152,806,881 to 152,818,975), and roughly 10 kb upstream and downstream of this region. Single nucleotide polymorphisms (SNPs) genotyped by HapMap are identified in blue (minor alleles) and red (major alleles). Gene regions with the direction of their respective reading frame, exons, and introns, are also given. The 13 SNPs in boxes are the tagSNPs selected in this gene region. rs2072658 and rs2072661 have no HapMap or LD information. The r2 LD color scheme is depicted. Two blocks are delineated using the default block definitions from Haploview.
Table 4.
Haplotype analysis of the 3′-UTR region in CHRNB2
| Haplotypea | Frequency | End of treatment, OR (95% CI) | 6-month follow-up, OR (95% CI) |
|---|---|---|---|
| 00000000 | 0.23 | 1.00 | 1.00 |
| 00010100 | 0.27 | 1.17 (0.76–1.80) | 1.06 (0.68–1.66) |
| 00000100 | 0.19 | 1.00 (0.62–1.60) | 0.86 (0.52–1.42) |
| 11001011 | 0.14 | 0.64 (0.38–1.07) | 0.50 (0.28–0.89) |
| 11101011 | 0.07 | 0.43 (0.20–0.92) | 0.29 (0.12–0.73) |
| 00001011 | 0.07 | 1.31 (0.67–2.53) | 0.94 (0.46–1.93) |
| **–*–**b | 0.98 | LRT P-value = 0.03c | LRT P-value = 0.007c |
aThe second block shown in Figure 1. Common alleles in each haplotype are designated by ‘0’, and variant alleles designated by ‘1’. The haplotype carrying the common alleles for all the SNPs was used as the referent allele.
bFrom Table 3, those SNPs with adjusted P-values <0.05 are indicated by asterisks. The first two SNPs are rs2072660 and rs2072661, respectively.
cThe likelihood ratio test (LRT) P-value is from a global test of the full haplotype model versus a covariate-only model. Covariates include age, gender, FTND, and treatment.
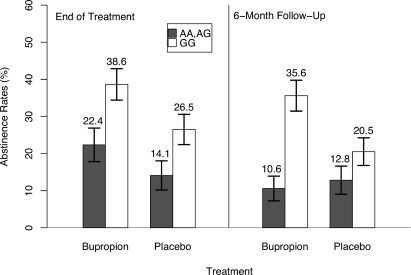
To demonstrate more clearly the impact of SNP rs2072661 on the abstinence rates, we examined in more detail the patterns of relapse from EOT to 6-month follow-up. Figure 2 presents the abstinence rates for all the individuals and stratified by rs2072661. As reflected in the odds ratio estimates for the genetic effect, the abstinence rates are lower for those individuals who carry the minor allele (AA or AG). This impact is greater at 6-month follow-up. Interestingly, from EOT to 6-month follow-up, individuals carrying the minor allele who had received bupropion during the treatment phase had the greatest reduction in abstinence once treatment ended. While not statistically significant (P = 0.10), for all the individuals carrying the minor allele who were treated with bupropion, the abstinence rate was 22.4% at EOT and decreased to 10.6% at 6-month follow-up. To further quantify this impact, we performed an analysis examining the abstinence rates at 6-month follow-up restricted to only those individuals abstinent at EOT (n = 112). For rs2072661, the P-value from the 2 degree-of-freedom LRT was 0.047. Of note, for those individuals who received bupropion, the impact of rs2072661 was substantial for decreasing the odds of quitting, once the individuals were no longer on treatment (OR = 0.25; 95% CI 0.08–0.78).
Figure 2.
Abstinence rates by CHRNB2 rs2072661 and by bupropion treatment. Abstinence rates comparing individuals with at least one variant allele for CHRNB2 rs2072661 (filled bars) to those with both common alleles (unfilled bars) were stratified by treatment (buproprion or placebo) and estimated at each time point (end of treatment and 6-month follow-up).
Time to relapse
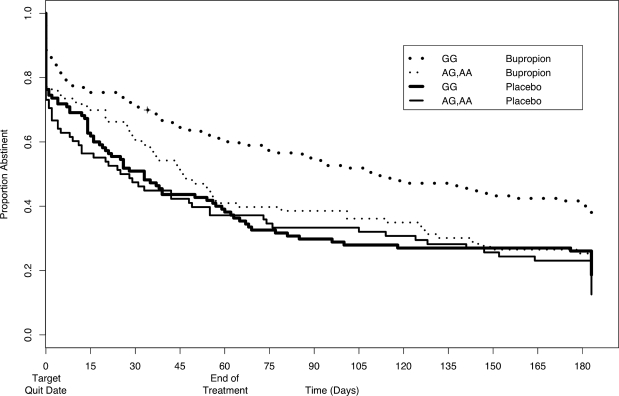
The pattern described above is also reflected when examining time to first relapse (Figure 3).
Figure 3.
Time to relapse to 6-month follow-up for CHRNB2 rs2072661. Time to relapse to 6-month follow-up in individuals with at least one variant allele for CHRNB2 rs2072661 (thin lines) was compared with those having both alleles common (thick lines), stratifying by treatment group: buproprion (dotted lines) and placebo (solid lines).
We note that relapse rates used in the time to relapse analysis are not directly comparable to the relapse rates presented in Figure 2, as the time-dependent information on relapse is via self-report and daily use can not be biochemically verified. Throughout the course of the trial and follow-up period, individuals with the WT genotype and received bupropion had the highest abstinence rates. Those individuals carrying the minor allele and who received buporpion had higher abstinence rates during the trial as compared to placebo, but those rates dropped to those of placebo by 6-month follow-up. A 2-df LRT for the gene and gene-treatment interaction from a Cox regression yielded a P-value of 0.0002.
Withdrawal symptoms
We examined the impact of rs2072661 on withdrawal symptoms measured using the withdrawal symptom severity index at TQD, coinciding with the peak symptoms, for those individuals who were abstinent (CO levels <10 ppm) for the week post-TQD. Because bupropion has been shown to impact withdrawal symptoms (26), we stratified our analysis by treatment from a linear regression on all the individuals receiving placebo, there was a 3.8 unit greater magnitude of withdrawal severity for individuals carrying the minor allele compared with those having the WT genotype (P = 0.0009). In contrast, for those who received bupropion, there was only a 0.45 unit difference between the two groups (P = 0.71).
Interaction with CHRNA4
Finally, as the α4β2 nAChR is the most widely distributed subtype nAChR, we investigated the interaction effect of the 11 SNPs genotyped within the CHRNA4 gene and rs2072661 at EOT and at 6-month follow-up. None of the interaction tests were significant at an α-level of 0.05. For example, within individuals with the WT genotype for rs2236196, a known functionally significant SNP in CHRNA4 (14), the CHRNB2 SNP rs2072661 effect reduced quit rates (OR = 0.39; 95% CI 0.24–0.65). In contrast, for those individuals with a minor allele for rs2236196, the impact on quit rates was negligible (OR = 0.89; 95% CI 0.31–2.50). While qualitatively suggestive, the P-value for the interaction test was only 0.11. At 6-month follow-up, the effect of rs2072661 did not differ by CHRNA4 rs2236196 carrier status.
DISCUSSION
Using a systems-based candidate gene approach we have identified polymorphisms within the β2 nicotinic acetylcholine receptor (CHRNB2) that exhibits significant association with the abstinence rates at EOT and at 6-month follow-up in a placebo-controlled trial of bupropion for smoking cessation. The association with abstinence was observed for two highly correlated (r2∼0.96) SNPs (rs2072661 and rs2072660) within the 3′UTR. Although the effects were independent of treatment, there was an indication of potential effect modification by bupropion. Specifically, although there was a difference in relapse rates at EOT between carriers and non-carriers for individuals who received bupropion, there was a substantial increase in relapse rates for those individuals carrying the minor allele after they went off treatment (Figure 2). Haplotype analysis capturing the genetic variability within the region confirmed the association across multiple SNPs and further indicated the independent role of the two SNPs. However, because of the high correlation between these SNPs, joint regression modeling was unable to discern the independent effect of each. Follow-up analyses on the top SNP (rs2072661) indicated a role in the time to relapse within the 6-month follow-up period and an impact on withdrawal symptoms at TQD. Investigation of a functionally significant SNP within CHRNA4, a biologically relevant interaction since the α4β2 nAChRs form a common subtype, demonstrated a suggestive, albeit non-significant, interaction.
Evidence for the important role of CHRNB2 in smoking cessation is consistent with the animal studies demonstrating the involvement of β2 subunit-containing nAChRs in nicotine-mediated release of dopamine and in the nicotine withdrawal syndrome. Nicotine reduces, and withdrawal increases the brain stimulation reward-thresholds in rodents (27,28), effects which are mediated largely via α4β2 nAChRs (29). Further, compared with WT mice, knockout mice lacking the β2 subunit exhibit attenuated nicotine self-administration and reduced nicotine stimulated dopamine release in the ventral striatum (30), as well as reductions in conditioned nicotine reinforcement (31,32). Effects of nicotine, and nicotine withdrawal, on cognitive function are also attenuated in β2 knockout mice (33,34).
Human imaging studies indicate that nicotine from a single cigarette nearly completely saturates α4β2 nAChRs (35), and abstinence from nicotine is associated with an increase in the number of unbound β2-containing nAChRs, and thus increasing urge to smoke (36). Based on these observations, genes encoding nAChR subunits have been a focus of a number of previous genetic studies of nicotine dependence. A recent study reported associations of initial subjective responses to nicotine with an SNP immediately upstream of CHRNB2 and one of the two 3′ UTR SNPs (rs2072660) identified by the present study (37). Other candidate gene studies that have examined one or both of the two CHRNB2 3′ UTR SNPs, along with additional SNPs in CHRNB2, have not found association with nicotine dependence at CHRNB2 (15,38–40) in a variety of community- and population-based samples of smokers. In addition, an analysis using smokers and non-smokers that included the two CHRNB2 3′ UTR SNPs and rs2236196 at CHRNA4 to detect gene–gene interactions associated with nicotine dependence failed to detect significant interaction (41). However, nicotine dependence only modestly predicts smoking cessation in response to bupropion (4) and twin studies of the genetic relationship between the two phenotypes suggest that the two phenotypes may have differing genetic contributions [reviewed by Lessov-Schlaggar et al. (42)]. Of note, neither the genes in the dopamine pathway examined here, including those implicated in prior studies, nor the CHRNA3/CHRNA5 gene cluster associated with nicotine dependence in recent reports (15,43–46) reached the threshold of significance for association with smoking cessation or treatment response.
The present study has both strengths and limitations. Strengths include the prospective evaluation of abstinence in the setting of a placebo-controlled trial and collection of DNA samples from all participants, rather than retrospective DNA collection that may result in bias. With regards to the molecular genetic analysis, strengths include conservative SNP selection, robust genotyping, and extensive quality control. Because we performed our SNP selection on an early version of HapMap, we opted to use a high r2 cutoff (0.95) in selecting tagSNPs and including all singleton SNPs. This proved to be a prudent strategy as our set of SNPs captured on an average 85% of the existing common SNPs in the current version of HapMap. While this strategy resulted in a higher number of correlated SNPs within each gene, this did not have a detrimental impact on the analysis as we accounted for the correlation in our multiple testing procedures. Furthermore, a slight level of redundancy in SNP coverage allowed for a more stringent criteria for the removal of SNPs with low call rates, resulting in 97% of the analyzed SNPs having call rates >95%. SNPs with low call rates were driven mostly by genotyping failures for individuals with whole genome-amplified DNA for that particular SNP only. Thus, when there was a discrepancy between call rates for individuals with genomic DNA and individuals with WGA DNA, we limited our analysis to only those individuals with genomic DNA. Finally, for the top SNP rs2072661, there was a 97% concordance between the genotypes obtained from the Illumina assay versus genotypes obtained using TaqMan® (Applied Biosystems, Foster City, CA, USA) assay (data not shown). When we compared carriers versus non-carriers, i.e. a dominant model, we observed 100% concordance.
Another strength of the current study was the genotyping of 233 ancestry informative SNPs to assess intercontinental admixture within individuals of the same self-identified ethnicity (47–49). These SNPs were genotyped on individuals from multiple ethnic groups and analyses investigating the structure used the frequency of SNPs within each self-identified group to more accurately defined ancestry proportions for any given individual. These ancestry estimates were not only used to visually inspect the levels of admixture within Caucasians, but also to empirically compare unadjusted estimates to those adjusted by ancestry proportions. Although we demonstrated that the structure did not impact the analyses, we limited our final analyses to only Caucasians, as LD patterns can vary substantially across ethnic groups and may lead to heterogeneity in effect estimates, thus negatively impacting power despite the inclusion of additional samples.
Finally, our rankings and determination of interesting SNPs was based on an adjusted P-value obtained using a method that accounts for multiple correlated tests when first determining the genetic model and then across all SNPs within each candidate gene region. This resulting adjusted P-value is less conservative than a uniform adjustment across the number of SNPs that assumes independence (i.e. Bonferroni correction), but more stringent than simply ignoring the evaluation of multiple SNPs. This analysis does not yield evidence for an independent effect for each SNP. Here, we used both haplotype and joint regression modeling. However, we do view this adjusted P-value to be the evidence for both a particular SNP and for the gene since the SNPs were originally chosen to capture the common variation within the gene. These adjusted P-values may then be compared with the appropriate significance level for the type of hypothesis of interest. Since the genes were chosen to capture the important components within the entire system, we use a conservative significance α-level of 0.0009 across the 54 gene regions to indicate significance at the system-level. We did not adjust the P-values in our presentation because we believe a transparent presentation of the results is necessary to emphasize associations that are consistent with previous knowledge and to highlight the need for replication of unexpected results. For example, several additional genes (TDO2, ADCYAP1, HTR1B, CDK5, and FOSB) have at least one SNP achieving an adjusted P-value of <0.05, but they are not significant at the systems level. These genes are still of interest as there are varying amount of prior evidence for each (50–53). In addition, there are several recent reports of genes (e.g. DRD1, DRD2, DRD3, ANKK1, BDNF, and NTRK2) within the dopamine system being involved in nicotine dependence (13,54–56). While there are some SNPs within these genes that have observed P-values <0.05 (e.g. DRD1 and DRD2) in the current study, none of these genes have SNPs with significantly adjusted P-values. Furthermore, none of the reported SNPs are significant in the current study (see Supplementary Material, Tables S1 and S2 for complete results). While this may be owing to the difference in phenotypes, confirmation of results by further studies is the key to valid conclusions in this area of research (57). In addition, systems-based analyses may be performed, which more formally integrates prior knowledge via ontologies into the identification and testing of relevant associations (58–61).
For our top SNP we were able to perform specific additional analyses motivated by prior knowledge to refine its potential mechanism of impact via time to relapse, withdrawal symptoms, and interaction effects with a key biologically relevant gene, CHRNA4. Having a detailed phenotypic information prospectively gathered from a randomized placebo-controlled trial was crucial for this further analysis. However, the collection of detailed information limited the original size of the study and thus we were unable to conclusively determine via statistical criteria SNP-treatment or SNP–SNP interactions, although effect estimates were suggestive in some instances. Although we included individuals from several ethnic groups for the investigation of population structure, we were unable to confirm our findings across all the self-identified ethnic groups because of the limited sample size for groups other than Caucasians.
While independent replication will ultimately be required, the results of the present study may have important implications for the treatment of nicotine dependence. We found strong and consistent evidence for the association of two CHRNB2 3′ UTR SNPs with multiple phenotypes assessed in the current trial, including abstinence at both EOT and 6-month follow-up, days to relapse, and nicotine withdrawal symptoms. While the literature provides four independent examples of the lack of association of these two CHRNB2 SNPs with nicotine dependence, these SNPs may be robust markers for identifying smokers most likely to relapse and those who may benefit from bupropion therapy. In addition, these SNPs should be examined within pharmacogenetic studies of varenicline, a new α4β2 nAChR partial agonist medication for smoking cessation. Future studies should also extend molecular genetic analysis to include the large 3′ UTR of CHRNB2 (39) and a novel set of nAChR-interacting proteins that regulate β2 nAChR signaling (62). For example, the 3′ UTR of CHRNB2, extends some 4 kb 3′ of the coding region, and contains seven predicted human micro-RNA targets, including a target for human miR-432 located 13 base pairs 5′ of rs2072660 (63).
MATERIALS AND METHODS
Study sample and design
The individuals in the study were enrolled in a bupropion double-blind placebo-controlled pharmacogenetic smoking cessation trial conducted by the University of Pennsylvania Transdisciplinary Tobacco Use Research Center. All appropriate IRB approvals were obtained by participating institutions as part of the Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Smokers were recruited from April 1999 to October 2001 at Georgetown University (Washington, DC, USA) and SUNY Buffalo (New York, USA). Details of the eligibility criteria and flow of participants through the enrollment, treatment, and follow-up phases of the trial can be found elsewhere (20). Briefly, trial participants included 600 smokers who were >18 years of age, and reported smoking more than 10 cigarettes a day for the prior 12 months. Exclusion criteria included pregnancy, a history of DSMIV axis I psychiatric disorder, seizure disorder, and current use of antidepressants or other psychotropic medications. All participants in the trial provided informed consent for both genotyping and treatment; however, at the time of this genotyping analysis, samples remained for 534 subjects. Analyses were limited to individuals of European ancestry with both phenotype and genotype data (n = 412). Although we examine population structure within all 534 individuals of differing self-identified ethnicities, we limit our primary analyses to only those individuals who self-identified as Caucasian because of the potential for differential linkage disequilibrium across ethnic groups to lead to heterogeneity of effect estimates.
Participants from both the sites received identical assessments and treatments. At an initial visit to the smoking clinic, participants provided a 40 mL blood sample and completed a set of standardized self-report questionnaires. Baseline nicotine dependence was assessed via the FTND (64). All participants received 10 weeks of either placebo or bupropion. Bupropion treatment was delivered according to the standard therapeutic dose (150 mg/day for the first 3 days, followed by 300 mg/day). All participants also received seven sessions of standardized behavioral group counseling, focusing on self-monitoring and behavioral modification approaches. All participants were instructed to quit smoking on a TQD 2 weeks after initiating medication and counseling. Smoking status was assessed by telephone interview at the EOT (8 weeks post-TQD) and at 6 months after the TQD using a validated timeline followback method (65). Interviewers were blind to study group assignment. Participants who reported complete abstinence (not even a puff of a cigarette) for at least the 7 days prior to the assessment were asked to complete an in-person visit for biochemical verification of abstinence. Saliva cotinine testing was performed for participants who reported abstinence at a given time-point using a gas–liquid chromatography method (66). Cotinine is the major proximate metabolite of nicotine and has a much longer half-life than nicotine, making it the preferred biomarker for tobacco use.
Phenotype assessment
The primary outcome was 7-day point prevalence abstinence (biochemically confirmed) at the EOT and 6-month follow-up. Participants were classified as abstinent if (1) they self-reported abstinence and (2) they had a cotinine reading of ≤15 ng/ml. Of the total 412 self-identified Caucasian individuals successfully genotyped, 187 participants self-reported abstinence at EOT (45%) and 133 of these individuals (71%) provided a sample for cotinine verification. At 6-months, 118 participants reporting abstinence (29%) and 89 (75%) of these individuals provided a sample. Those who failed to complete the follow-up or failed to provide a biosample for biochemical verification were presumed to have relapsed, as per convention (67). Of the 412 individuals, 112 were biochemically verified as abstinent at EOT and 90 during 6-month follow-up.
Secondary outcomes included days to relapse following the TQD (using Cox proportional hazards modeling) and withdrawal symptoms. For the time to relapse analysis, self-reported abstinence for the past 7 days at weekly intervals up to 6-month follow-up was used. We assessed withdrawal symptoms on the TQD using a self-report checklist assessing the severity (in the past 7 days) of common symptoms, including irritability, difficulty in concentrating, anxiousness, insomnia, drowsiness, nausea and general physical complaints (e.g. sweating, dizziness). Responses to each item was scored from 0= not at all to 3 = severe and were summed to create a withdrawal severity index (total scores could range from 18 to 60).
Gene and single nucleotide polymorphism selection
We selected 58 candidate genes using a comprehensive bioinformatic approach incorporating expert opinion, automated literature searches, and public pathway databases. The candidate gene list includes several neuronal nicotinic acetylcholine receptors (nAchRs) and genes in the dopamine-mediated reward system, including dopamine receptors, dopamine transporter, and enzymes involved in biosynthesis and metabolism. Additional addiction-related secondary messenger pathway genes, and genes hypothesized to play a role in modulating the dopamine pathway owing to upstream signaling (e.g. serotonin pathway) and those with direct physical interaction with dopamine receptors were also included. The complete list of genes is presented in Table 1.
Of our total of 1528 SNPs genotyped, 233 were ancestry informative markers (AIMs) and 1295 were used to capture the underlying genetic structure within our candidate genes. The AIMs were specifically selected to differentiate four parental populations (African, European, American Indian, and East Asian) (68) within each individual and for making precise estimates of coefficient of ancestry (69,70) and principal components (71) for each individual. We have previously genotyped these AIMs in a multi-ethnic sample from the Multiethnic Cohort Diversity Panel (72) and demonstrated the ability of these polymorphisms to identify levels of ancestral populations within a single individual.
For SNP selection within each candidate gene, we used a beta version of the Snagger software (73). We used HapMap SNP data from the CEPH and Chinese populations [Genome Build 35 (74)]. Because several of our selected candidate genes are located adjacent to each other on a chromosome, we performed our tagSNP selection across 54 regions of interest, expanding each region 10 kb upstream and downstream. Of the 1295 SNPs selected, 118 were of interest because of their putative function and, conditional on including these, an additional 1185 SNPs were selected to capture the underlying genetic structure. For each SNP in a gene, an LD bin was created containing all SNPs in the gene region meeting an r2 threshold of 0.95 with that specific SNP. From these bins, we preferentially selected tag SNPs using several criteria: Illumina design scores (quantifying how well a SNP can be genotyped), validation status (indicating how many platforms validate a SNP), minor allele frequency (MAF), and location (i.e. coding region). To ensure efficient Illumina genotyping, a potential tagSNP would not be selected if it was within 60 bp of another chosen SNP or 35 bp of any other known SNP in dbSNP. Because of the potential for some SNPs to fail in the genotyping, redundant tagSNPs were selected if the number of SNPs in a bin was large. Those SNPs that could not be tagged (i.e. singletons) were also included if their design score was greater than 0.4. Tag SNPs were first chosen in the CEPH population. If necessary to capture the diversity within the Chinese HapMap population sample, additional SNPs were selected. Genetic diversity within Chinese individuals was captured as part of a companion study within the USC Transdisciplinary Tobacco Use Research Center (75).
Genotyping
We performed all genotyping of the SNPs using the GoldenGate™ assay, which uses the Illumina platform (Illumina, San Diego, CA, USA) at the Genomics Core Facility at USC/Norris Comprehensive Cancer Center. We had various levels of quality control. This included automated protocols for the entire genotyping process utilizing robotics and barcoding, and the inclusion of replicates and CEPH trios to aid in genotyping and identification of errors. After initial auto-clustering, each SNP is edited to define three genotypes with the intent of maximizing call rate and minimizing possible error rate. Genotype calls are then evaluated for five criteria: (1) HapMap is queried for the results of the CEPH trios for each SNP and compared with our genotype calls; (2) replicate and trio errors are calculated for all plates/samples in a project; (3) non-informative frequency is calculated for each SNP; (4) deviation from the expected heterozygosity is determined; and (5) MAF is calculated. These criteria are used to identify any SNPs with genotyping errors that may require additional visual inspection, re-calling, or determination that a ‘no call’ be made.
Among the 1528 total SNPs, we excluded 41 SNPs with a call rate of zero and 57 SNPs with an observed MAF <0.01. There were a total of 1430 SNPs available for analysis (1198 SNPs within the candidate genes and 232 AIMs). Of the remaining SNPs, 97% had a SNP call rate ≥95%. Of the 1198 SNPs within the candidate genes, a total of eight SNPs had a P-value of <0.0001 from an exact test of Hardy–Weinberg proportions. The genotyping calls for these SNPs were re-examined, confirmed, and included in the analysis (76–79).
After exclusion of 41 SNPs, individual call rates were calculated using the entire multi-ethnic sample (n = 548) and after stratifying by DNA type, i.e. genomic (gDNA) and whole genome amplified (wgaDNA) DNA. Individuals with a call rate <90% were removed from the analysis (n = 14) for a total of 500 individuals with gDNA and 34 individuals with wgaDNA. While there was a substantial difference between the call rate distributions for the gDNA samples (99.6% of individuals with a call rate ≥95%) in comparison with the wgaDNA samples (85.3% of individuals with a call rate ≥95%), inspection of Illumina data for SNPs with drastically discordant SNP call rates between the two DNA types revealed reliable genotyping calls for SNPs with a SNP call rate calculated using only wgaDNA samples of ≥80%. Thus, for 51 SNPs in which the SNP call rate was ≤80% for wgaDNA samples, we only report analyses using the individuals with gDNA samples.
Statistical analysis
Single nucleotide polymorphism-specific association
We used a generalized linear model to evaluate the individual SNPs:
Here, g(μY) is a link function defining the specific generalized linear model. We used a logistic link for relapse outcomes at both EOT and 6-month follow-up. Thus, the corresponding effect estimates (e.g. β, γ, and δ) represent the log OR and the estimated parameter values are used to calculate the OR for the treatment of the WT genotype and the ORs for an additional risk allele. A Cox proportional hazards model was used for the time to relapse analysis. XG defined the genetic model as either additive or dominant and was mean-centered after determination of the genetic model. For all SNPs, the more common genotype was chosen as the referent. Because of the potential for small data bias, we did not examine the recessive model. For a given SNP, individuals with missing genotypes were excluded from the analysis. XTx was mean-centered and indicated if an individual received Bupropion for treatment. W defined the covariates used in the analysis with all covariates mean-centered. For our basic analysis, covariates included gender, age, and FTND. All analyses were performed among smokers who initiated therapy; consistent with an intent-to-treat analysis, those who withdrew were included in the analysis and were assumed to have returned to smoking.
We used a joint 2-df LRT to combine information from the genetic marginal effect (β) and the gene–treatment interaction effect (δ) as the primary test. Specifically, this test constrained both β ≡ 0 and δ ≡ 0 under the null hypothesis and was shown previously to provide good power across a wide range of underlying true causal models (80). Using this model, we first determined the most appropriate genetic model by testing both dominant and additive genetic models. The best fitting genetic model as determined by the model yielding the lowest P-value, was then used in all the subsequent analyses. Regression analyses were performed using the R Statistical Program (81).
Correcting for correlated tests within a gene and determining system-level significance
To account for the correlated tests from the determination of a genetic model for a given SNP and for the multiple correlated tests from the SNPs within each gene region, we performed a P-value adjustment for correlated tests. Specifically, the tests statistics are modeled as an asymptotically distributed multivariate normal with a co-variance structure estimated from the correlation of observed SNPs (82). This adjustment was performed first on the test statistics obtained by modeling dominant and additive genetic models for each SNP. The resulting adjusted P-values were then further adjusted to account both for the correlation and the number of tests performed across the SNPs within a gene region. The final adjusted P-value is reported and was used to rank the SNPs. System-level significance is determined using a Bonferroni correction across the 54 gene regions. Thus, for a family-wise or system-wide α-level we use 0.05/54 = 0.0009.
Investigation of confounding by population stratification
The entire study sample of 534 smokers was pooled with an additional 577 individuals from the University of Pennsylvania Nicotine Replacement Trial (20) and 355 individuals from the Multi-Ethnic Cohort Diversity Panel (72) that included African-Americans, Asians, Native Hawaiians, Latinos, and Caucasians. These individuals provide reference ethnic groups and help to better estimate the admixture within a given individual. The program STRUCTURE was used to estimate the coefficient of ancestry for each individual (69) and principal component analysis was used to estimate the principal components for each individual (81). For the STRUCTURE analysis, the best fitting analysis estimated four ancestral populations within the combined sample: African, Caucasian, Asian, and Amerindian. The ancestral populations and estimated amounts of admixture within each individual corresponded well with the first four principal components—components that explained 39% of the variation with all the other components explaining 1% or less of the variation each. Visual inspection showed very little individual admixture within each self-identified ethnic group, indicating a small potential for confounding because of population stratification. In addition, we empirically tested the impact of population structure on the analysis by running the base model adjusted for coefficients of ancestry or principal components within the self-identified ethnic groups. The effect estimates obtained from the adjusted analysis were neither substantially different in comparison with the unadjusted analysis nor did the corresponding P-values change considerably. Thus, we did not adjust for the variability captured by the coefficients of ancestry or principal components within the self-reported ethnicity when analyzing the Caucasian-only sample. However, we limited the analyses to only the individuals who self-identified as Caucasian because of the potential for differential linkage disequilibrium across different ethnic groups leading to heterogeneity in effect estimates.
SUPPLEMENTARY MATERIAL
FUNDING
The Pharmacogenetics of Nicotine Addiction Treatment Center is funded by the National Institute on Drug Abuse (U01 DA020830). Funding for the clinical trial was provided by NCI/NIDA P5084718 and NCI RO163562.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of the Pharmacogenetics of Nicotine Addiction Treatment Center, especially Huijun Ring for her early contributions to the center.
Conflict of Interest statement. Dr D.V.C. is a paid consultant to Pfizer, Inc. Dr R.F.T. hold shares in Nicogen Research Inc., a company that is focused on novel smoking cessation treatment approaches. None of the data contained in this manuscript alters or improves any commercial aspect of Nicogen. No funds of Nicogen were used in this work and Nicogen has not reviewed the manuscript or data. Dr N.L.B. is a paid consultant for several pharmaceutical companies that market smoking cessation medications. Dr C.L. has been a paid consultant and has conducted research sponsored by Pfizer, Astra Zeneca, and GlaxoSmithKline. GlaxoSmithKline provided study medication for the clinical trial, but had no role in the study design, interpretation, or funding.
REFERENCES
- 1.Scholl R.A., Lerman C. Current and emerging pharmacotherapies for treating tobacco dependence. Expert Opin. Emerg. Drugs. 2006;11:429–444. doi: 10.1517/14728214.11.3.429. [DOI] [PubMed] [Google Scholar]
- 2.Swan G.E., Jack L.M., Javitz H.S., McAfee T., McClure J.B. Predictors of 12-month outcome in smokers who received bupropion sustained-release for smoking cessation. CNS Drugs. 2008;22:239–256. doi: 10.2165/00023210-200822030-00004. [DOI] [PubMed] [Google Scholar]
- 3.Swan G.E., Javitz H.S., Jack L.M., Curry S.J., McAfee T. Heterogeneity in 12-month outcome among female and male smokers. Addiction. 2004;99:237–250. doi: 10.1111/j.1360-0443.2003.00629.x. [DOI] [PubMed] [Google Scholar]
- 4.Swan G.E., McAfee T., Curry S.J., Jack L.M., Javitz H., Dacey S., Bergman K. Effectiveness of bupropion sustained release for smoking cessation in a health care setting: a randomized trial. Arch. Intern. Med. 2003;163:2337–2344. doi: 10.1001/archinte.163.19.2337. [DOI] [PubMed] [Google Scholar]
- 5.Dale L.C., Glover E.D., Sachs D.P., Schroeder D.R., Offord K.P., Croghan I.T., Hurt R.D. Bupropion for smoking cessation: predictors of successful outcome. Chest. 2001;119:1357–1364. doi: 10.1378/chest.119.5.1357. [DOI] [PubMed] [Google Scholar]
- 6.Hurt R.D., Sachs D.P., Glover E.D., Offord K.P., Johnston J.A., Dale L.C., Khayrallah M.A., Schroeder D.R., Glover P.N., Sullivan C.R., et al. A comparison of sustained-release bupropion and placebo for smoking cessation. N. Engl. J. Med. 1997;337:1195–1202. doi: 10.1056/NEJM199710233371703. [DOI] [PubMed] [Google Scholar]
- 7.Benowitz N.L. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin. Pharmacol. Ther. 2008;83:531–541. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- 8.Paterson N.E., Balfour D.J., Markou A. Chronic bupropion attenuated the anhedonic component of nicotine withdrawal in rats via inhibition of dopamine reuptake in the nucleus accumbens shell. Eur. J. Neurosci. 2007;25:3099–3108. doi: 10.1111/j.1460-9568.2007.05546.x. [DOI] [PubMed] [Google Scholar]
- 9.Portugal G.S., Gould T.J. Bupropion dose-dependently reverses nicotine withdrawal deficits in contextual fear conditioning. Pharmacol. Biochem. Behav. 2007;88:179–187. doi: 10.1016/j.pbb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerman C., Roth D., Kaufmann V., Audrain J., Hawk L., Liu A., Niaura R., Epstein L. Mediating mechanisms for the impact of bupropion in smoking cessation treatment. Drug Alc. Depend. 2002;67:219–223. doi: 10.1016/s0376-8716(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 11.West R., Baker C.L., Cappelleri J.C., Bushmakin A.G. Effect of varenicline and bupropion SR on craving, nicotine withdrawal symptoms, and rewarding effects of smoking during a quit attempt. Psychopharmacology (Berlin) 2008;197:371–377. doi: 10.1007/s00213-007-1041-3. [DOI] [PubMed] [Google Scholar]
- 12.Gelernter J., Yu Y., Weiss R., Brady K., Panhuysen C., Yang B.Z., Kranzler H.R., Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum. Mol. Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- 13.Huang W., Payne T.J., Ma J.Z., Beuten J., Dupont R.T., Inohara N., Li M.D. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African- American sample. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.37. Epub ahead of print 19 March, doi:10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- 14.Hutchison K.E., Allen D.L., Filbey F.M., Jepson C., Lerman C., Benowitz N.L., Stitzel J., Bryan A., McGeary J., Haughey H.M. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch. Gen. Psychiatry. 2007;64:1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- 15.Saccone S.F., Hinrichs A.L., Saccone N.L., Chase G.A., Konvicka K., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O., et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Ye Y., Wang X., Gelernter J., Ma J.Z., Li M.D. DOPA decarboxylase gene is associated with nicotine dependence. Pharmacogenomics. 2006;7:1159–1166. doi: 10.2217/14622416.7.8.1159. [DOI] [PubMed] [Google Scholar]
- 17.Berrettini W.H., Wileyto E.P., Epstein L., Restine S., Hawk L., Shields P., Niaura R., Lerman C. Catechol-O-methyltransferase (COMT) gene variants predict response to bupropion therapy for tobacco dependence. Biol. Psychiatry. 2007;61:111–118. doi: 10.1016/j.biopsych.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 18.David S.P., Strong D.R., Munafo M.R., Brown R.A., Lloyd-Richardson E.E., Wileyto P.E., Evins E.A., Shields P.G., Lerman C., Niaura R. Bupropion efficacy for smoking cessation is influenced by the DRD2 Taq1A polymorphism: analysis of pooled data from two clinical trials. Nicotine. Tob. Res. 2007;9:1251–1257. doi: 10.1080/14622200701705027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heitjan D.F., Guo M., Ray R., Wileyto E.P., Epstein L.H., Lerman C. Identification of pharmacogenetic markers in smoking cessation therapy. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007 doi: 10.1002/ajmg.b.30669. Epub ahead of print 28 Dec, doi:10.1038/sj.tpj.6500492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerman C., Jepson C., Wileyto E.P., Epstein L.H., Rukstalis M., Patterson F., Kaufmann V., Restine S., Hawk L., Niaura R., et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology. 2006;31:231–242. doi: 10.1038/sj.npp.1300861. [DOI] [PubMed] [Google Scholar]
- 21.Swan G.E., Jack L.M., Valdes A.M., Ring H.Z., Ton C.C., Curry S.J., McAfee T. Joint effect of dopaminergic genes on likelihood of smoking following treatment with bupropion SR. Health Psychol. 2007;26:361–368. doi: 10.1037/0278-6133.26.3.361. [DOI] [PubMed] [Google Scholar]
- 22.Swan G.E., Valdes A.M., Ring H.Z., Khroyan T.V., Jack L.M., Ton C.C., Curry S.J., McAfee T. Dopamine receptor DRD2 genotype and smoking cessation outcome following treatment with bupropion SR. Pharmacogen. J. 2005;5:21–29. doi: 10.1038/sj.tpj.6500281. [DOI] [PubMed] [Google Scholar]
- 23.Gotti C., Moretti M., Gaimarri A., Zanardi A., Clementi F., Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochem. Pharmacol. 2007;74:1102–1111. doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Hughes J.R., Stead L.F., Lancaster T. Antidepressants for smoking cessation. Cochrane Database Syst. Rev. 2007 doi: 10.1002/14651858.CD000031.pub2. CD000031. [DOI] [PubMed] [Google Scholar]
- 25.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Mooney M.E., Sofuoglu M. Bupropion for the treatment of nicotine withdrawal and craving. Expert. Rev. Neurother. 2006;6:965–981. doi: 10.1586/14737175.6.7.965. [DOI] [PubMed] [Google Scholar]
- 27.Cryan J.F., Bruijnzeel A.W., Skjei K.L., Markou A. Bupropion enhances brain reward function and reverses the affective and somatic aspects of nicotine withdrawal in the rat. Psychopharmacology (Berlin) 2003;168:347–358. doi: 10.1007/s00213-003-1445-7. [DOI] [PubMed] [Google Scholar]
- 28.Kenny P.J., Markou A. Nicotine self-administration acutely activates brain reward systems and induces a long-lasting increase in reward sensitivity. Neuropsychopharmacology. 2006;31:1203–1211. doi: 10.1038/sj.npp.1300905. [DOI] [PubMed] [Google Scholar]
- 29.Kenny P.J., Markou A. Conditioned nicotine withdrawal profoundly decreases the activity of brain reward systems. J. Neurosci. 2005;25:6208–6212. doi: 10.1523/JNEUROSCI.4785-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picciotto M.R., Zoli M., Rimondini R., Lena C., Marubio L.M., Pich E.M., Fuxe K., Changeux J.P. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391:173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 31.Brunzell D.H., Chang J.R., Schneider B., Olausson P., Taylor J.R., Picciotto M.R. β2-Subunit-containing nicotinic acetylcholine receptors are involved in nicotine-induced increases in conditioned reinforcement but not progressive ratio responding for food in C57BL/6 mice. Psychopharmacology (Berlin) 2006;184:328–338. doi: 10.1007/s00213-005-0099-z. [DOI] [PubMed] [Google Scholar]
- 32.Walters C.L., Brown S., Changeux J.P., Martin B., Damaj M.I. The beta2 but not alpha7 subunit of the nicotinic acetylcholine receptor is required for nicotine-conditioned place preference in mice. Psychopharmacology (Berlin) 2006;184:339–344. doi: 10.1007/s00213-005-0295-x. [DOI] [PubMed] [Google Scholar]
- 33.Portugal G.S., Kenney J.W., Gould T.J. Beta2 subunit containing acetylcholine receptors mediate nicotine withdrawal deficits in the acquisition of contextual fear conditioning. Neurobiol. Learn. Mem. 2008;89:106–113. doi: 10.1016/j.nlm.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehner J.M., Keller J.J., Keller A.B., Picciotto M.R., Paylor R., Booker T.K., Beaudet A., Heinemann S.F., Balogh S.A. Role of neuronal nicotinic receptors in the effects of nicotine and ethanol on contextual fear conditioning. Neuroscience. 2004;129:11–24. doi: 10.1016/j.neuroscience.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Brody A.L. Functional brain imaging of tobacco use and dependence. J. Psychiatry Res. 2006;40:404–418. doi: 10.1016/j.jpsychires.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley J.K., Krishnan-Sarin S., Cosgrove K.P., Krantzler E., Frohlich E., Perry E., Dubin J.A., Estok K., Brenner E., Baldwin R.M., et al. Human tobacco smokers in early abstinence have higher levels of beta2* nicotinic acetylcholine receptors than nonsmokers. J. Neurosci. 2006;26:8707–8714. doi: 10.1523/JNEUROSCI.0546-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehringer M.A., Clegg H.V., Collins A.C., Corley R.P., Crowley T., Hewitt J.K., Hopfer C.J., Krauter K., Lessem J., Rhee S.H., et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144:596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- 38.Li M.D., Beuten J., Ma J.Z., Payne T.J., Lou X.Y., Garcia V., Duenes A.S., Crews K.M., Elston R.C. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum. Mol. Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 39.Lueders K.K., Hu S., McHugh L., Myakishev M.V., Sirota L.A., Hamer D.H. Genetic and functional analysis of single nucleotide polymorphisms in the beta2-neuronal nicotinic acetylcholine receptor gene (CHRNB2) Nicotine. Tob. Res. 2002;4:115–125. doi: 10.1080/14622200110098419. [DOI] [PubMed] [Google Scholar]
- 40.Silverman M.A., Neale M.C., Sullivan P.F., Harris-Kerr C., Wormley B., Sadek H., Ma Y., Kendler K.S., Straub R.E. Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor beta2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am. J. Med. Genet. 2000;96:646–653. [PubMed] [Google Scholar]
- 41.Lou X.Y., Chen G.B., Yan L., Ma J.Z., Zhu J., Elston R.C., Li M.D. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lessov-Schlaggar C.N., Pergadia M.L., Khroyan T.V., Swan G.E. Genetics of nicotine dependence and pharmacotherapy. Biochemical. Pharm. 2008;75:178–195. doi: 10.1016/j.bcp.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amos C.I., Wu X., Broderick P., Gorlov I.P., Gu J., Eisen T., Dong Q., Zhang Q., Gu X., Vijayakrishnan J., et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:612–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berrettini W., Yuan X., Tozzi F., Song K., Francks C., Chilcoat H., Waterworth D., Muglia P., Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychitary. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P., et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 46.Thorgeirsson T.E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K.P., Manolescu A., Thorleifsson G., Stefansson H., Ingason A., et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas D.C., Witte J.S. Point: population stratification: a problem for case–control studies of candidate-gene associations? Cancer Epidemiol. Biol. Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- 48.Wacholder S., Rothman N., Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J. Natl. Cancer Inst. 2000;92:1151–1158. doi: 10.1093/jnci/92.14.1151. [DOI] [PubMed] [Google Scholar]
- 49.Wacholder S., Rothman N., Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol. Biol. Prev. 2002;11:513–520. [PubMed] [Google Scholar]
- 50.Hamada M., Hendrick J.P., Ryan G.R., Kuroiwa M., Higashi H., Tanaka M., Nairn A.C., Greengard P., Nishi A. Nicotine regulates DARPP-32 (dopamine- and cAMP-regulated phosphoprotein of 32 kDa) phosphorylation at multiple sites in neostriatal neurons. J. Pharmacol. Exp. Ther. 2005;315:872–878. doi: 10.1124/jpet.105.090852. [DOI] [PubMed] [Google Scholar]
- 51.Lerer E., Kanyas K., Karni O., Ebstein R.P., Lerer B. Why do young women smoke? II. Role of traumatic life experience, psychological characteristics and serotonergic genes. Mol. Psychiatry. 2006;11:771–781. doi: 10.1038/sj.mp.4001855. [DOI] [PubMed] [Google Scholar]
- 52.Soderstrom K., Qin W., Williams H., Taylor D.A., McMillen B.A. Nicotine increases FosB expression within a subset of reward- and memory-related brain regions during both peri- and post-adolescence. Psychopharmacology (Berlin) 2007;191:891–897. doi: 10.1007/s00213-007-0744-9. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan P.F., Neale B.M., van den Oord E., Miles M.F., Neale M.C., Bulik C.M., Joyce P.R., Straub R.E., Kendler K.S. Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;126:23–36. doi: 10.1002/ajmg.b.20138. [DOI] [PubMed] [Google Scholar]
- 54.Huang W., Ma J.Z., Payne T.J., Beuten J., Dupont R.T., Li M.D. Significant association of DRD1 with nicotine dependence. Hum. Genet. 2008;123:133–140. doi: 10.1007/s00439-007-0453-9. [DOI] [PubMed] [Google Scholar]
- 55.Huang W., Payne T.J., Ma J.Z., Li M.D. A functional polymorphism, rs6280, in DRD3 is significantly associated with nicotine dependence in European-American smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008 doi: 10.1002/ajmg.b.30731. [DOI] [PubMed] [Google Scholar]
- 56.Li M.D., Lou X.Y., Chen G., Ma J.Z., Elston R.C. Gene–gene interactions among CHRNA4, CHRNB2, BDNF, and NTRK2 in nicotine dependence. Biol. Psychiatry. 2008 doi: 10.1016/j.biopsych.2008.04.026. Epub ahead of print 3 June, doi: 10.1016/j.biopsych.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 58.Conti D.V., Cortessis V., Molitor J., Thomas D.C. Bayesian modeling of complex metabolic pathways. Hum. Hered. 2003;56:83–93. doi: 10.1159/000073736. [DOI] [PubMed] [Google Scholar]
- 59.Conti D.V., Lewinger J.P., Swan G.E., Tyndale R.F., Benowitz N.L., Thomas P.D. Using Ontolgies in Hierarchical Modeling of Genes and Exposures in Biologic Pathways. In National Cancer Institute, Phenotypes and Endophenotypes: Foundations for Genetic Studies of Nicotine use and Dependence, Tobacco Control Monograph No. 20. Bethesda, MD: U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute; in press. [Google Scholar]
- 60.Thomas C.E., Ganji G. Integration of genomic and metabonomic data in systems biology–are we ‘there’ yet? Curr. Opin. Drug Discov. Devel. 2006;9:92–100. [PubMed] [Google Scholar]
- 61.Thomas D.C. The need for a systematic approach to complex pathways in molecular epidemiology. Cancer Epidemiol. Biol. Prev. 2005;14:557–559. doi: 10.1158/1055-9965.EPI-14-3-EDB. [DOI] [PubMed] [Google Scholar]
- 62.Kabbani N., Woll M.P., Levenson R., Lindstrom J.M., Changeux J.P. Intracellular complexes of the beta2 subunit of the nicotinic acetylcholine receptor in brain identified by proteomics. Proc. Natl Acad. Sci. USA. 2007;104:20570–20575. doi: 10.1073/pnas.0710314104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heatherton T.F., Kozlowski L.T., Frecker R.C., Fagerstrom K.O. The Fagerstrom test for nicotine dependence: a revision of the Fagerstrom tolerance questionnaire. Br. J. Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 65.Brown R., Burgess E., Sales S., Whiteley J. Reliability and validity of a smoking timeline follow-back interview. Psychol. Addictive. Behav. 1998;12:101–112. [Google Scholar]
- 66.Feyerabend C., Russell M.A. A rapid gas–liquid chromatographic method for the determination of cotinine and nicotine in biological fluids. J. Pharm. Pharmacol. 1990;42:450–452. doi: 10.1111/j.2042-7158.1990.tb06592.x. [DOI] [PubMed] [Google Scholar]
- 67.SRNT Subcommittee on Biochemical Verification. Biochemical verification of tobacco use and cessation. Nictotine. Tob. Res. 2002;4:149–159. doi: 10.1080/14622200210123581. [DOI] [PubMed] [Google Scholar]
- 68.Smith M.W., Patterson N., Lautenberger J.A., Truelove A.L., McDonald G.J., Waliszewska A., Kessing B.D., Malasky M.J., Scafe C., Le E., et al. A high-density admixture map for disease gene discovery in african americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pritchard J.K., Stephens M., Rosenberg N.A., Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 72.Haiman C.A., Stram D.O., Pike M.C., Kolonel L.N., Burtt N.P., Altshuler D., Hirschhorn J., Henderson B.E. A comprehensive haplotype analysis of CYP19 and breast cancer risk: the multiethnic cohort. Hum. Mol. Genet. 2003;12:2679–2692. doi: 10.1093/hmg/ddg294. [DOI] [PubMed] [Google Scholar]
- 73.Edlund C.K., Lee W.H., Li D., Van Den Berg D.J., Conti D.V. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC Bioinformat. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thorisson G.A., Smith A.V., Krishnan L., Stein L.D. The international HapMap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chou C.P., Li Y., Unger J.B., Xia J., Sun P., Guo Q., Shakib S., Gong J., Xie B., Liu C., et al. A randomized intervention of smoking for adolescents in urban Wuhan, China. Prev. Med. 2006;42:280–285. doi: 10.1016/j.ypmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Cox D.G., Kraft P. Quantification of the power of Hardy–Weinberg equilibrium testing to detect genotyping error. Hum. Hered. 2006;61:10–14. doi: 10.1159/000091787. [DOI] [PubMed] [Google Scholar]
- 77.Kang S.J., Gordon D., Finch S.J. What SNP genotyping errors are most costly for genetic association studies? Genet. Epidemiol. 2004;26:132–141. doi: 10.1002/gepi.10301. [DOI] [PubMed] [Google Scholar]
- 78.Leal S.M. Detection of genotyping errors and pseudo-SNPs via deviations from Hardy–Weinberg equilibrium. Genet. Epidemiol. 2005;29:204–214. doi: 10.1002/gepi.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zou G.Y., Donner A. The merits of testing Hardy-Weinberg equilibrium in the analysis of unmatched case-control data: a cautionary note. Ann. Hum. Genet. 2006;70:923–933. doi: 10.1111/j.1469-1809.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 80.Kraft P., Yen Y.C., Stram D.O., Morrison J., Gauderman W.J. Exploiting gene–environment interaction to detect genetic associations. Hum. Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 81.R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: [Google Scholar]
- 82.Conneely K.N., Boehnke M. So many correlated tests, so little time! Rapid adjustment of P-values for multiple correlated tests. Am. J. Hum. Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.