Summary
We show that a molecular scaffold can be utilized to convert a receptor binding aptamer into a receptor agonist. Many receptors (including tumor necrosis receptor family members) are activated when they are multimerized on the cell surface. Molecular scaffolds have been utilized to assemble multiple receptor-binding peptide ligands to generate novel activators of such receptors. We demonstrate that an RNA aptamer that recognizes OX40, a member of the tumor necrosis factor (TNF) receptor superfamily, can be converted into a receptor-activating aptamer by assembling two copies on an olignucleotide-based scaffold. The OX40 receptor-activating aptamer is able to induce nuclear localization of NFκB, cytokine production and cell proliferation as well as enhance the potency of dendritic cell-based tumor vaccines when systemically delivered to mice.
Introduction
RNA ligands or aptamers that bind protein targets can be isolated from a combinatorial library of RNA sequences through the SELEX process [1–3]. Aptamers are an emerging class of therapeutic agents that are potentially useful in a wide variety of therapeutic settings, including tumor immunotherapy [4, 5]. RNA aptamers offer several advantages over traditionally used therapeutic agents including the ability to inhibit protein-protein interactions [6], an avoidance of immunogenicity [7] and the opportunity to rapidly reverse the aptamer’s activity via an “antidote” oligonucleotide [8–10]. Here we sought to determine if a molecule scaffold could be employed to convert a receptor binding aptamer into a receptor agonist, targeting the cell surface receptor OX40.
The modulation of receptor signaling during immune responses has tremendous potential for the treatment of a wide range of diseases including inflammation, autoimmune disease, heart disease and cancer. In particular, the development of therapeutic agents that can modulate the function of the tumor necrosis factor receptor superfamily has received much attention. Because these receptors are activated by ligand-induced multimerization on the cell surface, the development of multimeric receptor binding ligands has been a particular focus. For example, Fournel and colleagues recently demonstrated that a molecular scaffold could be decorated with peptide ligands that recognize the tumor necrosis factor (TNF) receptor family member CD40 and this multivalent ligand could activate CD40 receptor function [11]. Here we evaluate whether such a scaffold approach can be employed to convert an aptamer that recognizes the OX40 receptor into a receptor agonist.
OX40 (CD134, TNFRSF4) is also member of the TNF family of receptors. OX40 is expressed on the surface of activated T cells and interaction with its ligand, OX40 ligand, leads to increased immune function manifested by T cell proliferation and cytokine production [12–14]. As with many other receptors involved in modulating immune cell function (e.g. CD28, CD40, 4-1BB) [15], agonistic antibodies targeting OX40 have been developed [16]. In vitro and in vivo studies have demonstrated that such antibodies can enhance tumor immune responses by inducing dimerization of the OX40 receptor on the cell surface. The promise of the monoclonal antibody pre-clinical data led to a phase I clinical trial using OX40 agonistic antibodies as potential cancer therapeutics [17]. Unfortunately, the murine origin of the antibody used in this trial generated concern about the possibility of anti-murine immune responses following a single administration. Thus in the only clinical trial that has been reported, the safety and efficacy of OX40 antibody treatment could not be established through multiple administrations [18]. More recently a trimeric OX40 ligand fused to the human IgG Fc has been developed as an alternative OX40 agonist for use in patients but its in vitro and in vivo functionality remains to be determined [18]. As an alternative to such protein-based agents, we sought to determine if RNA aptamers could be utilized to stimulate murine OX40 function. Here we describe how a malleable oligonucleotide-based molecular scaffold can be employed to convert an RNA aptamer against murine OX40 into a receptor-activating aptamer.
Results
Isolation of OX40 aptamers and identification of aptamer sequences
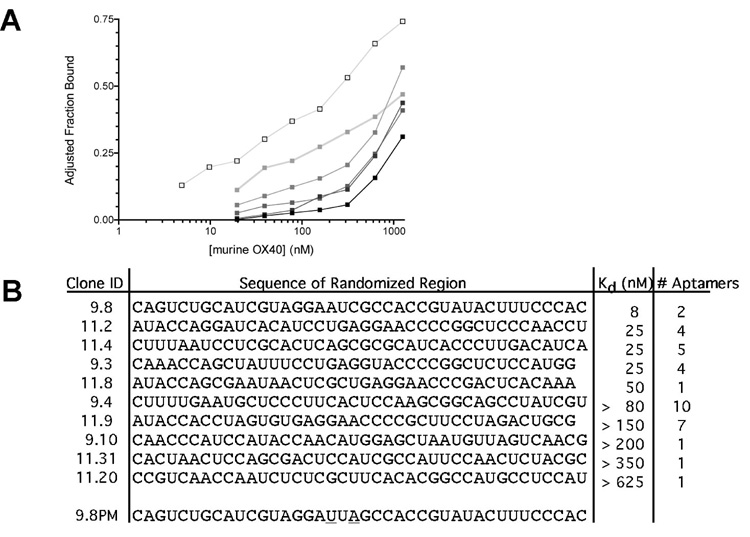
RNA aptamers specific to the T cell costimulatory receptor OX40 were isolated using the SELEX method [1–3]: Murine OX40 human IgG Fc fusion protein was coupled to protein G coated beads to facility RNA partitioning. To avoid amplification of RNA binding undesired portions of this construct, RNA capable of interacting with the human IgG Fc or protein G alone were removed from the randomized RNA library via preincubation with these proteins. This “precleared” RNA pool was then added to the immobilized murine OX40 fusion protein. The OX40 binding RNA was reverse transcribed and amplified using RT & PCR and a secondary enriched RNA library was created by in vitro transcription. This “selection round” was repeated ten times with enhancing stringency, defined as increasing RNA: protein ratios. Selection progress was monitored by comparing binding affinities of the SELEX round RNA to the protein after each round of selection (Figure 1A). After binding affinity no longer increased following additional selection rounds, the sequences of the isolated aptamers were determined.
Figure 1. Isolated OX40 aptamers and binding to recombinant and cell surface OX40.
(A) The binding affinities for the indicated SELEX rounds to a murine OX40 –IgG Fc fusion protein were determined by differential filter binding (RNA pool (■), Round 3 ( ), Round 5 (
), Round 5 ( ), Round 7 (
), Round 7 ( ), Round 9 (
), Round 9 ( ), and Round 11 (□)).
), and Round 11 (□)).
(B) Summary of aptamer sequences against murine OX40 including a point mutant version of aptamer 9.8 (9.8 PM) and their binding affinities The sequence of the 5’ (GGGAGGACGATGCGG) and 3’ fixed regions (CAGACGACTCGCTGAGGATCCGAGA) have been excluded for brevity.
As shown in Figure1B, this process yielded a number of RNA aptamers that bound OX40 with varying affinities (kDs 8–625nM). Aptamer 9.8 was chosen for further study since it had the highest affinity for the OX40 fusion protein. To ensure that RNA aptamer 9.8 bound OX40 and not the Fc portion of IgG or protein G that were used in the selection, we performed additional binding studies. Such analysis demonstrates that aptamer 9.8 binds the extracellular domain of murine OX40 (Supplemental Figure S1).
To control for non-specific activity, we created a mutant aptamer containing two point mutations (aptamer 9.8PM, Figure 1B) that disrupt the aptamer-OX40 interaction. To generate the mutant aptamer we utilized the RNA structure prediction algorithm m-fold [19]. The lowest resulting free energy structure is depicted in Supplemental Figure S2. We chose to mutate two nucleotides in a conserved loop of the aptamer. These nucleotide substitutions maintain the overall predicted structure of the aptamer but abolish its ability to bind OX40 (Figure 2D). This mutant aptamer served as a control in all of our cell and in vivo studies.
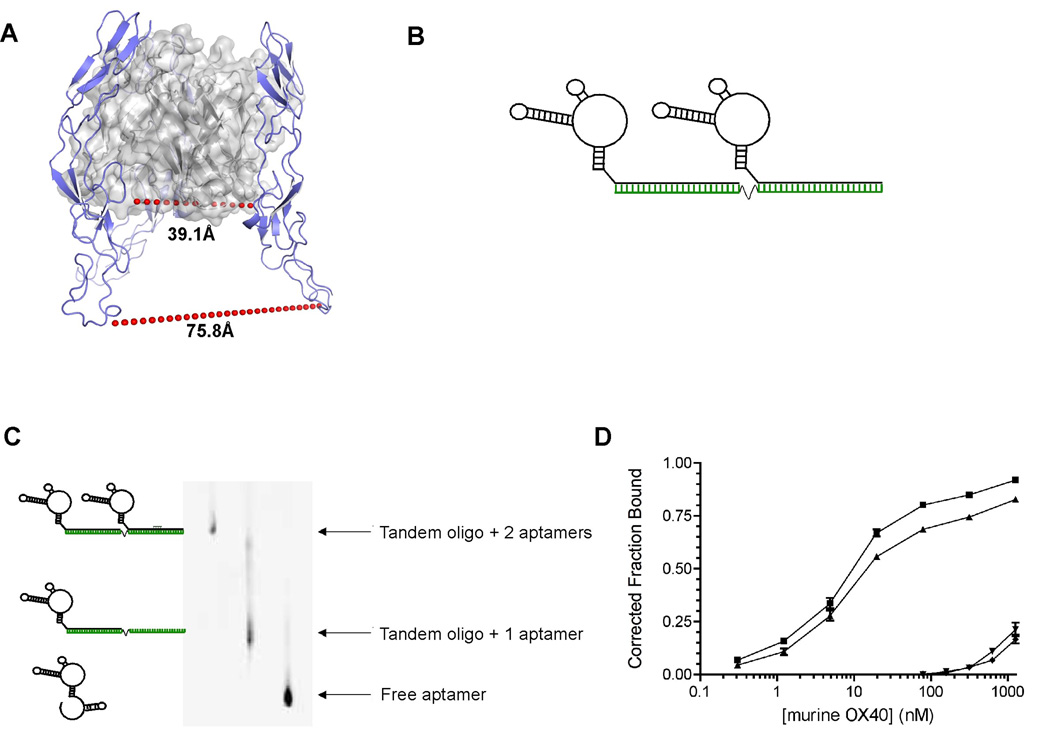
Figure 2. Aptamer dimerization approach.
(A) Threading-based structural model of murine OX40L bound to murine OX40. Shown is a cartoon of a trimeric murine OX40L complex (grey ribbon with transparent surface) liganded with murine OX40 (blue). Each OX40L protomer is liganded stoichiometrically 1:1 to OX40. The shortest (39.1Å) and longest (75.8Å) inter-OX40 distances to equivalent residues are denoted by the red dashed line.
(B) Aptamer 9.8 or point mutant were dimerized by annealing to a oligonucleotide (green) containing 2 sites for aptamer hybridization separated by a polyethylene spacer.
(C) Dimerization of RNA aptamers using a DNA scaffold. RNA aptamers were dimerized by heat annealing to a DNA scaffold at a 1:1 ratio of binding sites. The resulting mixture of dimer (2 aptamer + scaffold) and monomer (1 aptamer + scaffold) were PAGE purified using an 8% native polyacrylamide gel. The identity of gel-eluted dimers was verified by re-running the dimerized RNA on a PAGE gel and visualization using ethidium bromide. The image colors have been inverted for ease of viewing.
(D) Aptamer dimers retain binding affinity to purified OX40 protein. Gel purified aptamer dimers were radioactively labeled and their binding affinities to murine OX40-IgG Fc fusion protein were determined by differential filter binding. Shown are binding curves of OX40-Fc fusion protein to aptamer monomer (■), aptamer dimer (▲), point mutant monomer (▼) or point mutant dimer (◆) (n=3). Error bars indicate SEM.
Converting the OX40 aptamer into a receptor-activating aptamer
Unfortunately, the monomeric aptamer 9.8 is unable to stimulate OX40 function (Supplementary Fig. S3). In an attempt to convey OX40 agonistic activity to aptamer 9.8, we considered characteristics of known OX40 agonists. Agonists known to functionally activate the OX40 receptor include antibody formulations [16] and multimerized versions (dimerized/trimerized [18]) of OX40’s natural ligand (OX40 Ligand) (Figure 2A). These proteins share the common feature of possessing multiple binding sites for OX40. They have the capacity to crosslink receptor subunits, leading to receptor activation and signaling. Therefore, we developed a DNA scaffold that is able to bind two copies of the aptamer 9.8 (Figure 2B). DNA was chosen for the scaffold as we wanted to avoid dsRNA to address concerns about toll like receptor activation due to the fact that double-stranded RNAs have been shown to bind and activate cell surface expressed toll-like-receptors [20]. The aptamer annealing sites on the scaffold were separated by a polyethylene spacer. This spacer was chosen to provide flexibility to the assembled complex as a model of the structure of OX40 complexed with the OX40 ligand indicated that the receptors in the natural receptor-ligand complex are positioned between 39Å and 76Å apart from one another. Incubation of aptamer 9.8 with the DNA scaffold at a ratio of 2:1 resulted in three different species, a DNA scaffold with 2 bound aptamers, a scaffold with a single bound aptamer and unbound aptamer. The three different species were purified via native PAGE followed by elution (Figure 2C). When two copies of the aptamer are assembled on the DNA scaffold they retain their ability to bind the murine OX40 with high affinity (Figure 2D). Furthermore, the aptamer dimers were shown to be capable of binding to two OX40 molecules by gel shift analysis (Supplementary Fig. S4).
OX40 activation is known to serve as costimulatory signal and enhance T cell proliferation [21], induce cytokine secretion [22], and initiate T cell signaling events [23]. We next determined if the fully assembled DNA scaffold-dimeric aptamer complex could elicit these effects. We primed murine T cells with the antigen Staphyloccocal enterotoxin B (SEB) in vivo (Figure 3A) and mice were sacrificed and their lymph nodes removed. Single cell suspensions of the lymph node derived cells were labeled with Carboxy Fluorescein Succinimidyl Ester (CFSE) as outlined in the experimental procedures section [21]. The fluorescently labeled cells were restimulated with the SEB antigen in vitro in the presence of the OX40 bivalent aptamer-DNA scaffold. As shown in Figure 3B and Supplemental Figure S5, treatment of the cells with SEB in addition to either the scaffold-assembled bivalent aptamer or an agonisitic OX40 antibody (OX86) led to an increase in cell proliferation. By contrast, the mutant aptamer containing scaffold and an isotype control antibody had no effect. Thus a DNA scaffold can be utilized to assemble two copies of aptamer 9.8 in close proximity to one another and convert the OX40 aptamer into a receptor-activating aptamer.
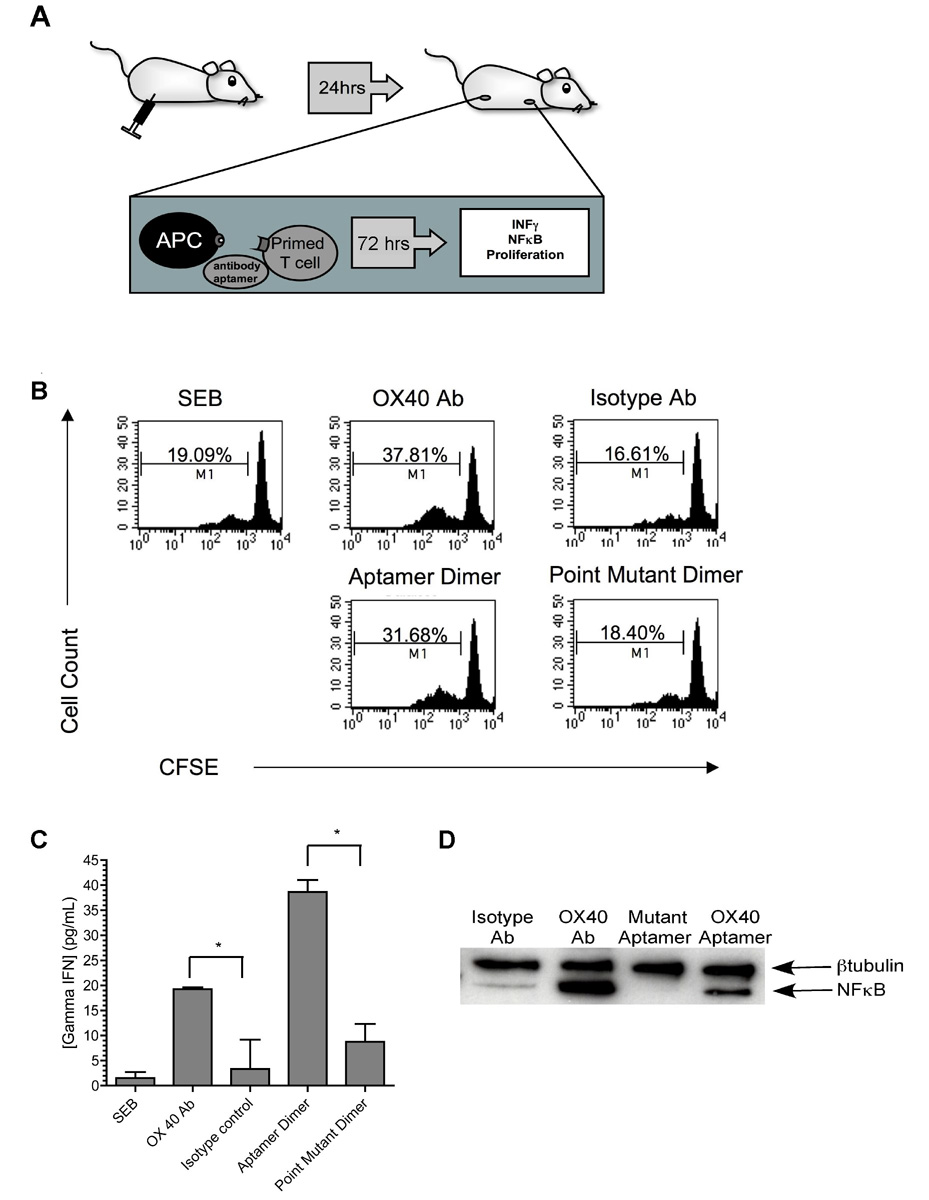
Figure 3. RNA aptamer dimers are capable of enhancing Staphylococcal Enterotoxin B induced proliferation, IFNγ production, and nuclear translocation of NFκB.
(A) Schematic of the experimental setup: Murine T cells were primed in vivo by i.p. injection of Staphylococcal enterotoxin B (SEB). After 24 hours, ingual, auxiliary, and mesenteric lymph nodes were harvested. Cells were restimulated with SEB ex vivo, in the absence or presence of OX40 signaling through the agonistic OX40 aptamer or agonistic antibody. Effects on cell proliferation, IFNγ secretion and nuclear translocation of NFκB were determined.
(B) The scaffold-dimerized OX40 aptamer’s effect on proliferation of SEB primed lymph node cells was assessed by flow-cytometric analysis of the CFSE labeled cells. An OX40 antibody agonist served as positive control. The percentage of proliferating cells is noted. The summary of three independent experiments are depicted in Supplemental Figure S5).
(C) OX40 activation leads to increased IFNγ release. Concentrations of IFNγ in the culture supernatants of the previous experiment were determined by ELISA. Depicted are the mean values of three measurements. Error bars indicate SEM ; *p<0.05.
(D) OX40 activation results in the nuclear translocation of NFκB. To measure the effect of the OX40 receptor-activating aptamer on NFκB localization, cells were cultured as above in absence of CFSE labeling. After 72 hours of culture, nuclei were isolated and subjected to western blot analysis. NFκB and the loading control β-tubulin were detected using specific antibodies and visualized by chemiluminescence
Activation of the OX40 receptor also results in the induction of IFNγ secretion [23],[22]. Therefore, to confirm our cell proliferation results, we evaluated the levels of IFNγ secreted from receptor activating aptamer treated lymph node cells. As shown in Figure 3C, addition of the DNA scaffold-aptamer dimer resulted in a dramatic increase in the production of this cytokine. By contrast the aptamer-scaffold complex did not induce TNFα expression, indicating that the toll like receptors were not being stimulated by the aptamer treatment (Supplemental Figure S6). Finally, since activation of OX40 is known to result in the increased nuclear translocation of NFκB [23, 24], we isolated and evaluated the nuclear fractions from cells for NFκB following treatment with the aptamer-DNA scaffold complex [25]. As shown in Figure 3D, treatment with either the aptamer-DNA complex or an agonistic OX40 antibody resulted in NFκB nuclear localization. By contrast, treatment with the mutant aptamer-containing complex had no effect on NFκB localization. Collectively these results indicate that when two copies of aptamer 9.8 are dimerized by a DNA scaffold, the complex is able to activate the OX40 receptor on primed T cells in culture.
OX40 receptor-activating aptamer function in vivo
To determine if the OX40 receptor-activating aptamer can also act as an agonist in vivo, we evaluated the compounds’ ability to induce OX40 function in a tumor immunotherapy setting. More precisely, we assessed its ability to enhance antitumor responses generated by dendritic cells (DC) transfected with a tumor antigen [26]. DC are antigen-presenting cells that can be modified to present antigens to activate T cells. One approach to supply antigens to dendritic cells is through messenger RNA transfection. Vaccination with antigen pulsed DCs has been previously shown to induce tumor immune responses. To evaluate the OX40 receptor-activating aptamer’s activity in vivo, we evaluated its ability to enhance a DC-based anti-tumor vaccine in mice.
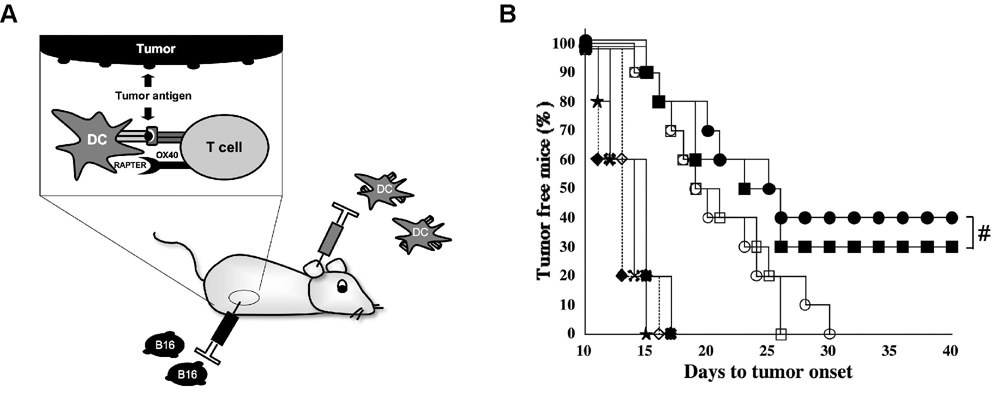
Female C57/BL6 mice were implanted with B16-F10.9 melanoma tumor cells and vaccinated with DCs pulsed with either the melanoma antigen tyrosinase-related protein 2 (TRP-2) or actin (control) mRNA (Figure 4A). The vaccine was systemically administered in the presence of OX40 receptor-activating aptamer, mutant OX40 aptamer, OX40 agonistic antibody or an isotype control antibody [27]. As shown in Figure 4B, administration of DCs containing the TRP-2 antigen alone delayed the development of a palpable tumor compared to control antigen treated animals but did not lead to a cure with all animals developing tumors by weeks 25–30. However, administration of either the OX40 receptor-activating aptamer or the OX40 agonistic antibody to animals receiving the DC-TRP-2 vaccination resulted in tumor free survival in 30–40% of the animals for over 40 days. Thus systemic administration of the OX40 activating aptamer significantly enhanced the potency of a DC tumor vaccine in vivo (DC-TRP-2 + control aptamer versus DC-TRP-2 + OX40 aptamer, P≤ 0.05).
Figure 4. The agonistic OX40 aptamer enhances tumor immunity in mice immunized with TRP-2 RNA transfected DCs.
(A) Mice (5–10/group) were subcutaneously implanted with 2.5 × 104 F10.9 cells in the flank and immunized with 105 RNA transfected DCs at the base of each ear pinna. DCs were generated and electroporated with TRP-2 RNA as described in methods. Mice were injected with OX40 antibody, control antibody, agonistic OX40 aptamer or control point mutant aptamer.
(B) Results of the tumor vaccination experiment. Reported are the days to tumor onset after implantation of tyrosinase related protein transfected DCs in the presence or absence of OX40 induction (DC/TRP-2 + OX40 receptor-activating aptamer (■), DC/TRP-2 + Control aptamer (□), DC/TRP-2 + OX40 antibody (●), DC/ TRP-2 + isotype control (○)), or control antigen transfected DCs (DC/Actin + OX40 receptor-activating aptamer (◆), DC/ Actin + Control aptamer (★), DC/Actin + OX40 antibody (◇), DC/ Actin + Isotype control ( )). The experiment was repeated 3 times with similar results. Enhancement in DC-TRP-2 immunotherapy is not significantly different (#, P>0.05) between OX40 Ab and OX40 receptor-activating aptamer.
)). The experiment was repeated 3 times with similar results. Enhancement in DC-TRP-2 immunotherapy is not significantly different (#, P>0.05) between OX40 Ab and OX40 receptor-activating aptamer.
Discussion
Using the tumor necrosis factor (TNF) receptor family member OX40 as an example, our results demonstrate that RNA aptamers dimerized through annealing to a molecular scaffold can act as receptor agonists. RNA aptamers represent a class of molecules with inherent advantages over the use of antibodies as therapeutic agonists. In contrast to many protein based therapeutic agents, aptamers can be safely administered repeatedly without eliciting compound specific antibodies [7]. The OX40 agonistic antibody currently in clinical development was developed in a murine host and therefore its administration is limited to a single dose because of the potential to develop human anti-mouse antibodies (HAMA) as a reaction to the protein of murine origin [18]. This shortcoming is particularly limiting for treatment of chronic diseases such as cancer. Moreover, the potential exists for patients to develop neutralizing antibodies to humanized monoclonal antibodies following repeated administration.
Recently, we described the first example of an aptamer agonist [28]. However, the approach taken to generate this aptamer agonist is restrictive. Two aptamers were cross-linked through the addition of two complementary sequence extensions to form an aptamer dimer. Each stretch of 21 RNA nucleotides formed a base paired linker that is approximately 65Å in length and is fairly rigid (Supplementary Fig. S7A). This method of aptamer cross-linking is limiting due to the difficulty in varying cross-linker length and the restriction to the number [29] of conformations the cross-linked molecule can adopt. If the paired section is too short, stability is compromised, while further increases in aptamer length greatly decrease aptamer yields through in vitro transcription or chemical synthesis. Moreover the length of the complex is critical for the formation of an aptamer agonist as the goal is to crosslink multiple receptor monomers to induce signaling.
In a paper by Santulli-Marotto et al. [5], multimerization of two CTLA-4 dimers through aptamers that were arrayed on a complementary scaffold was used as a means to increase avidity, as illustrated in Supplementary Fig. S7B. However, the mode of dimerization in this case does not alter the functionality of the aptamers as it is not designed to affect the dimerization of individual CTLA-4 receptors but merely increase the apparent binding affinity through increased valency.
This study is the first demonstration of using a scaffold to arrange two aptamers to crosslink receptor monomers and thereby induce receptor function. The scaffold approach is based on a duplex formation allowing for more flexibility as the scaffold contains a polyethylene glycol linker (Supplementary Fig. S7C), that conveys flexibility much like the hinge region in an antibody (Supplementary Fig. S7D). This approach could allow for application to many other cell surface receptors as the scaffold can accommodate a variety of distances and conformations much like molecular calipers (Supplementary Fig. S7C). Furthermore, the scaffold-based approach taken in this study has the advantage of being amenable to optimization. For example, to enhance the activity of this particular agonistic aptamer, a variety of modifications to the scaffold could be explored: The length of the spacer that is currently 18 carbons in length could be varied to enhance this particular aptamers’ ability to crosslink two OX40 receptor monomers and induce signaling. Moreover, the crystal structure of the OX40-OX40 Ligand complex revealed that the receptor and ligand appear to interact as two trimers [30]. Trimerization of other tumor necrosis family members has previously been shown to induce OX40 signaling [11]. Thus, trimerization of the aptamer on a tripartide scaffold may yield an even more effective agonist.
Finally, the current work is a significantly advance over our previously published work with the 4-1BB aptamer with regard to clinical relevance. In the 4-1BB aptamer case, the in vivo activity of the aptamer complex was demonstrated following local delivery of the aptamer into a the highly immunogenic, easily treatable mastocytoma cell line p815-derived tumor [29]. In this paper, the aptamer agonist was directly injected into the tumor. By contrast, the studies described in this paper on the OX40 receptor-activating aptamer demonstrate that an agonistic aptamer can be used to treat a highly metastatic, poorly immunogenic, and aggressive type of melanoma (F10.9) [31] following systemic administration. Since most cancer patient applications will likely require systemic administration, we believe that the model employed in this study is more clinically relevant.
The large-scale synthesis of the full-length compound on a scale relevant for use as a drug would be difficult and chemical synthesis would be low yield due to the aptamers length (80nt). For clinical application, however, the length of the aptamer can be truncated to its functional core. The truncated aptamer can then be manufactured in large scale by chemical synthesis. Finally, we anticipate that smaller amounts of agonistic aptamers will be required to elicit phenotypic effects compared to antagonistic aptamers. Decrease is the dosage of aptamer required should lower the cost of goods.
Significance
The development of safe and effective receptor agonists have become the focus of much biomedical research. In particular, agonists that can enhance immune responses for treatment of various diseases, including cancer, are being developed and tested in preclinical and clinical studies at an ever-increasing rate. The traditional compounds used as receptor agonists have been derived from protein and peptide based chemistries. In this paper, we demonstrate that assembly of a nucleic acid aptamer upon an oligonucleotide scaffold can mimic the properties of these proteinaceous agents and yield a nucleic acid-based receptor agonist. Previously, it has been shown that dimerization or multimerization of receptor subunits through multivalent ligands can induce tumor necrosis factor receptor function, a receptor family that include the receptors costimulatory receptors OX40 and CD40. We therefore generated a bivalent aptamer construct by annealing monomeric aptamers to an oligonucleotide scaffold. We included a flexible hinge in the scaffold to allow for adaptation to a span of inter-molecular distances between different OX40 receptor monomers. Just as protein based agonists, the generated receptor-activating aptamer can induce receptor function ex vivo and in vivo. Beyond merely mimicking the receptor signaling properties of protein-based agents, the agonistic aptamer has all the advantages of RNA. Due to these inherent advantages of nucleic acid-based agonists and the generalizability of the approaches we describe, we anticipate that agonistic aptamers will emerge as a useful and safe class of therapeutic agents.
Experimental Procedures
Aptamer isolation using the SELEX method
2’ Fluoro pyrimidine RNA aptamers specific to the extracellular portion of murine OX40 were isolated using the SELEX method [1, 2]. A 80 nucleotide combinatorial RNA library was created by transcription of a partially randomized DNA oligonucleotide (5’GGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGGN40 CAGACGACTCGCTGAGGATCCGAGA3’) as described [32]. This library was subjected to two “preclearing” steps to remove RNAs specific to human IgG Fc as well as protein G. To this end, the randomized RNA library was incubated with 1 nmol of human IgG1 (Sigma) at 37°C for 30 minutes. IgG-bound RNA was removed by centrifugation over a 0.4 micron nitrocellulose Centrex column (Whatman). The preclearing step was subsequently followed by incubation with magnetic protein G coated beads (Dynal). After bead pelleting through exposure to a magnet, the supernatant was applied to a nitrocellulose Centrex column (Whatman). All binding reactions were carried out in 150mM NaCl, 2mM CaCl2, 20mM Hepes (pH 7.4), 0.01% BSA buffer.
To enrich for OX40 binding RNAs, murine OX40 human IgG Fc fusion protein (R&D systems) was immobilized by coupling to protein G coated magnetic beads (Dynal) according to manufacturers instructions. The bead coupled OX40 fusion protein was incubated with the “precleared” RNA pool and washed three times with a 20 fold excess volume wash buffer (150mM NaCl, 2mM CaCl2, 20mM Hepes (pH 7.4)) to remove non-interacting RNA. RNA bound to OX40 was extracted by a 30 minute incubation in phenol: chloroform: isoamyl alcohol (25:24:1). The RNA was amplified by reverse transcription followed by PCR.
A secondary, enriched RNA pool was created with transcription using a 2’OH purine, 2’F pyrimidine nucleotide mixture using T7 polymerase. Transcripts were gel purified and eluted into TE, pH 7.5 (10 mM Tris pH 7.5, 0.1mM EDTA). Following overnight elution, RNA was washed three times in TE, pH 7.5 using Centricon 30 columns (Millipore).
Eleven rounds of selection were performed with increasing stringency throughout the selection process by increasing the RNA: Protein ratio in the selection reaction. Aptamers from rounds 9 and 11 DNA were cloned into the EcoRI/ BamHI (New England Biolabs) sites in a pUC18 plasmid. Single colonies were sequenced and amplified by low cycle PCR amplification following by in vitro transcription.
Monomeric aptamer binding affinity
Binding constants were determined using filter binding assays in buffer composed of 20mM Hepes pH 7.4, 150mM NaCl, 2mM CaCl2. To determine the affinities of monomeric aptamers, serial dilutions of murine OX40 IgG Fc fusion protein (R&D systems), human IgG1 (Sigma) or protein G (Zymed) were incubated with monomeric 5’ 32P radiolabeled aptamers at 2000 cpm/µL. The mixture was passed over a stack of membranes consisting of a Protran nitrocellulose and GeneScreen Plus nylon membrane through application of a vacuum. The membranes were exposed to a phosphoimager screen, scanned and quantitated using a Molecular Dynamics Storm 840 phosphoimager. Finally, differential fractions of RNA bound were calculated and graphed using Prism [32].
Aptamer structure prediction
The predicted secondary structure of generated aptamers was determined by utilizing the algorithm m-fold (http://bioweb.pasteur.fr/seqanal/interfaces/mfold.html) using default settings for folding parameters.
Generation of point mutant aptamer
The point mutant RNA aptamer was generated by in vitro transcription of DNA template produced by annealing two oligonucleotides. One nmole of each oligonucleotide (5′GGGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGGCAGTCTGCATCGTAGGATTAGC3′and 5′TCTCGGATCCTCAGCGAGTCGTCTGGTGGGAAAGTATACGGTGGCTAATCCTACGATGCAGACTG3′) were heated to 95°C for 5 minutes and annealed by cooling to 4°C. A double stranded DNA transcription template was created using treatment with Exo-Klenow fragment (New England Biolabs). The reaction was stopped by the additon of 2mM EDTA followed by phenol: chloroform and chloroform extraction. The template was purified by triplicate washing using a centricon 30 column and 10 mM Tris pH 7.5, 0.1mM EDTA buffer. 2’F modified point mutant RNA aptamer was generated through in vitro transcription using T7 polymerase.
Aptamer dimerization
A DNA scaffold consisting of a 20 nucleotide repeat of the complementart sequence of 3’ fixed region of the aptamer separated by an 18 carbon spacer (Operon) (5’ TCTCGGATCCTCAGCGAGT carbon spacer TCTCGGATCCTCAGCGAGT 3’) was used to dimerize RNA aptamers. RNA aptamers were mixed with this scaffold at a 2:1 molar ratio of RNA to scaffold. The mixture was heated to 95°C for 5 minutes followed by slow cooling to room temperature.
For binding affinity determination, aptamer dimers were purified using 8% native PAGE purification followed by overnight elution into 2mL of TE, pH 7.5 buffer at 4°C followed by extensive washing as described above.
Quantification of dimer aptamer binding affinity
Radioactive labelling of monomeric RNA normally involves a dephosphorylation step at 65°C using bacterial alkaline phosphatase (Invitrogen) followed by radioactive labeling using T4 kinase and 32P gamma labeled ATP. However, gel purified RNA dimers are heat labile and therefore could not be heated to 65°C. Therefore, dimers were 3’ radiolabelled by incubation with T4 RNA ligase (Ambion) according to manufacturers instructions at 4°C. This method leads to a lower incorporation efficiency but has the advantage of avoiding exposure of the dimer to heat. Binding affinities were determined as described for the aptamer monomer.
Proliferation assay
Activation of OX40 leads to increased T cell proliferation. We therefore tested the effect of the dimerized aptamers on activation of OX40 and the consequent increase in lymph node cell proliferation [21]. 50µg of Staphyloccocal enterotoxin B (Sigma) resuspended in PBS (Gibco) was administered to female Balb/c mice intraperitoneally. Auxillary, inguinal and mesenteric lymph nodes were harvested after 24 hours. Cells were teased into single cell suspension and labelled with carboxyfluoroscein succinimidyl ester (CFSE/ Pierce) by incubating cells at a concentration of 1 million cells/mL in PBS (Gibco) containing 5% fetal bovine serum (HyClone) and 2mM CFSE at room temperature for 5 minutes. Cells were washed twice using phosphate buffered saline with 5% fetal bovine serum followed by a final wash with RPMI containing 10% fetal bovine serum.
105 cells were seeded in wells of a round bottom 96 well plate well (Corning) and were cultured for 72 hours in complete RPMI (Gibco) containing 10% fetal bovine serum (HyClone), in the presence of 0.5ng/mL Staphyloccocal enterotoxin B in a humidified chamber at 37°C/ 5% CO2. Experimental groups also included 33nM OX40 agonistic antibody (OX86), isotype control (ebiosciences), 66nM of aptamer dimer or point mutant aptamer dimer. Groups were set up in five replicates and pooled for analysis. Cell proliferation data was collected using flow cytometry using a FACScalibur and evaluated using CellQuest software (Becton Dickson).
Determination of IFNγ concentration in tissue culture supernatants
Supernatants of proliferation assay replicates were pooled after 72 hours of culture. Interferon γ secretion was measured in triplicate using the Ready Set Go Elisa kit (ebiosciences) following manufacturers instructions.
Nuclear NFκB detection
NFκB is translocated to the nucleus as a result of OX40 signaling. As a measure of OX40 activation, we therefore determined presence of nuclear NFκB in murine lymph node cells incubated with the aptamer or point mutant dimer compared to the agonisitic OX40 antibody. Mice were injected with staphylococcal enterotoxin B and lymph nodes harvested as described in the proliferation assay. Cells were teased into single cell suspension and 105 cells per 96 well plate well were seeded in complete RPMI containing 0.5ng/mL staphylococcal enterotoxin B. Aptamer dimers or antibodies were added at a concentration of 66 nM. After 72 hour culturing, cells were pelleted and nuclei were isolated using to the Sigma CelLytic NuCLEAR Extraction kit according to manufacturer’s instructions for hypotonic nuclear isolation [25]. The absorbance of the generated protein fractions at A280 was determined. Equivalent amounts of protein were loaded onto a 4–15% denaturing PAGE gel (Biorad) and transferred to a polyvinylidene difluoride (PDVF) membrane by electroblotting. NFκB was detected using a specific primary antibody (Santa Cruz) followed by incubation with a horseradish peroxidase labeled secondary antibody (goat anti rabbit Santa Cruz). Protein was visualized using the ECL plus chemiluminescence detection kit (GE Amersham) and captured through exposure to film. Antibodies were removed from the membranes by a 15-minute incubation with Restore Western Blotting Stripping Buffer (Pierce). Successful stripping was verified by treatment with chemiluminescence reagents and exposure to film. The nuclear protein loading control beta tubulin was detected through incubation with a primary followed by a secondary HRP conjugated antibody.
Murine bone marrow precursor-derived DC
Marrow from tibias and femurs of C57BL/6 mice were harvested followed by treatment of the precursors with ammonium chloride Tris buffer for 3 min at 37°C to deplete the red blood cells. The precursors were plated in RPMI with 5% FCS and GM-CSF (15 ng/ml) and IL-4 (10 ng/ml). GM-CSF and IL-4 were obtained from Peprotech (Rocky Hill, NJ). Cells were plated at 106/ml and incubated at 37°C and 5% CO2. 3 days later the floating cells (mostly granulocytes) were removed and the adherent cells replenished with fresh GM-CSF and IL-4 containing medium. 4 days later the non-adherent cells were harvested, washed and electroporated with RNA.
Electroporating murine DC with RNA
DC were harvested on day 7, washed and gently resuspended in Opti-MEM (GIBCO, Grand Island, NY) at 2.5 × 107/ ml. The used DC culture media was saved as conditioned media for later use. Cells were electroporated in 2 mm cuvettes (200 µl of DC (5 × 106 cells) at 300 V for 500 µs using an Electro Square Porator ECM 830, BTX, San Diego, CA). The amount of TRP-2 or actin RNA used was 3 µg, per 106 DC. Cells were immediately transferred to 6-well plates containing a 1:1 combination of conditioned DC growth media and fresh RPMI with GMCSF and IL4. Transfected cells were incubated at 37°C, 5% CO2 for 4h in the presence of 100 ng/ml LPS (Sigma product # L265L, E.coli 026:B6), washed two times in PBS and then injected into mice.
Statistical analysis
Statistical analysis was performed using the graphing software Prism. Two-tailed, nonparametric t-tests were carried out using the default parameters. Statistical analysis of in vivo data was completed using the logrank (Mantel-Haenszel test). Confidence intervals equal to or less than 0.05 were considered to constitute statistical significance.
Supplementary Material
Acknowledgements
This publication was made possible by Grant Number 1UL1 RR024128-01 from the National Institutes for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and roadmap for Medical Research to S. Nair as well as grants from the National Institutes of Health (NIH) to B. Sullenger (HL065222). We would like to thank D. Snyder, J. Mi, and Y. Liu for their technical assistance and D. E. Dollins as well as M. Kierlin-Duncan for critical reading of this manuscript. Thanks to B. Ramsay Shaw for critical discussions.
Competing Interest Statement
Dr. Sullenger and Dr. Gilboa are scientific founders of Argos Biosciences a company focused on the clinical development of DC-based vaccines and Dr. Sullenger is a scientific founder of b3 Bio Inc. a biotechnology company focused upon using aptamers as delivery agents. Neither company provided any support for the studies performed in the manuscript. The work was supported through grants from the NIH to B.A.S. and S.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 4.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 5.Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 6.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Direct in vitro selection of a 2'-O-methyl aptamer to VEGF. Chem Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Macugen Pegaptanib 1-Year Systemic Safety Results from a Safety-Pharmacokinetic Trial in Patients with Neovascular Age-Related Macular Degeneration. Ophthalmology. 2007 doi: 10.1016/j.ophtha.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 9.Dyke CK, Steinhubl SR, Kleiman NS, Cannon RO, Aberle LG, Lin M, Myles SK, Melloni C, Harrington RA, Alexander JH, Becker RC, Rusconi CP. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- 10.Nimjee SM, Keys JR, Pitoc GA, Quick G, Rusconi CP, Sullenger BA. A novel antidote-controlled anticoagulant reduces thrombin generation and inflammation and improves cardiac function in cardiopulmonary bypass surgery. Mol Ther. 2006;14:408–415. doi: 10.1016/j.ymthe.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Fournel S, Wieckowski S, Sun W, Trouche N, Dumortier H, Bianco A, Chaloin O, Habib M, Peter JC, Schneider P, Vray B, Toes RE, Offringa R, Melief CJ, Hoebeke J, Guichard G. C3-symmetric peptide scaffolds are functional mimetics of trimeric CD40L. Nat Chem Biol. 2005;1:377–382. doi: 10.1038/nchembio746. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg AD. OX40: targeted immunotherapy--implications for tempering autoimmunity and enhancing vaccines. Trends Immunol. 2002;23:102–109. doi: 10.1016/s1471-4906(01)02127-5. [DOI] [PubMed] [Google Scholar]
- 13.Sugamura K, Ishii N, Weinberg AD. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat Rev Immunol. 2004;4:420–431. doi: 10.1038/nri1371. [DOI] [PubMed] [Google Scholar]
- 14.Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 15.Melero I, Hervas-Stubbs S, Glennie M, Pardoll DM, Chen L. Immunostimulatory monoclonal antibodies for cancer therapy. Nat Rev Cancer. 2007;7:95–106. doi: 10.1038/nrc2051. [DOI] [PubMed] [Google Scholar]
- 16.al-Shamkhani A, Birkeland ML, Puklavec M, Brown MH, James W, Barclay AN. OX40 is differentially expressed on activated rat and mouse T cells and is the sole receptor for the OX40 ligand. Eur J Immunol. 1996;26:1695–1699. doi: 10.1002/eji.1830260805. [DOI] [PubMed] [Google Scholar]
- 17.Weinberg AD, Thalhofer C, Morris N, Walker JM, Seiss D, Wong S, Axthelm MK, Picker LJ, Urba WJ. Anti-OX40 (CD134) administration to nonhuman primates: immunostimulatory effects and toxicokinetic study. J Immunother (1997) 2006;29:575–585. doi: 10.1097/01.cji.0000211319.00031.fc. [DOI] [PubMed] [Google Scholar]
- 18.Morris NP, Peters C, Montler R, Hu HM, Curti BD, Urba WJ, Weinberg AD. Development and characterization of recombinant human Fc:OX40L fusion protein linked via a coiled-coil trimerization domain. Mol Immunol. 2007;44:3112–3121. doi: 10.1016/j.molimm.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. [Google Scholar]
- 20.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali SA, Ahmad M, Lynam J, McLean CS, Entwisle C, Loudon P, Choolun E, McArdle SE, Li G, Mian S, Rees RC. Anti-tumour therapeutic efficacy of OX40L in murine tumour model. Vaccine. 2004;22:3585–3594. doi: 10.1016/j.vaccine.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 22.Kawamata S, Hori T, Imura A, Takaori-Kondo A, Uchiyama T. Activation of OX40 signal transduction pathways leads to tumor necrosis factor receptor-associated factor (TRAF) 2- and TRAF5-mediated NF-kappaB activation. J Biol Chem. 1998;273:5808–5814. doi: 10.1074/jbc.273.10.5808. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 24.Arch RH, Thompson CB. 4-1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappaB. Mol Cell Biol. 1998;18:558–565. doi: 10.1128/mcb.18.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi J, Zhang X, Rabbani ZN, Liu Y, Su Z, Vujaskovic Z, Kontos CD, Sullenger BA, Clary BM. H1 RNA polymerase III promoter-driven expression of an RNA aptamer leads to high-level inhibition of intracellular protein activity. Nucleic Acids Res. 2006;34:3577–3584. doi: 10.1093/nar/gkl482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair S, Boczkowski D. RNA-transfected dendritic cells. Expert Rev Vaccines. 2002;1:507–513. doi: 10.1586/14760584.1.4.507. [DOI] [PubMed] [Google Scholar]
- 27.Nair S, Boczkowski D, Fassnacht M, Pisetsky D, Gilboa E. Vaccination against the forkhead family transcription factor Foxp3 enhances tumor immunity. Cancer Res. 2007;67:371–380. doi: 10.1158/0008-5472.CAN-06-2903. [DOI] [PubMed] [Google Scholar]
- 28.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8 T cells and inhibit tumor growth in mice. J Clin Invest. 2007 doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talmage DW, Woolnough JA, Hemmingsen H, Lopez L, Lafferty KJ. Activation of cytotoxic T cells by nonstimulating tumor cells and spleen cell factor(s) Proc Natl Acad Sci U S A. 1977;74:4610–4614. doi: 10.1073/pnas.74.10.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Compaan DM, Hymowitz SG. The crystal structure of the costimulatory OX40-OX40L complex. Structure. 2006;14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Porgador A, Gansbacher B, Bannerji R, Tzehoval E, Gilboa E, Feldman M, Eisenbach L. Anti-metastatic vaccination of tumor-bearing mice with IL-2-geneinserted tumor cells. Int J Cancer. 1993;53:471–477. doi: 10.1002/ijc.2910530320. [DOI] [PubMed] [Google Scholar]
- 32.Layzer JM, Sullenger BA. Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides. 2007;17:1–11. doi: 10.1089/oli.2006.0059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.