Abstract
Advances in NMR spectroscopy have enabled the study of larger proteins that typically have significant overlap in their spectra. Specific 15N-amino acid incorporation is a powerful tool for reducing spectral overlap and attaining reliable sequential assignments. However, scrambling of the label during protein expression is a common problem. We describe a rapid method to evaluate the fidelity of specific 15N-amino acid incorporation. The selectively labeled protein is proteolyzed, and the resulting peptides are analyzed using MALDI mass spectrometry. The 15N incorporation is determined by analyzing the isotopic abundance of the peptides in the mass spectra using the program DEX. This analysis determined that expression with a 10-fold excess of unlabeled amino acids relative to the 15N-amino acid prevents the scrambling of the 15N label that is observed when equimolar amounts are used. MALDI TOF-TOF MS/MS data provide additional information that shows where the “extra” 15N labels are incorporated, which can be useful in confirming ambiguous assignments. The described procedure provides a rapid technique to monitor the fidelity of selective labeling that does not require a lot of protein. These advantages make it an ideal way of determining optimal expression conditions for selectively labeled NMR samples.
Keywords: MALDI mass spectrometry, tandem mass spectrometry, specific 15N-amino acid labeling, proteolytic digestion, NMR spectroscopy
Almost as soon as isotopic labeling became useful for NMR spectroscopy, researchers desired reliable ways to incorporate labels at only certain amino acids to ease the assignment process or to study dynamics of certain residues (Griffey et al. 1985). Specific labeling can be achieved by expressing the protein in Escherichia coli grown in medium containing the specific 15N-labeled amino acid(s) (Torchia et al. 1988). However, metabolism of the 15N-amino acid and subsequent amino acid biosynthesis in E. coli can result in scrambling of the label. Scrambling can be prevented by feedback inhibition of these processes, where the medium is supplemented with the other 14N-amino acids (Torchia et al. 1989; Yamazaki et al. 1991; Roth et al. 1992; Ramesh et al. 1994). However, the amount of supplementation required is variable, and it is often difficult to determine whether scrambling was eliminated, especially if spectral assignments are unavailable. Alternatively, the protein can be expressed in E. coli auxotrophs, which lack the enzymes that metabolize the amino acids that are being labeled (Waugh 1996). However, E. coli auxotrophs may not be available with the genotype that is optimal for expression of some proteins. In addition, auxotrophs tend to grow more slowly, so selectively labeling a protein that requires deuteration is often very difficult. Finally, cell-free systems have been developed for production of specifically labeled proteins, but production of large quantities of protein is difficult (Etezady-Esfarjani et al. 2007).
We describe a method for assessing the fidelity of specific labeling using an IκBα mutant as a test case. IκBα is an ankyrin repeat protein inhibitor of the NF-κB family of transcription factors. We have shown that regions of IκBα are highly flexible (Croy et al. 2004; Truhlar et al. 2006), and we decided to pursue NMR studies of the AR domain. The HSQC spectrum of IκBα has many missing and overlapping resonances, which are both common challenges for attaining sequential assignments. Therefore, we turned to stabilizing mutations and selective labeling to assist in the assignment process.
Results and Discussion
The HSQC spectrum of the ankyrin repeat domain of the C186P/A220P mutant of IκBα (IκBαCP/AP) shows only 141 of the 209 expected peaks. Due to the missing resonances and overlap in the HSQC spectrum, assignment of the backbone resonances is difficult, so selective 15N amino acid labeling was used to improve confidence in the sequential assignments. Interestingly, the HSQC spectrum of 15N-Leu IκBαCP/AP showed the expected number of peaks (37), but comparison of this spectrum with the HSQC spectrum of 15N-Ala IκBαCP/AP showed that several resonances in the 15N-Leu spectrum were also present in the 15N-Ala spectrum (Supplemental Fig. 1). Therefore, we suspected that scrambling of the 15N label was occurring during protein expression.
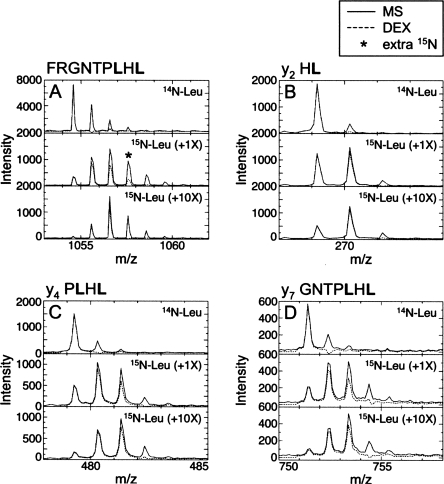
We developed a mass spectrometry (MS)-based technique to monitor the fidelity of 15N incorporation under different expression conditions. Once the system is set up, data collection and analysis can be completed in just a few hours. This procedure utilizes the program DEX that was developed to quantify the number of deuterons incorporated in peptides during amide H/2H exchange experiments (Hotchko et al. 2006). Peptic digestion of IκBαCP/AP yields 10 peptides, covering 40% of the sequence, with sufficient signal-to-noise ratios to confidently interpret the isotopic abundance in the peptide. Tryptic digestion could also be used to generate peptides; however, tryptic digestion of IκBαCP/AP yielded fewer peptides than peptic digestion (data not shown). We prepared 15N-Leu IκBαCP/AP by supplementing the expression medium with 15N-Leu and equimolar concentrations of all other unlabeled amino acids, hereafter 15N-Leu (+1X) IκBαCP/AP (see Materials and Methods). The isotopic abundance in each 15N-Leu IκBαCP/AP peptide was analyzed using DEX (Hotchko et al. 2006). DEX is a Fourier deconvolution method that analyzes isotopically labeled peptide mass envelopes. The deconvolution process allows removal of the natural isotopic abundance component and subsequent determination of the amount of isotope incorporation in the peptide. For each peptide analyzed, DEX calculates the population containing 0, 1, …, n 15Ns incorporated, where n is the maximum number incorporated. In our analysis, populations greater than 10% were considered significant. Since these signals are expected to be visible in the NMR spectra, we compared n in each peptide with the expected number of 15N incorporation events. We found that only two overlapping peptides in 15N-Leu (+1X) IκBαCP/AP show the expected number of 15N incorporation events. The mass spectra of the eight remaining peptides show either one or two “extra” 15N incorporation events (Fig. 1A; Supplemental Table 1). These data clearly show that 15N incorporation is not limited to Leu in 15N-Leu (+1X) IκBαCP/AP. Some of the IκBαCP/AP sequential assignments have now been attained. Consistently, the HSQC spectrum of the 15N-Leu (+1X) IκBαCP/AP shows peaks that are assigned to Arg, Ala, Val, and Glu (data not shown). This scrambling of the label results from 15N-Leu metabolism to 15N-Glu, which is a precursor to a variety of amino acids in E. coli, including Arg, Ala, and Val (Waugh 1996).
Figure 1.
MS and MS/MS spectra reveal 15N scrambling. (A) DEX analysis (– –) of the mass spectrum (—) of the 142–150 IκBαCP/AP peptide (MH+ 1054.6) shows no 15N in the unlabeled control sample (top), one extra 15N label in the 15N-Leu IκBαCP/AP sample prepared with 1X unlabeled amino acids (middle), and faithful incorporation in the 10X sample (bottom). The MS/MS spectra of the y2-ion (B), the y4-ion (C), and the y7-ion (D) of the 142–150 IκBαCP/AP peptide all show faithful incorporation of 15N-Leu with both 1X and 10X unlabeled amino acids, indicating that the extra 15N in the 1X sample is at either F142 or R143.
Feedback inhibition of 15N scrambling during expression has been successfully utilized to prepare selectively labeled proteins; however, very different amounts of the unlabeled amino acids were used to suppress the scrambling of the 15N label (Torchia et al. 1989; Yamazaki et al. 1991; Roth et al. 1992; Ramesh et al. 1994). We expressed 15N-Leu IκBαCP/AP with a 10-fold excess of all other unlabeled amino acids, hereafter 15N-Leu (+10X) IκBαCP/AP. This protein shows faithful 15N incorporation in all 10 peptides (Fig. 1A; Supplemental Table 1). Thus, an excess of all other unlabeled amino acids suppresses the scrambling of the label. Interestingly, the HSQC spectrum of 15N-Leu (+10X) IκBαCP/AP shows 29 of the 37 expected resonances (all with leucine-specific chemical shifts), which is not unexpected given the 68 peaks missing from the 15N-IκBαCP/AP spectrum. Analysis of 15N-Leu (+5X) IκBαwild-type also shows faithful 15N incorporation, suggesting that a fivefold excess is sufficient (data not shown). Similar experiments using 15N-Ala and 15N-Val (+1X) IκBαCP/AP also show scrambled 15N incorporation, but specific incorporation when expressed with a 10-fold excess of unlabeled amino acids. Overall, these data show that this procedure readily identifies whether or not the 15N-amino acid is faithfully incorporated under a given set of expression conditions.
If MALDI MS confirms the fidelity of the specific 15N-amino acid incorporation in the experimental samples, it is sufficient to only analyze the MS spectra. However, in some cases, additional information from the scrambled samples may be useful. Collision-induced dissociation (CID) of peptides in tandem mass spectrometry (MS/MS) experiments can be used to determine the peptide sequence (Medzihradszky 2005). Fragments containing one to n-1 amino acids, where n is the number of amino acids in the peptide, may be observed in the MS/MS spectrum. Thus, MALDI MS/MS analysis of peptides that are overlabeled could be used to identify which regions of the peptide contain the extra 15N label(s). DEX is not designed to analyze MS/MS data. However, y-ions3 have the same chemical formula as an MH+ peptide ion with the equivalent remaining amino acids. Therefore, the natural isotopic abundance calculated by DEX for the sequence of a y-ion will be accurate. Hence, we created a list of y-ions that do not overlap with other potential ions, and we analyzed the isotopic abundance of those y-ions in the MS/MS spectra. In the 142–150 peptide, the y2-ion (HL, MH+ 269.2) shows incorporation of one 15N, suggesting that only the Leu is labeled (Fig. 1B). The y4-ion of the same peptide (PLHL, MH+ 479.3) shows incorporation of two 15N, suggesting that both Leu residues are labeled (Fig. 1C). The y5-, y6-, and y7-ions (y7 GNTPLHL, MH+ 751.4) of the same peptide also show incorporation of only two 15N, suggesting that only the two Leu residues are labeled (Fig. 1D; Supplemental Table 1). However, the full peptide (FRGNTPLHL, MH+ 1054.6) shows incorporation of three 15N labels (Fig. 1A). Together, these data suggest that one of the two N-terminal amino acids (FR) incorporate the 15N label. Alternatively, there may be fractional labeling of multiple amino acids in the peptide that is not distinguished from the noise in short fragments. However, comparison of the MS and MS/MS data with the partially assigned HSQC spectrum of 15N-Leu (+1X) IκBαCP/AP shows that Arg143, which is covered by the 142–150 peptide, does incorporate a 15N label. Furthermore, comparison of the 15N incorporation in all peptides and y-ions analyzed (Supplemental Table 1) indicates that the overlabeling in 15N-Leu (+1X) IκBαCP/AP appears to be localized to discrete regions. In fact, one of the 209–213 residues (VNAQE), which are in the middle of the 202–220 and 203–220 peptides, contains an extra 15N label, but the N- and C-terminal residues in these peptides show faithful 15N incorporation. Interestingly, while some Ala, Val, and Glu residues in 15N-Leu (+1X) IκBαCP/AP incorporate 15N, not all Ala, Val, and Glu residues are labeled. Additionally, two separate preparations of 15N-Leu (+1X) IκBαCP/AP show identical overincorporation of the 15N labels. Overall, these data clearly show that MS/MS analysis of overlabeled samples provides additional information regarding which residues incorporate the 15N label. By extracting from the MS/MS data the identities of the overlabeled regions, one can further narrow down the resonance assignments in the HSQC spectra of scrambled samples.
We have demonstrated that analysis of proteolyzed selectively labeled proteins by MS, combined with a measurement of isotopic abundance using DEX, provides a rapid procedure by which the fidelity of specific 15N-amino acid incorporation can be determined. This procedure requires only a small amount of protein (3 nmol), and it does not require multiple samples or sequential NMR assignments. Therefore, this technique can be used as an initial quality control check for selectively labeled NMR samples, which will increase confidence in the resonance assignments.
Materials and Methods
Expression, specific labeling, and purification of IκBα
IκBα(67–287)C186P/A220P [pET11a vector (Novagen)] was transformed into E. coli BL21 (DE3) cells. Unlabeled protein expression was done in M9 minimal medium as previously described (Croy et al. 2004). Specific 15N-amino acid growths (1X) were grown in M9 minimal medium with the 15N-labeled amino acid and all other 14N amino acids added to the medium in proportion to their abundance in the amino acid sequence to a final total amount of 1g/L culture. For 10X growths, the amount of the 14N amino acids was increased 10-fold. The protein was expressed and purified as previously described, except that induction was for 16 h at 18°C (Croy et al. 2004).
Proteolysis and mass spectrometry
Unlabeled or 15N-Leu IκBαCP/AP (13 μM in 50 μL) in 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM DTT was diluted 10-fold with 0.1% trifluoroacetic acid (final pH 2.2). IκBαCP/AP (150 μL) was digested with 25 μL immobilized pepsin (Pierce) on ice for 5 min, with vortexing every 0.5 min. Immobilized pepsin was pelleted by centrifugation for 5 s at 16,100g. Aliquots (10 μL) of digested IκBαCP/AP (supernatant) were analyzed or stored at –80°C. Digested IκBαCP/AP was mixed with an equal volume of α-cyano-4-hydroxycinnamic acid (Agilent Technologies) and spotted on MALDI target. As described, this procedure used 10 nmol of protein; however, if the initial sample volume is reduced to 15 μL, the procedure requires only 3 nmol of protein.
Two MS spectra (5000 shots) were collected for each sample on a 4800 TOF/TOF MALDI mass spectrometer (Applied Biosystems) using single-shot protection. Laser power was selected for each sample to give maximal intensity with no shots suppressed. Peptic IκBαCP/AP peptides were previously sequenced by MS/MS (Ferreiro et al. 2007). MS/MS spectra (5000 shots, resolution 200) were collected, with the tallest peak in the MS peptide envelope selected as the precursor and a maximum of two spectra collected using a single spot.
Data analysis
The extent of 15N incorporation in each peptide or fragment of interest in the MS or MS/MS spectra, respectively, was calculated using DEX (www.sdsc.edu/CCMS/DEX/) (Hotchko et al. 2006) (see Supplemental material for detailed protocol). MS and MS/MS spectra were exported as ASCII files using Data Explorer (Applied Biosystems). A list of peptides was generated for MS spectral analysis. A “fragment list” for MS/MS spectral analysis of y-ions with no potential overlaps from other ion types was generated using MS-Product in Protein Prospector (http://prospector.ucsf.edu). Profiles for the natural isotopic abundance in each peptide or fragment were generated using the isotopic-fast-profiles script (0% deuterium, mode 1). DEX-master-script, using the resulting profiles, analyzed the isotope incorporation. The centroid program in the HD-analysis-script output the weights for 15N incorporation, where consecutive weights >10 were considered significant. Only peptides or fragments that showed no 15N incorporation in the analysis of 14N-Leu proteins were quantified.
Acknowledgments
S.M.E.T. was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute. C.F.C. was supported by a Ruth L. Kirschstein NRSA Predoctoral Fellowship Award F31 GM81897. Research funding was provided by NIH grant GM071862.
Footnotes
Fragmentation at the C–N peptide bond results in b- and y-ions, which retain the N- or C-terminal amino acids, respectively. The number of amino acids in the resulting fragment is denoted as a subscript, starting with the N- or C terminus for b- or y-ions, respectively.
Supplemental material: see www.proteinscience.org
Reprint requests to: Elizabeth A. Komives, Department of Chemistry and Biochemistry, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA; e-mail: ekomives@ucsd.edu; fax: (858) 534-6174.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036418.108.
References
- Croy, C.H., Bergqvist, S., Huxford, T., Ghosh, G., Komives, E.A. Biophysical characterization of the free IκBα ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etezady-Esfarjani, T., Hiller, S., Villalba, C., Wüthrich, K. Cell-free protein synthesis of perdeuterated proteins for NMR studies. J. Biomol. NMR. 2007;39:229–238. doi: 10.1007/s10858-007-9188-0. [DOI] [PubMed] [Google Scholar]
- Ferreiro, D.U., Cervantes, C.F., Truhlar, S.M., Cho, S.S., Wolynes, P.G., Komives, E.A. Stabilizing IκBα by “consensus” design. J. Mol. Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffey, R.H., Redfield, A.G., Loomis, R.E., Dahlquist, F.W. Nuclear magnetic resonance observation and dynamics of specific amide protons in T4 lysozyme. Biochemistry. 1985;24:817–822. doi: 10.1021/bi00325a001. [DOI] [PubMed] [Google Scholar]
- Hotchko, M., Anand, G.S., Komives, E.A., Ten Eyck, L.F. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15:583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky, K.F. Peptide sequence analysis. Methods Enzymol. 2005;402:209–244. doi: 10.1016/S0076-6879(05)02007-0. [DOI] [PubMed] [Google Scholar]
- Ramesh, V., Frederick, R.O., Syed, S.E., Gibson, C.F., Yang, J.C., Roberts, G.C. The interactions of Escherichia coli trp repressor with tryptophan and with an operator oligonucleotide. NMR studies using selectively 15N-labelled protein. Eur. J. Biochem. 1994;225:601–608. doi: 10.1111/j.1432-1033.1994.00601.x. [DOI] [PubMed] [Google Scholar]
- Roth, S.M., Schneider, D.M., Strobel, L.A., Van Berkum, M.F., Means, A.R., Wand, A.J. Characterization of the secondary structure of calmodulin in complex with a calmodulin-binding domain peptide. Biochemistry. 1992;31:1443–1451. doi: 10.1021/bi00120a022. [DOI] [PubMed] [Google Scholar]
- Torchia, D.A., Sparks, S.W., Bax, A. NMR signal assignments of amide protons in the α-helical domains of staphylococcal nuclease. Biochemistry. 1988;27:5135–5141. doi: 10.1021/bi00414a028. [DOI] [PubMed] [Google Scholar]
- Torchia, D.A., Sparks, S.W., Bax, A. Staphylococcal nuclease: Sequential assignments and solution structure. Biochemistry. 1989;28:5509–5524. doi: 10.1021/bi00439a028. [DOI] [PubMed] [Google Scholar]
- Truhlar, S.M., Torpey, J.W., Komives, E.A. Regions of IκBα that are critical for its inhibition of NF-κB.DNA interaction fold upon binding to NF-κB. Proc. Natl. Acad. Sci. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh, D.S. Genetic tools for selective labeling of proteins with α-15N-amino acids. J. Biomol. NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- Yamazaki, T., Yoshida, M., Kanaya, S., Nakamura, H., Nagayama, K. Assignments of backbone 1H, 13C, and 15N resonances and secondary structure of ribonuclease H from Escherichia coli by heteronuclear three-dimensional NMR spectroscopy. Biochemistry. 1991;30:6036–6047. doi: 10.1021/bi00238a030. [DOI] [PubMed] [Google Scholar]