Abstract
Protein aggregation is a hallmark of several neurodegenerative diseases and also of cataracts. The major proteins in the lens of the eye are crystallins, which accumulate throughout life and are extensively modified. Deamidation is the major modification in the lens during aging and cataracts. Among the crystallins, the βA3-subunit has been found to have multiple sites of deamidation associated with the insoluble proteins in vivo. Several sites were predicted to be exposed on the surface of βA3 and were investigated in this study. Deamidation was mimicked by site-directed mutagenesis at Q42 and N54 on the N-terminal domain, N133 and N155 on the C-terminal domain, and N120 in the peptide connecting the domains. Deamidation altered the tertiary structure without disrupting the secondary structure or the dimer formation of βA3. Deamidations in the C-terminal domain and in the connecting peptide decreased stability to a greater extent than deamidations in the N-terminal domain. Deamidation at N54 and N155 also disrupted the association with the βB1-subunit. Sedimentation velocity experiments integrated with high-resolution analysis detected soluble aggregates at 15%–20% in all deamidated proteins, but not in wild-type βA3. These aggregates had elevated frictional ratios, suggesting that they were elongated. The detection of aggregates in vitro strongly suggests that deamidation may contribute to protein aggregation in the lens. A potential mechanism may include decreased stability and/or altered interactions with other β-subunits. Understanding the role of deamidation in the long-lived crystallins has important implications in other aggregation diseases.
Keywords: lens, βA3-crystallin, deamidation, protein aggregation, cataract, sedimentation velocity
Cataract is the most common cause of preventable blindness in the world (Resnikoff et al. 2004). Age-related cataract is a protein aggregation disease associated with the insolubization of modified proteins in the lens (Harrington et al. 2004). The lens contains a high concentration of proteins that are mostly structural proteins called crystallins. To enable transparency, crystallins form hetero-oligomers that assemble into ordered structures (Delaye and Tardieu 1983). Modifications that disrupt this order most likely contribute to aggregation and precipitation.
The proteins from older lenses contain many modifications including truncation, methylation, oxidation, disulfide bond formation, glycation, racemization, and deamidation (Groenen et al. 1993; Fujii et al. 1994; Ahmed et al. 1997; Lampi et al. 1998; Takemoto and Boyle 1998; Hanson et al. 2000; Wilmarth et al. 2006). The large accumulation of post-translationally modified crystallins occurs during normal aging because of the low turnover of proteins in the lens. Deamidation is the most abundant modification in the lens accounting for >60% of the total reported modifications, and was the only modification increased in the insoluble proteins (Wilmarth et al. 2006).
Nonenzymatic deamidation is initiated by formation of an imide intermediate between the terminal amide and the carboxyl group of Asn or Gln residues. Rapid hydrolysis completes deamidation. The five-member succinimide ring of Asn forms more readily than the six-member ring of Gln (Robinson and Robinson 2004a). Because of the much slower rate of deamidation of Gln, the Asn content can dictate the overall levels of deamidation in a protein (Robinson 1974). Deamidation can also occur enzymatically. Enzymatic deamidation by transglutaminase leads to deamidation of several Gln on the extensions of βA3 (Boros et al. 2008). There is no known enzyme for deamidation of Asn.
Every deamidation site has a preprogrammed rate of deamidation based on the neighboring amino acids. Rapid hydrolysis completes deamidation, resulting in a negative charge at asparaginyl and glutaminyl residues, which can induce structural changes. The deamidation rates of all asparagine and glutamine amides in a large number of peptides have been determined (Robinson and Robinson 2004a) and fit well with the reported in vivo data. For example, in vivo deamidation at N120 in the linker region of βA3-crystallin was estimated at 70% (Tsur et al. 2005). This site has a very rapid in vitro rate of deamidation on the order of days.
In vivo deamidation levels in the long-lived crystallins from adult lenses are actually lower than would be predicted from the in vitro peptide data, indicating suppression by the higher ordered structure (Robinson and Robinson 2001). The high protein concentrations and tight packing in the lens appear to be molecular determinants of deamidation. Environmental factors, such as changes in pH, also influence deamidation (Robinson and Robinson 2004b). Limited data is available regarding in vivo rates of deamidation throughout life (Dasari et al. 2007; Hains and Truscott 2007), but when compared with rates in peptides, would provide information regarding the contribution of higher ordered structure to deamidation.
Deamidation introduces a negative charge at asparaginyl and glutaminyl residues, which may induce structural changes. These structural changes may lead to exposure of new residues, making additional sites susceptible to deamidation, as has been shown for ribonuclease A in vitro (Zabrouskov et al. 2006). However, since deamidation can occur even under mild conditions in vitro (Nonaka et al. 2008), the use of high pH and high temperature may artifactually introduce deamidation. The purpose of the experiments in this study was to determine which physiological deamidation sites have the greatest affect on structure and stability as a first step in determining the overall relevance of deamidation during aging and cataracts.
Of the crystallins, the α-crystallins, which also have properties of chaperones, are a major species in the insoluble proteins, along with several of the β- and γ-crystallins, including βA3, βB1, and γS (Hanson et al. 2000; David et al. 2005; Harrington et al. 2007). βA3 and βA1, derived from an alternate start codon, comprise ∼8% of the total crystallins in the young lens (Robinson et al. 2006) and were found in significant amounts by spectral counting in the insoluble proteins of aged and cataractous lenses (David et al. 2005; Wilmarth et al. 2006).
βA3 is extensively deamidated during aging and cataracts (Lampi et al. 1998; Zhang et al. 2003) and the extent of deamidation increases in the insoluble proteins (Wilmarth et al. 2006; Dasari et al. 2007). Deamidations most associated with the insoluble proteins appear to be preferentially located on the surface (Lapko et al. 2002; Wilmarth et al. 2006). Therefore, deamidations on the surface of βA3 were investigated in this study. Several of these sites were also deamidated at homologous sites in other β-subunits, making these sites potential “hot spots” for deamidation (Wilmarth et al. 2006). The sites to investigate were further chosen because they were reported to be extensively deamidated at 10%–35% in vivo.
Surface deamidations on βA3-crystallin investigated in this study were predicted to be on the N-terminal domain (Q42, N54) or the C-terminal domain (N133, N155). We also investigated deamidation in the linker region (N120), as we had previously reported deamidation in the βB1-linker altered oligomerization (Harms et al. 2004). We found deamidation on the surface of βA3 led to aggregation and that specific sites were associated with destabilization or altered oligomer associations. Our results support the hypothesis that the accumulation of extensively deamidated β-crystallins during one's life contributes to insolubilization of crystallins. These results have implications in other aggregation diseases, where deamidation may also play a role (Watanabe et al. 1999).
Results
The effect of surface deamidations on the structure of human βA3-crystallin
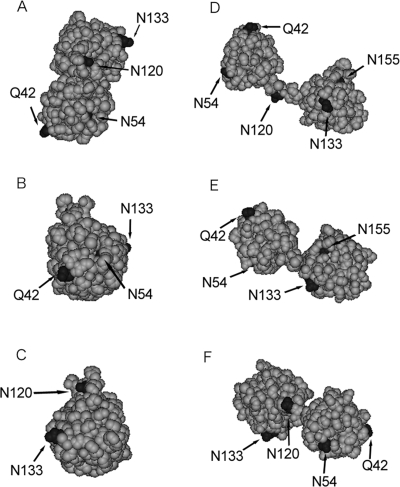
Since the crystal structure for βA3 is not known, homology modeling was performed to predict the effect of specific deamidations on βA3 structure. βA3 was modeled to the closed monomer of βB1 (Fig. 1A–C) or to the open monomer of βB2 (Fig. 1D–F) (Bax et al. 1990; Sergeev et al. 2000; Van Montfort et al. 2003). In both models, Q42, N120, and N133 were visible and exposed. Asn54 was partially buried in the closed model (visible in Fig. 1A,B) and exposed in the open model (visible in Fig. 1D–F). Asn155 was partially buried in the open model (visible in Fig. 1D,E) and buried in the closed model (not visible in Fig. 1A–C).
Figure 1.
Three-dimensional models of βA3-crystallin. Closed monomer structure (A–C), based on βB1-crystallin (PDB:1OKI). Open monomer structure (D–F), based on βB2-crystallin (PDB: 1BLB). Top view of models (top panels), right side (middle), and left side (bottom). Positions of deamidation sites denoted in black.
Wild-type (WT) βA3 and the five mutants in Figure 1 were expressed in the soluble proteins of Escherichia coli. After purification, only a single protein band was observed on SDS-PAGE, indicating high purity (data not shown). Site-directed mutagenesis was confirmed by digestion with trypsin or with glu-C for N155D, followed by mass spectrometry. Only peptide fragments belonging to βA3 were detected in significant amounts by mass spectrometry, further indicating high purity of the proteins.
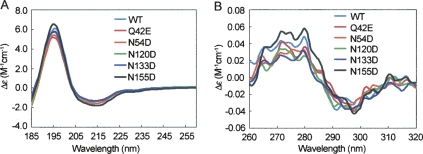
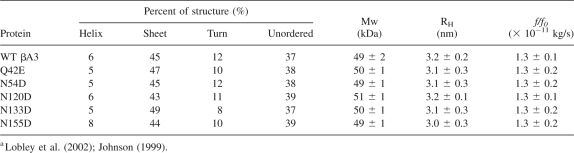
To examine the effect of deamidation on βA3 structure, mutants were analyzed by far- or near-UV circular dichroism (CD). WT βA3 and βA3 deamidated mutants showed typical β-strand-rich structures, characteristic of β-crystallins, with strong maxima at 195 nm and minima at 215 nm in the far-UV range (Fig. 2A). All proteins folded with the expected secondary conformation in vitro, and deamidation did not alter secondary structure. The predicted β-sheet content of 43%–49% and helical content of 5%–8% was similar to previously reported structures (Takata et al. 2007; Table 1). In contrast, the near-UV CD spectra of the five mutants differed from WT, particularly in the heights of the spectral bands between 290 and 260 nm, indicating differences in tertiary structure (Fig. 2B).
Figure 2.
Far-UV CD (A) and near-UV CD spectra (B) of WT, Q42E, N54D, N120D, N133D, and N155D βA3-crystallins.
Table 1.
Secondary structure prediction by the variable selection method, molar mass, and size of recombinant βA3-crystallinsa
To investigate the dimerization, homogeneity, and the overall shape of βA3, size-exclusion chromatography in line with multiangle light scattering (SEC-MALS) and analytical ultracentrifugation (AUC) analyses were performed. The weight-average molar masses derived from light-scattering experiments were ∼51 ± 3 kDa for all proteins, at all concentrations between 1.0 and 10 mg/mL (Table 1). This closely matches the size of a dimer at 50,300 Da without the N-terminal acetylation (Lampi et al. 1997). Each protein eluted in a single symmetrical peak with a 5 kDa difference in molar masses across the peak, indicating little polydispersity of the samples.
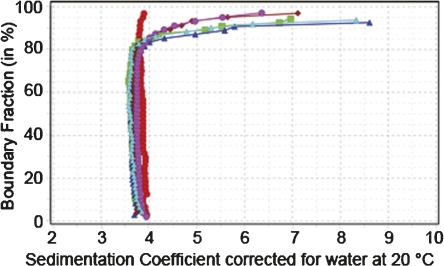
Sedimentation analysis showed coefficients between 3.6 and 4.0 (Fig. 3). The van Holde-Weischet integral distributions indicated a homogeneous composition of WT βA3 sedimenting at 4 s, while all mutants sedimented slightly slower at around 3.9 s and showed the presence of about 15%–20% aggregation.
Figure 3.
Enhanced van Holde-Weischet integral distribution plots of WT βA3 (red circles), N54D (green squares), N133D (blue triangles), N120D (maroon diamonds), N155D (magenta circles), and Q42E (cyan triangles).
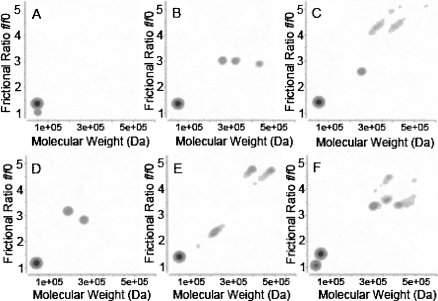
Two-dimensional spectrum analysis and genetic algorithm analysis of the distribution of sedimentation coefficients (Brookes and Demeler 2006, 2007; Brookes et al. 2006) revealed a major sedimenting species with Mw in excellent agreement with the βA3 dimer Mw, and frictional ratios, f/f o, between 1.4 and 1.5 for the dimer species for all proteins (Fig. 4). In addition, we detected that deamidation led to several minor species with significantly elevated frictional ratios, suggesting aggregation. The aggregates were dispersed in size, ranging from 200 to 700 kDa and had frictional coefficient ratios, suggesting a slightly nonglobular shape, consistent with an elongated protein.
Figure 4.
Genetic algorithm/Monte Carlo analysis of WT βA3 and mutants, WT βA3 (A), N155D (B), Q42E (C), N133D (D), N54D (E), N120D (F).
The effect of deamidations on the stability of human βA3-crystallin in urea
The relative stabilities of the deamidated mutants were compared with WT βA3 during unfolding in urea. Protein unfolding and refolding were detected by measuring fluorescence emission dominated by nine tryptophan residues in βA3, of which only four are buried (Bateman et al. 2003). The emission spectra for the βA3 mutants were indistinguishable from WT in the absence of urea (data not shown), supporting the above CD results that deamidation did not significantly alter the overall structure of βA3.
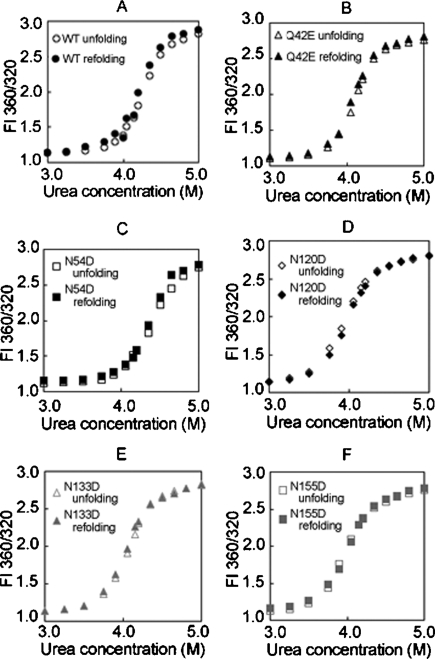
Unfolding of WT βA3 and all deamidated mutants in urea showed high cooperativity, without an observable transition in the unfolding curve that would indicate an intermediate (Fig. 5), as was apparent for other β-crystallins (Koteiche et al. 2007). Unfolding was reversible down to 2 M urea for all proteins. Refolding proceeded along a similar path to that observed during unfolding. At urea concentrations <2 M, a slight increase in fluorescence intensity was observed, due to light-scattering aggregates, indicating incomplete refolding.
Figure 5.
Unfolding and refolding of βA3-crystallins in urea, WT (A), Q42E (B), N54D (C), N120D (D), N133D (E), and N155D (F).
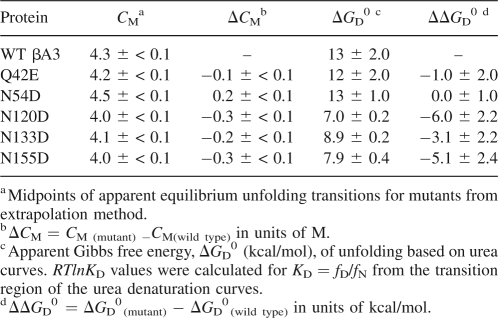
Without a visible intermediate in the unfolding curves, the data were fit assuming two states, native and denatured. The midpoints of transition curves, Cm, were used to rank stability. The Cm for WT βA3 was observed at 4.3 M urea (Table 2). Deamidation at N120, N133, and N155 shifted unfolding to the left, and at Q42, shifted unfolding to the left only in the second part of the curve. This implied deamidation at Q42 had a greater destabilization effect on the intermediate to the unfolded protein, than on the native to the intermediate. Deamidation at N54 did not alter stability in urea.
Table 2.
Equilibrium apparent unfolding free energy for WT, Q42E, N54D, N120D, N133D, and N155D of βA3-crystallin
Apparent free energies were approximated for the unfolding and refolding curves in Figure 5 (Table 2). The relative apparent ΔGD 0's and the unfolding curves of all deamidated mutants were lower than for WT βA3 and N54D. Similar results were obtained at either excitation at 283 or 293 nm. Relative stabilities from both the concentration of the midpoint of the unfolding curve and the apparent free energies were from most stable to least; N54D ≥ WT > Q42E > N133D > N155D ≥ N120D.
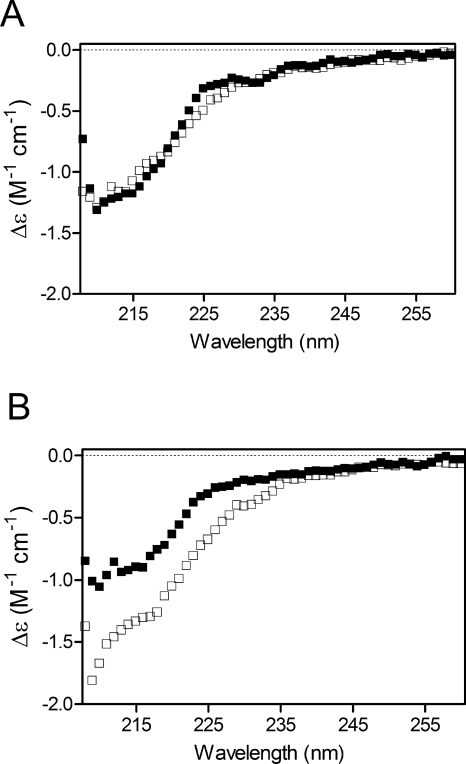
For the mutant most destabilized by deamidation, N120D, unfolding and refolding were also monitored by far-UV CD and compared with WT. In Figure 6, spectra are shown for proteins in 3.6 M urea and demonstrate the overlapping spectra for unfolding/refolding of WT (Fig. 6A), but not for N120D (Fig. 6B). At higher urea concentrations unfolding/refolding spectra were similar for both proteins. The refolding spectrum of N120D at 3.6 M urea (Fig. 6, open squares) has a greater signal than the unfolding spectrum (Fig. 6, closed squares). This difference between unfolding and refolding of N120D suggests the presence of an aggregate or a nonnative intermediate.
Figure 6.
Stability of WT βA3 and N120D in 3.6 M urea measured by CD. Unfolding (■) and refolding (□) of (A) WT and (B) N120D.
The effect of deamidations on the stability of human βA3-crystallin during heating
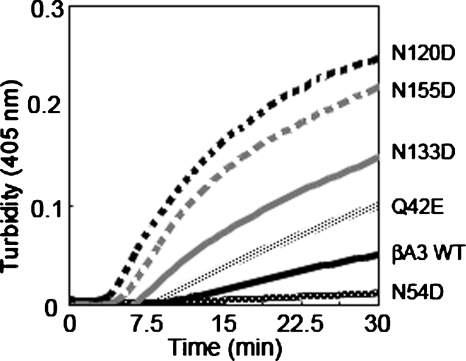
WT βA3 and deamidated mutants of βA3 were heated to 55°C, and the change in turbidity measured at 405 nm (Fig. 7; Takata et al. 2007). All deamidated βA3 mutants showed greater turbidity, except for N54D. Deamidation in the linker region and on the C-terminal domain (N120D, N133D, and N155D) led to a rapid increase in turbidity within 30 min, after which samples started to precipitate.
Figure 7.
Turbidity at 405 nm during heating at 55°C for WT, Q42E, N54D, N120D, N133D, N155D βA3-crystallins.
The effects of deamidation on the interactions of βA3-crystallin with βB1-crystallin
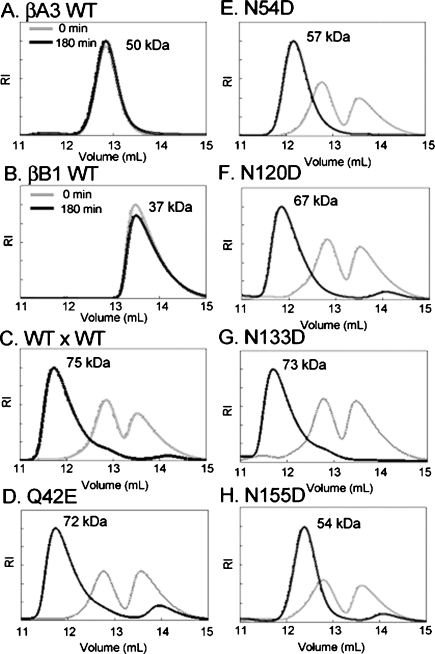
βA3- and βB1-crystallins form hetero-oligomers (Bateman et al. 2003; Liu and Liang. 2007; Dolinska et al. 2008). The βHigh fraction contains all of the β-subunits in approximately equal amounts (Lampi et al. 1998). Therefore, equal amounts of WT βB1 and βA3-subunits were mixed at 37°C. Hetero-oligomers were resolved from homodimers by SEC-MALS.
The molar masses of WT βA3 and the βA3 mutants at 0.5 mg/mL averaged 50 kDa, suggesting dimer formation (Fig. 8A). The molar mass of βB1 at 0.5 mg/mL ranged from 60.1 ± 0.2 kDa to 27.3 ± 0.1 kDa across the peak, with 75% ≤ 35 kDa (Fig. 8B), suggesting mixed monomer dimers (Lampi et al. 2001). After 180 min incubation of both proteins together, a single peak eluted with a greater molar mass than for either βB1 or βA3 (Fig. 8C). The molar masses across this peak ranged from 80 to 50 kDa at the tailing edge, suggesting a mixture of βB1:βA3 hetero-oligomers. This is in agreement with reports of a tetramer-dimer equilibrium of βA3:truncated βB1 by Bateman et al. (2003) at similar concentrations.
Figure 8.
Size exclusion chromatograms of βB1–βA3 hetero-oligomers after 180 min incubation at 37°C. WT βA3 (A), WT βB1 (B). Equal molar amounts of βB1 mixed with βA3 WT (C), Q42E (D), N54D (E), N120D (F), N133D (G), or N155D (H).
Deamidation at Q42, N120, or N133 did not alter the molar masses of the hetero-oligomers formed with WT βB1 (Fig. 8D,F,G). In contrast, deamidation at N54 or N155 led to hetero-dimers with WT βB1 of 54–57 kDa (Fig. 8E,H). The peaks were symmetrical with similar molar masses across the peak, suggesting a single species with Mw of dimers. The rates of hetero-oligomer formation differed, with the hetero-oligomer of N120D or N155D with WT βB1 being the major species within 30–60 min compared with 90–180 min for all other βA3s.
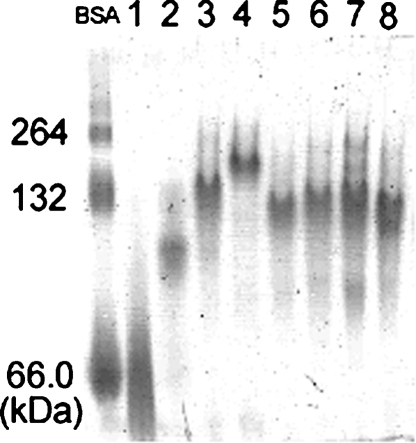
The composition of the peaks was confirmed by electrophoresis. The presence of both WT βB1 and WT βA3 or a deamidated βA3-subunit in the early eluting peak was confirmed by SDS-PAGE (data not shown). Protein fractions under the peaks were subjected to Blue Native PAGE and a single band confirmed formation of hetero-oligomers (Fig. 9). However, the hetero-oligomers resolved as broad bands, similar to WT βB1 alone, suggesting a possible mixture of different size oligomers. Mixtures of βB1 with βA3-subunits resolved at higher molecular weights than the BSA dimer of 132 kDa. This suggested a higher Mw than the BSA standard, and would be in disagreement with the Mw obtained from MALS above. This difference in Mw more likely reflects an unusual migration of the β-crystallins compared to the globular BSA during electrophoresis.
Figure 9.
Blue Native PAGE of proteins from size exclusion chromatography in Figure 8. Native albumin standard (left), WT βB1 (lane 1), WT βA3 (lane 2), WT βA3:WT βB1 (lane 3), WT βB1:Q42E (lane 4), WT βB1:N54D (lane 5), WT βB1:N120D (lane 6), WT βB1:N133D (lane 7), and WT βB1:N155D (lane 8).
Discussion
Deamidation is directly associated with crystallin aggregation
The major finding of this study was that deamidations associated with insolubilization in vivo led to aggregation in vitro. Our findings strongly suggest that deamidation may directly contribute to insolubilization of βA3 in the aged and cataractous lens.
Our use of high-resolution finite element models integrated with supercomputing allowed for the accurate detection of different sedimenting species and their frictional ratios by sedimentation velocity. The aggregates detected by these methods were not detected by classical light-scattering methods. The wide range of molecular weight species predicted from sedimentation velocity would have eluted in multiple fractions during chromatography and may have been below the concentration detection of the MALS instrument. In the compact environment of the lens, even a low level of aggregation may disrupt the tight packing of crystallins and scatter light.
Aggregation detected in this study is especially significant in light of the overwhelming amount of deamidation in the lens, and in particular, βA3, during normal aging and cataract formation (Lampi et al. 1998; Wilmarth et al. 2006). The accumulation of multiple deamidations may further increase the level of soluble aggregates in vivo and lead to insolubilization. For example, at least eight sites of deamidation in βA3, of which five were investigated here, were greater in the insoluble proteins than the soluble proteins of aged and cataractous lenses (Wilmarth et al. 2006). Investigating the effect of deamidation at individual sites is the first step in elucidating the role of deamidation in the lens.
While our data suggests that site-specific deamidations induce soluble aggregation, multiple modifications are most likely needed for insolubilization. By altering protein structure, deamidation may expose buried residues that are then susceptible to further deamidations, or other modifications such as oxidation or proteolysis. In support of this, deamidated βA3 fragments have been selectively associated with the insoluble proteins from cataractous lenses (Harrington et al. 2004).
Deamidation-induced aggregation may have significance in other diseases as well. In Alzheimer's, deamidation was identified in τ protein isolated from paired helical filaments from Alzheimer's diseased brain tissue (Watanabe et al. 1999). Deamidation may also play a role in tissue destruction during diabetes. Only 5% of deamidated amylin peptides in vitro were enough to mimic amyloid aggregate formation, similar to the amylin aggregates in the pancreas of patients with Type 2 diabetes mellitus (Nilsson et al. 2002). These studies suggest that low levels of deamidation may trigger aggregation and be cytotoxic.
Deamidation decreased protein stability and may decrease stability in vivo
A potential mechanism for deamidation-induced aggregation may be destabilization. We and others have previously reported that βA3 is prone to associate and aggregate upon perturbation of its structure by urea, heating, or low pH, even in the presence of DTT (Bateman et al. 2003; Takata et al. 2007). Deamidations at the interface between domains further decreased stability in βA3 (Takata et al. 2007), as has also been reported for γD (Flaugh et al. 2006), with a greater effect observed with multiple deamidations (Lampi et al. 2006; Takata et al. 2007). In this study, we showed that deamidation on the surface, except for at N54, also decreased stability of βA3.
It is not readily apparent why deamidation at N54 led to aggregation but not to decreased stability. We have previously reported that deamidation at N157 in βB1, located on an outside surface loop, also increased stability (Harms et al. 2004). Specific effects of deamidation may involve different interactions with nearby residues or solvent.
The effects of deamidation on stability are dependent on the site of deamidation. Deamidations in the C-terminal domain at N133 and N155 decreased βA3 stability more than deamidations in the N-terminal domain at Q42 and N54, regardless of the degree of exposure of the residue. For example, N54 and N155 are both buried or partially buried (Fig. 1), but deamidation at N54 increased stability, while deamidation at N155 decreased stability. Our results are consistent with reports that the C-terminal domain is less stable than the N-terminal domain (Gupta et al. 2006).
Surface deamidations shifted the unfolding of βA3 to lower urea concentrations equally from native to unfolded, except for deamidation at Q42 in the N-terminal domain, which affected the latter part of the curve. This indicated that the transition from an intermediate to the unfolded state was destabilized, consistent with the N-terminal domain unfolding after the less stable C-terminal domain (Gupta et al. 2006). Because a distinct transition in the unfolding curve was not apparent by fluorescence, the data were not fit to a three-state transition, but instead, a two-state transition was approximated. The difference in apparent free energy between the least stable mutants, N120D and N155D, and WT was 5–6 kcal/mol. This accounts for possible disruption of one to two hydrogen bonds.
When CD was used to monitor the unfolding of deamidated βA3, differences between unfolding and refolding were detected, suggesting a possible aggregate or nonnative intermediate. The nature of this intermediate appears to have an increased content of β-strand, as has previously been observed for a deletion mutant of βA3 (Reddy et al. 2004). We did not observe a similar effect upon refolding of WT. These results suggest that deamidation may increase the tendency of βA3 to unfold and to aggregate.
Deamidation altered subunit–subunit interactions
Modified β-crystallins are prone to associate (Zhang et al. 2003). Therefore, a potential mechanism that was explored in this study was deamidation on the surface of βA3, altering interactions with other β-crystallin subunits. Deamidation at N54 or N155 disrupted hetero-oligomer formation, resulting in hetero-dimers only. Because reducing agent was present during oligomer formation, it is not expected that disulfide bonds would have led to dimer formation. Complex hetero-oligomers of β-crystallin subunits are isolated from lens tissue, reflecting the complexity of these interactions in vivo (Lampi et al. 1998). Deamidation at specific sites may disrupt these in vivo interactions and is a potential mechanism for contributing to aggregation.
Conclusion
While other modifications occur during normal aging of the lens, deamidation appears to be the most extensive (Lampi et al. 1998; Wilmarth et al. 2006; Hains and Truscott 2007). Yet, the role of deamidation in insolubilization of lens crystallins and cataract formation has remained speculative. The detection of deamidated βA3-crystallin aggregates strongly suggests that deamidation may initiate formation of light-scattering aggregates in the lens. It is noteworthy that the formation of aggregates was detected prior to precipitation induced by perturbing in heat or urea and may be a precondition for cataracts.
The accumulation during one's life of extensively deamidated crystallins at the levels reported in vivo may trigger crystallin aggregation, leading to precipitation. A potential mechanism for aggregate formation due to deamidation is a decreased stability and altered interactions between β-subunits.
Understanding the role of deamidation in the crystallins has important implications in other aggregation diseases. The lens is, in many ways, a long-term incubator for studying the role of protein modifications on protein aggregation. Future studies are needed to identify the nature of the aggregate and to determine whether it is possible to prevent this aggregate before precipitation.
Materials and Methods
Mutagenesis, expression, and purification of recombinant proteins
Wild-type human βA3-crystallin (WT βA3) and βB1-crystallin (WT βB1) were recombinantly expressed in E. coli as previously described (Lampi et al. 2001; Takata et al. 2007). cDNA was translated from RNA from lenses <1 yr of age obtained anonymously from human donors (Lions Eye Bank of Oregon) (Lampi et al. 2001). In vivo deamidation sites in βA3 were mimicked by replacing glutamine 42 with glutamic acid and asparagines 54, 120, 133, and 155 with aspartic acid by site-directed mutagenesis using internal primers containing the desired mutation (QuikChange Mutagenesis kit, Stratagene). The mutation was introduced into the WT βA3 sequence in the pCR-Blunt II-TOPO vector and then subcloned into the pET-3a vector (Novagen). Clones containing the correct insert were confirmed by sequencing (Nevada Genomics Center).
Recombinant βA3 and βB1 were purified by ion exchange chromatography as reported previously (Lampi et al. 2001; Takata et al. 2007). Purity of the proteins was checked by SDS-PAGE. Proteins were digested with trypsin or glu-C followed by mass spectrometry (LTQ, ThermoFinnigan), to confirm the deamidation sites.
CD spectroscopy
CD measurements in the near- and far-UV ranges were performed using a JASCO J-810 spectropolarimeter (JASCO). Samples were exhaustively dialyzed with buffer (pH 6.8) containing 5 mM NaH2PO4, 5 mM Na2HPO4, and 100 mM NaF. Measurements were performed in a 1.0-cm cell for near-UV and in a 0.1-cm cell for far-UV at 4°C as previously described (Takata et al. 2007). Protein concentrations were 0.10–0.15 mg/mL for far-UV CD and 0.2–0.3 mg/mL for near-UV CD. Concentrations of all mutants were determined by amino acid analysis (Molecular Structure Facility, UC Davis). Spectra were processed using CDtool from Birkbeck College, London (Lees et al. 2004). Percent secondary structure was calculated using software available at the DICHROWEB website located at http://www.cryst.bbk.ac.uk/cdweb/html/home.html, which is part of the BBSRC Center for Protein and Membrane Structure and Dynamics (Lobley et al. 2002; Whitmore and Wallace 2004). A modification of the variable selection method, CDSSTR, was used to analyze data from 270 to 175 nm (Johnson 1999). Experiments were performed twice.
Association of expressed proteins
Multiangle laser light scattering (MALS) (DAWN HELEOS, Wyatt Technology) and Quasi-elastic laser light scattering (QELS, Wyatt) in line with size-exclusion chromatography (SEC) was performed to determine the association state of βA3 mutants. Samples were concentrated to between 1.0 and 10 mg/mL as previously described (Takata et al. 2007). A 100-μL sample was injected onto a Sepharose 12 10/300 GL column (Amersham Biosciences) equilibrated in buffer (pH 6.8) containing 29 mM Na2HPO4, 29 mM NaH2PO4, 100 mM KCl, 1 mM EDTA, and 1 mM DTT with a flow rate of 0.4 mL/min.
In order to estimate the globularity of the protein, sedimentation velocity experiments were performed. All samples were exhaustively dialyzed into 5 mM NaH2PO4, 5 mM Na2HPO4 (pH 6.8), and 100 mM NaCl. The reducing agent, TCEP at 2 mM, was added to all samples. Sedimentation velocity experiments were performed at a rotor speed of 60,000 rpm, 20°C, and monitored by UV intensity measurements at 280 nm using a Beckman Coulter Optima XL-I (Beckman Coulter). Sedimentation data was analyzed with UltraScan 9.4 (http://www.ultrascan.uthscsa.edu, developed by Dr. B. Demeler at The University of Texas Health Science Center at San Antonio [UTHSCSA]). In order to assess composition, model independent s-value distributions were determined using the enhanced van Holde-Weischet method in the UltraScan software (Demeler and van Holde 2004). Molecular weight and frictional ratios were determined with two-dimensional spectrum analysis (Brookes et al. 2006) and the genetic algorithm optimization method (Brookes and Demeler 2006, 2007). Hydrodynamic corrections for buffer conditions were made according to data published by Laue et al. (1992) as implemented in UltraScan. Data were fitted on the UTHSCSA Bioinformatics Core Facility Linux cluster and on the Lonestar cluster at the Texas Advanced Computing Center.
Differential stability of deamidated proteins
Stability of deamidated mutant proteins was measured by fluorescence spectrometry during unfolding/refolding in urea. A stock solution of 7.64 M urea in phosphate buffer was prepared according to the methods of Pace (1986) and Takata et al. (2007). The buffer (pH 7.0) contained 50 mM Na2HPO4, 50 mM NaH2PO4, 5 mM DTT, and 2 mM EDTA. Proteins at 1 μM were incubated in urea for 24 h at 22°C. For refolding experiments, proteins were incubated in 7.5 M urea for 5 h, then diluted to the desired urea concentrations and incubated for a total of 24 h.
Fluorescence intensities were measured on a Photon Technology International QM-2000-7 spectrometer using the manufacture's software, FeliX (Photon Technology International). Emission spectra were recorded between 300 and 400 nm with an excitation wavelength at 283 nm. Slit widths were set to 2 nm. Emission spectra were corrected for the buffer signal. The extent of unfolded protein was calculated from the normalized intensities (FI 360/320). Relative stabilities were determined in this study by fitting the data to a two-state model and measuring the denaturant concentration at the transition midpoint, CM, and approximating the apparent free energy, apparent ΔGD (Lampi et al. 2006).
Far-UV CD was also used to measure unfolding and refolding of WT and N120D at 6 μM in increasing urea concentrations. Experiments were performed as described above for CD, except 100 mM NaF was replaced with 100 mM NaCl. Samples were prepared as described above for fluorescence intensity measurements.
Heat stability
Heat-induced precipitation of βA3 was measured in a thermal-jacketed cuvette with constant stirring (Cary 4 Bio UV-Visible spectrophotometer, Varian). Heat incubation was performed in the same buffer as the MALS experiments described above. All samples were concentrated to 0.5 mg/mL, and the tendency of the proteins to aggregate was monitored at 405 nm at 55°C for 180 min.
Interaction of βA3- and βB1-subunits
For hetero-oligomer detection, recombinant βB1 and βA3 mutants were premixed in equilibrating buffer at equal molar amounts for a final concentration of 1.0 mg/mL. Mixtures were incubated for 30, 60, 90, and 180 min at 37°C. The mixtures were filtered and injected onto the MALS-QELS-SEC system. The weight-averaged molar masses were calculated from the refractive index with software provided by the manufacturer (ASTRA V, Wyatt Technology) as previously described (Harms et al. 2004). Eluted fractions were visualized by SDS-PAGE.
Electrophoresis
The same samples from the interaction analysis above were mixed with Blue Native PAGE containing PFO-Na sample buffer (20 mM Tris-HCl, 20 mM Tris-Base, 0.5 mM EDTA, 15% [v/v] glycerol, 0.05% PFO-Na, 0.5% Coommassie blue [G-250] at pH 7.0). Blue Native PAGE and SDS-PAGE electrophoresis were performed as previously described with some modifications using precast, 1.0-mm thick 8 × 8 cm, polyacrylamide Nu-PAGE 10% Bis-Tris gels (Invitrogen) (Ramjeesingh et al. 1999; Yang et al. 2002). Blue Native PAGE was performed at 200 V at 4°C until the blue dye reached the bottom of the gel. Nondenatured albumin was used as a standard. Proteins were visualized by destaining in 20% methanol.
Molecular modeling
βA3 has previously been modeled as “opened” and “closed” monomers predicted to exist at low concentrations (pdb: 1BLB) (Sergeev et al. 2000). Homology modeling was used in the present experiments to predict the position of the deamidation sites in both conformations. The human βA3 sequence was modeled using either the bovine βB2 dimer structure (pdb: 1BLB) as an open dimer model or the human βB1-crystallin (pdb: 1OKI) as a close dimer model. The models were made and refined by the homology modeling server, 3D-jigsaw (Cancer Research,UK). The models were represented as monomer subunits in CPK model representation using DS ViewerPro software (Fig. 1) (Accelrys Inc.).
Protein concentration
Concentrations were calculated from UV absorbance at 280 nm with extinction coefficients for βA3 at 2.62 (mg/mL)−1cm−1 and for βB1 at 2.07 (mg/mL)−1cm−1. Amino acid analysis was also performed (Molecular Structure Facility, UC Davis).
Acknowledgments
We acknowledge Daniel Flannery, Jason Lampi, Alaina Phillips, Virgil Schirf, Lionel Trujillio, and Luke Woodbury for their expert technical help. We also acknowledge funding provided by the NIH/NEI R01-EY012239 (K.J.L.), NIH R01-RR022200 (B.D.), and Lori and Duane Stueckle Dean's Distinguished Professorship (J.T.O.), and the following core facilities: Mass Spectrometry at Oregon Health and Science University (NIH-EY 10572), Nevada Genomics Center (P20RR01 6463), Biomedical Research Center, Protein Structure Core at Boise State University (NIH/NCRR P20RR016454), and Texas Advanced Computing Center (NSF Teragrid Allocation grant TGMCB070038).
Footnotes
Reprint requests to: Kirsten J. Lampi, Department of Integrative Biosciences, School of Dentistry, Oregon Health and Science University, 611 Southwest Campus Drive, Portland, OR 97239-3098, USA; e-mail: lampik@ohsu.edu; fax: (503) 494-8554.
Abbreviations: BBSRC, Biotechnology and Biological Sciences Research Council; DTT, dithiothreitol; EDTA, ehtylendiaminetetraacetic acid; FI, fluorescence intensity; GuHCl, guanidine hydrochloride; MW, molecular weight; MWCO, molecular weight cut off; PDB, Protein Data Bank; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophroresis; TCEP, tris(2-carboxyethyl)phosphine.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035410.108.
References
- Ahmed, M.U., Brinkmann Frye, E., Degenhardt, T.P., Thorpe, S.R., Baynes, J.W. N-ε-(carboxyethyl)lysine, a product of the chemical modification of proteins by methylglyoxal, increases with age in human lens proteins. Biochem. J. 1997;324:565–570. doi: 10.1042/bj3240565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, O.A., Sarra, R., van Genesen, S.T., Kappe, G., Lubsen, N.H., Slingsby, C. The stability of human acidic β-crystallin oligomers and hetero-oligomers. Exp. Eye Res. 2003;77:409–422. doi: 10.1016/s0014-4835(03)00173-8. [DOI] [PubMed] [Google Scholar]
- Bax, B., Lapatto, R., Nalini, V., Driessen, H., Lindley, P.F., Mahadevan, D., Blundell, T.L., Slingsby, C. X-ray analysis of βB2-crystallin and evolution of oligomeric lens proteins. Nature. 1990;347:776–780. doi: 10.1038/347776a0. [DOI] [PubMed] [Google Scholar]
- Boros, S., Wilmarth, P.A., Kamps, B., de Jong, W.W., Bloemendal, H., Lampi, K., Boelens, W.C. Tissue transglutaminase catalyzes the deamidation of glutamines in lens βB2- and βB3-crystallins. Exp. Eye Res. 2008;86:383–393. doi: 10.1016/j.exer.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Brookes, E., Demeler, B. Genetic algorithm optimization for obtaining accurate molecular weight distributions from sedimentation velocity experiments. Analytical ultracentrifugation VIII. In: Wandrey C., Cölfen H., editors. Progress in colloid polymer science. Vol. 131. Springer; Berlin, Germany: 2006. pp. 78–82. [Google Scholar]
- Brookes, E., Demeler, B. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. Proceedings of the 9th annual conference on genetic and evolutionary computation, Genetic and Evolutionary Computation Conference; London, UK. 2007. [Google Scholar]
- Brookes, E., Boppana, R.V., Demeler, B. Computing large sparse multivariate optimization problems with an application in biophysics. Proceedings of the 9th annual conference on genetic and evolutionary computation, Genetic and Evolutionary Computation Conference; London, UK. 2006. [Google Scholar]
- Dasari, S., Wilmarth, P.A., Rustvold, D.L., Riviere, M.A., Nagalla, S.R., David, L.L. Reliable detection of deamidated peptides from lens crystallin proteins using changes in reversed-phase elution times and parent ion masses. J. Proteome Res. 2007;6:3819–3826. doi: 10.1021/pr070182x. [DOI] [PubMed] [Google Scholar]
- David, L.L., Wilmarth, P.A., Rustvold, D.L., Riviere, M.A. Association for Research in Vision and Ophthalmology; 2005. Global proteomic strategy to quantify oxidized cysteines in human nuclear cataract. www.arvo.org. [Google Scholar]
- Delaye, M., Tardieu, A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983;302:415–417. doi: 10.1038/302415a0. [DOI] [PubMed] [Google Scholar]
- Demeler, B., van Holde, K.E. Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Dolinska, M.B., Chan, M.P., Sergeev, Y.V., Wingfield, P.T., Hejtmancik, F.J. Association for Research in Vision and Ophthalmology; 2008. Association properties of βB1- and βA3-crystallins: Ability to form heterotetramers. Abstract 4099. www.arvo.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugh, S.L., Mills, I.A., King, J. Glutamine deamidation destabilizes human γD-crystallin and lowers the kinetic barrier to unfolding. J. Biol. Chem. 2006;281:30782–30793. doi: 10.1074/jbc.M603882200. [DOI] [PubMed] [Google Scholar]
- Fujii, N., Ishibashi, Y., Satoh, K., Fujino, M., Harada, K. Simultaneous racemization and isomerization at specific aspartic acid residues in αB-crystallin from the aged human lens. Biochim. Biophys. Acta. 1994;1204:157–163. doi: 10.1016/0167-4838(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Groenen, P.J., van Dongen, M.J., Voorter, C.E., Bloemendal, H., de Jong, W.W. Age-dependent deamidation of α B-crystallin. FEBS Lett. 1993;322:69–72. doi: 10.1016/0014-5793(93)81113-e. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Srivastava, K., Srivastava, O.P. Truncation of motifs III and IV in human lens βA3-crystallin destabilizes the structure. Biochemistry. 2006;45:9964–9978. doi: 10.1021/bi060499v. [DOI] [PubMed] [Google Scholar]
- Hains, P.G., Truscott, R.J. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J. Proteome Res. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- Hanson, S.R., Hasan, A., Smith, D.L., Smith, J.B. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- Harms, M.J., Wilmarth, P.A., Kapfer, D.M., Steel, E.A., David, L.L., Bachinger, H.P., Lampi, K.J. Laser light-scattering evidence for an altered association of β B1-crystallin deamidated in the connecting peptide. Protein Sci. 2004;13:678–686. doi: 10.1110/ps.03427504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, V., McCall, S., Huynh, S., Srivastava, K., Srivastava, O.P. Crystallins in water soluble-high molecular weight protein fractions and water insoluble protein fractions in aging and cataractous human lenses. Mol. Vis. 2004;10:476–489. [PubMed] [Google Scholar]
- Harrington, V., Srivastava, O.P., Kirk, M. Proteomic analysis of water insoluble proteins from normal and cataractous human lenses. Mol. Vis. 2007;14:1680–1694. [PubMed] [Google Scholar]
- Johnson, W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- Koteiche, H.A., Kumar, M.S., McHaourab, H.S. Analysis of βB1-crystallin unfolding equilibrium by spin and fluorescence labeling: Evidence of a dimeric intermediate. FEBS Lett. 2007;581:1933–1938. doi: 10.1016/j.febslet.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampi, K.J., Ma, Z., Shih, M., Shearer, T.R., Smith, J.B., Smith, D.L., David, L.L. Sequence analysis of βA3, βB3, and βA4 crystallins completes the identification of the major proteins in young human lens. J. Biol. Chem. 1997;272:2268–2275. doi: 10.1074/jbc.272.4.2268. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Ma, Z., Hanson, S.R., Azuma, M., Shih, M., Shearer, T.R., Smith, D.L., Smith, J.B., David, L.L. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp. Eye Res. 1998;67:31–43. doi: 10.1006/exer.1998.0481. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Oxford, J.T., Bachinger, H.P., Shearer, T.R., David, L.L., Kapfer, D.M. Deamidation of human β B1 alters the elongated structure of the dimer. Exp. Eye Res. 2001;72:279–288. doi: 10.1006/exer.2000.0950. [DOI] [PubMed] [Google Scholar]
- Lampi, K.J., Amyx, K.K., Ahmann, P., Steel, E.A. Deamidation in human lens βB2-crystallin destabilizes the dimer. Biochemistry. 2006;45:3146–3153. doi: 10.1021/bi052051k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko, V.N., Purkiss, A.G., Smith, D.L., Smith, J.B. Deamidation in human γ S-crystallin from cataractous lenses is influenced by surface exposure. Biochemistry. 2002;41:8638–8648. doi: 10.1021/bi015924t. [DOI] [PubMed] [Google Scholar]
- Laue, T.M., Shah, B.D., Ridgeway, T.M., Pelletier, S.L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S.E., Rowe A.J., Horton J.C., editors. Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- Lees, J.G., Smith, B.R., Wien, F., Miles, A.J., Wallace, B.A. CDtool-an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 2004;332:285–289. doi: 10.1016/j.ab.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Liu, B.F., Liang, J.J. Protein–protein interactions among human lens acidic and basic β-crystallins. FEBS Lett. 2007;581:3936–3942. doi: 10.1016/j.febslet.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobley, A., Whitmore, L., Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- Nilsson, M.R., Driscoll, M., Raleigh, D.P. Low levels of asparagines deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: Implications for the study of amyloid formation. Protein Sci. 2002;11:342–349. doi: 10.1110/ps.48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, Y., Aizawa, T., Akieda, D., Yasui, M., Watanabe, M., Watanabe, N., Tanaka, I., Kamiya, M., Mizuguchi, M., Demura, M., et al. Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions. Proteins. 2008;72:313–322. doi: 10.1002/prot.21927. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Ramjeesingh, M., Huan, L.J., Garami, E., Bear, C.E. Novel method for evaluation of the oligomeric structure of membrane proteins. Biochem. J. 1999;342:119–123. [PMC free article] [PubMed] [Google Scholar]
- Reddy, M.A., Bateman, O.A., Chakarova, C., Ferris, J., Berry, V., Lomas, E., Sarra, R., Smith, M.A., Moore, A.T., Bhattacharya, S.S., et al. Characterization of the G91del CRYBA1/3-crystallin protein: A cause of human inherited cataract. Hum. Mol. Genet. 2004;13:945–953. doi: 10.1093/hmg/ddh110. [DOI] [PubMed] [Google Scholar]
- Resnikoff, S., Pascolini, D., Etya'ale, D., Kocur, I., Pararajasegaram, R., Pokharel, G.P., Mariotti, S.P. Global data on visual impairment in the year 2002. Bull. World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- Robinson, A.B. Evolution and the distribution of glutaminyl and asparaginyl residues in proteins. Proc. Natl. Acad. Sci. 1974;71:885–888. doi: 10.1073/pnas.71.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.E., Robinson, A.B. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc. Natl. Acad. Sci. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.E., Robinson, A.B. Althouse Press; Cave Junction, OR: 2004a. Molecular clocks. Chapter 6; pp. 51–83. [Google Scholar]
- Robinson, N.E., Robinson, A.B. Prediction of primary structure deamidation rates of asparaginyl and glutaminyl peptides through steric and catalytic effects. J. Pept. Res. 2004b;63:437–448. doi: 10.1111/j.1399-3011.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- Robinson, N.E., Lampi, K.J., Speir, J.P., Kruppa, G., Easterling, M., Robinson, A.B. Quantitative measurement of young human eye lens crystallins by direct injection Fourier transform ion cyclotron resonance mass spectrometry. Mol. Vis. 2006;12:704–711. [PubMed] [Google Scholar]
- Sergeev, Y.V., Wingfield, P.T., Hejtmancik, J.F. Monomer-dimer equilibrium of normal and modified β A3-crystallins: Experimental determination and molecular modeling. Biochemistry. 2000;39:15799–15806. doi: 10.1021/bi001882h. [DOI] [PubMed] [Google Scholar]
- Takata, T., Oxford, J.T., Brandon, T.R., Lampi, K.J. Deamidation alters the structure and decreases the stability of human lens βA3-crystallin. Biochemistry. 2007;46:8861–8871. doi: 10.1021/bi700487q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, L., Boyle, D. Deamidation of specific glutamine residues from α-A crystallin during aging of the human lens. Biochemistry. 1998;37:13681–13685. doi: 10.1021/bi981542k. [DOI] [PubMed] [Google Scholar]
- Tsur, D., Tanner, S., Zandi, E., Bafna, V., Pevzner, P.A. Identification of post-translational modifications via blind search of mass-spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- Van Montfort, R.L., Bateman, O.A., Lubsen, N.H., Slingsby, C. Crystal structure of truncated human βB1-crystallin. Protein Sci. 2003;12:2606–2612. doi: 10.1110/ps.03265903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, A., Takio, K., Ihara, Y. Deamidation and isoaspartate formation in smeared τ in paired helical filaments. Unusual properties of the microtubule-binding domain of τ. J. Biol. Chem. 1999;274:7368–7378. doi: 10.1074/jbc.274.11.7368. [DOI] [PubMed] [Google Scholar]
- Whitmore, L., Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmarth, P.A., Tanner, S., Dasari, S., Nagalla, S.R., Riviere, M.A., Bafna, V., Pevzner, P.A., David, L.L. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: Does deamidation contribute to crystallin insolubility? J. Proteome Res. 2006;5:2554–2566. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z.G., Liu, Y., Mao, L.Y., Zhang, J.T., Zou, Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry. 2002;41:13012–13020. doi: 10.1021/bi026064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabrouskov, V., Han, X., Welker, E., Zhai, H., Lin, C., vanWijk, K.J., Scheraga, H.A., McLafferty, F.W. Stepwise deamidation of ribonuclease A at five sites determined by top down mass spectrometry. Biochemistry. 2006;45:987–992. doi: 10.1021/bi0517584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Smith, D.L., Smith, J.B. Human β-crystallins modified by backbone cleavage, deamidation and oxidation are prone to associate. Exp. Eye Res. 2003;77:259–272. doi: 10.1016/s0014-4835(03)00159-3. [DOI] [PubMed] [Google Scholar]