Abstract
Numerous human disorders are associated with the formation of protein fibrils. The fibril-forming capacity of a protein has been found in recent studies to be determined by a short segment of residues that forms a dual β-sheet, called a steric zipper, in the spine of the fibril. The question arises as to whether a fibril-forming segment, when inserted within the sequence of a globular protein, will invariably cause the protein to form fibrils. Here we investigate this question by inserting the known fibril-forming segment NNQQNY into the globular enzyme, T7 endonuclease I. From earlier studies, we know that in its fibril form, NNQQNY is in an extended conformation. We first found that the inserted NNQQNY stimulates fibril formation of T7 endonuclease I in solution. Thus NNQQNY within T7 endonuclease I can exist in an extended conformation, capable of forming the steric zipper in the core of a fibril. We also found that T7 endonuclease I folds into a decamer that does not form fibrils. We determined the structure of the decamer by X-ray crystallography, finding an unusual oligomer without point group symmetry, and finding that the NNQQNY segments within the decamer adopt two twisted conformations, neither is apparently able to fibrillize. We conclude that twisting of fibril forming sequences from the fully extended conformation, imposed by the context of their placement in proteins, can interfere with fibril formation.
Keywords: amyloid, steric zipper, T7 endonuclease I, protein structure/folding, crystallography
Formation of amyloid fibrils is linked to a wide range of human disorders, including Alzheimer's disease, Parkinson's disease, type II diabetes mellitus, and the transmissible spongiform encephalopathies (Chiti and Dobson 2006; Surewicz et al. 2006; Westermark et al. 2007). Although formed by proteins with unrelated primary sequences and tertiary structures, the fibrils share some common characteristics such as elongated morphology, affinity to dyes Congo red and thioflavin T (ThT), and the cross-β X-ray diffraction pattern. Despite intensive research efforts, what determines a protein's ability to form fibrils has remained poorly understood.
Three major pieces of evidence suggest that the amyloidogenicity of a protein may be attributed to short amino acid segments. First, short segments identified from amyloid-forming proteins form amyloid-like fibrils in vitro. Several synthetic peptides homologous to Aβ protein were found to form fibrils (Kirschner et al. 1987; Halverson et al. 1990); A six-residue segment from tau protein and a seven-residue segment from yeast prion Sup35 protein also form fibrils in vitro (von Bergen et al. 2000; Balbirnie et al. 2001). Second, swapping of a short segment from one protein to a homolog can create a fibril-former from a non-former. Transplantation of a seven-residue segment from the amyloidogenic human β2 microglobulin to its homologous but non-amyloidogenic mouse β2 microglobulin created an amyloidogenic protein (Ivanova et al. 2004). Similarly, insertion of a six-residue fragment from the amyloidogenic SH3 domain of bovine phosphatidyl-inositol-3′-kinase to the non-amyloidogenic SH3 domain of α-spectrin caused it to form amyloid-like fibrils (Ventura et al. 2004). Third, insertion of non-homologous short amino acid segments into a stable globular protein can create a fibril-former. For example, insertion of ten glutamine residues into RNase A resulted in fibril formation (Sambashivan et al. 2005). Insertion of non-homologous peptide STVIIE and the fragment KLVFFA of the Aβ peptide to the α-spectrin SH3 domain also leads to fibril formation (Esteras-Chopo et al. 2005).
More than a dozen short segments have recently been crystallized and their structures in the crystalline form have been determined (Nelson et al. 2005; Sawaya et al. 2007). The structures of these segments contribute insights into the structure and assembly of the amyloid fibrils at atomic resolution. These segments can be categorized into two groups: The first group includes sequences derived from proteins with a stable globular structure such as insulin, RNase A, and prion protein, and the second group includes sequences derived from proteins that have been reported to be natively unfolded, including Sup35, tau protein, and Aβ (Sawaya et al. 2007). For the segments in the first group, it is evident that these peptides can adopt both a fibrillization-competent conformation as seen in the crystal structure and a conformation that is compatible with the globular fold in the non-fibrillar form. It has been shown that partial unfolding of the globular fold containing these fibril-forming segments can lead to fibril formation (Bouchard et al. 2000; Lee and Eisenberg 2003). For the segments in the second group, however, it is not clear whether the fibril-forming proclivity of these peptides is compatible with their incorporation into structures of globular proteins. This work addresses this question by investigating the structure of a fibril-forming segment from a natively disordered protein in the context of a globular fold.
The segment GNNQQNY from the yeast prion protein Sup35 in Saccharomyces cerevisiae was found to exhibit the amyloid properties of the full-length Sup35 (Balbirnie et al. 2001). The structures of GNNQQNY and NNQQNY in microcrystals reveal a tightly packed β-sheet structure, termed a steric zipper, which appears to be the general cross-β feature of amyloid fibrils (Nelson et al. 2005). In Sup35 protein, NNQQNY is located in the N-terminal domain that has been found to be natively disordered (Glover et al. 1997). Therefore, it remains an open question whether insertion of NNQQNY into the sequence of a globular protein will invariably cause the protein to form fibrils. In this work, we inserted the segment NNQQNY into a globular protein T7 endonuclease I (T7EI), a DNA junction-resolving enzyme from bacteriophage T7. T7EI exists naturally as a domain-swapped (Bennett et al. 1994) dimer as shown from its crystal structure (Hadden et al. 2001). We have previously used T7EI as a model system to study the mechanism of amyloid formation (Guo and Eisenberg 2006; 2007). Here we investigate the amyloid formation of T7EI with the NNQQNY insert (T7EI−NNQQNY) and determined its crystal structure. The structure of T7EI−NNQQNY reveals two twisted conformations of NNQQNY, which are different from the fibrillization-compelling extended conformation. Our results show that fibril-forming sequences can be incorporated into globular folds, which deter fibril formation.
Results and Discussion
T7EI−NNQQNY forms short fibrils with amyloid-like properties
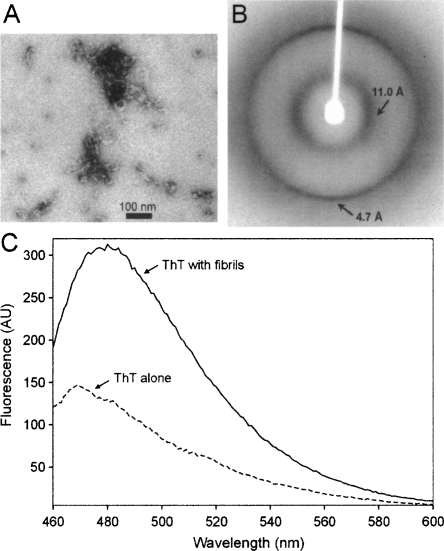
The segment NNQQNY was inserted into the hinge region of T7EI, which connects the N- and C-terminal domains of the domain-swapped wild type T7EI dimer (Supplemental Fig. S1A). Supplemental Figure S1B shows the changes in amino acid sequence as the result of insertion. T7EI−NNQQNY was expressed in Escherichia coli and purified from the soluble fraction. Insertion of NNQQNY does not change the expression level and solubility of T7EI. On native gel electrophoresis, T7EI−NNQQNY runs at a similar migration rate as the T7EI protein without insert (Supplemental Fig. S2A), suggesting T7EI−NNQQNY stays as a soluble dimer in solution. Incubation of T7EI−NNQQNY at 4°C for about four weeks leads to formation of short worm-like fibrils with a diameter of ≈10 nm as shown by electron microscopy (EM) (Fig. 1A). The morphology of these fibrils remains unchanged even after six months (data not shown). X-ray diffraction studies of the aligned fibrils reveal a characteristic cross-β diffraction pattern with a strong meridional reflection at 4.7 Å and an equatorial reflection at 11.0 Å (Fig. 1B), suggesting that these fibrils possess a cross-β structure. Dye binding assays show that the fibrils bind to the amyloid-specific dye ThT (Fig. 1C). These results suggest that the fibrils of T7EI−NNQQNY have amyloid-like properties.
Figure 1.
Amyloid-like properties of the T7EI−NNQQNY fibrils. (A) Electron micrograph of the T7EI−NNQQNY fibrils. (B) X-ray diffraction pattern of oriented T7EI−NNQQNY fibrils. A strong meridional reflection at ≈4.7 Å and equatorial reflection at ≈11 Å are characteristics of cross-β structure. (C) Fluorescence emission spectra of ThT alone and with T7EI−NNQQNY fibrils. Binding of ThT to amyloid fibrils gives rise to an emission peak at 482 nm. AU, arbitrary unit.
Our results show that insertion of NNQQNY into T7EI accelerates fibril formation. T7EI without the NNQQNY insert remains soluble at 4°C for several months (Guo and Eisenberg 2006), but T7EI with NNQQNY insert forms fibrils within four weeks at 4°C (Fig. 1). Insertion of other fibril-forming segments in globular proteins has also been reported to produce a fibril-former from a non-former (Esteras-Chopo et al. 2005; Sambashivan et al. 2005). These results support the idea that the fibril-forming capacity of a protein is determined by small segments of amino acids.
The structure of NNQQNY in the context of T7EI differs from its structure in segment crystals
We took a crystallographic approach to investigate the structure of T7EI−NNQQNY. The full-length T7EI−NNQQNY did not yield any crystals in our crystallization screens, so we made an N-terminal truncation construct of T7EI−NNQQNY, which lacks the His-tag sequence and residues 2 − 17 of T7EI. Native gel electrophoresis shows that this construct forms oligomers (Supplemental Fig. S2B). The size of the oligomers could not be determined from gel electrophoresis due to lack of appropriate molecular weight markers. Using ammonium sulfate as precipitant, N-terminal truncated T7EI−NNQQNY forms needle-like crystals within a couple of days. After several weeks, some cube-like crystals appear. The needle-like crystals diffract poorly, but the cube-like crystals diffracted to 3.0 Å at the Advanced Light Source. The structure of the cube-like crystals is reported below.
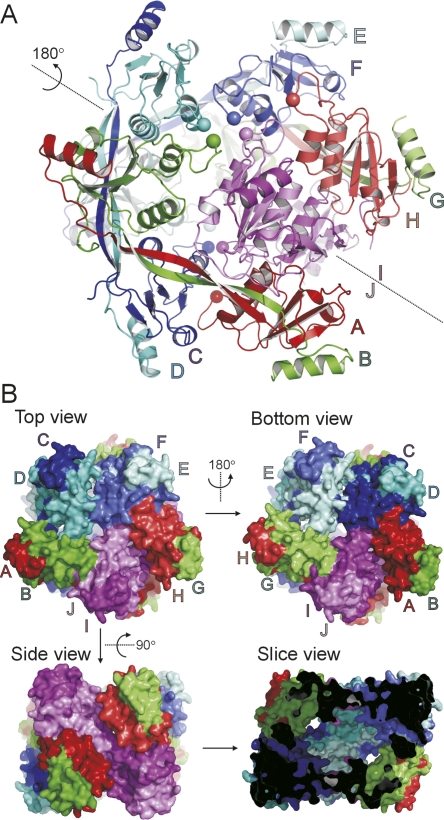
The structure of T7EI−NNQQNY was determined and refined to 3.0 Å with Rwork = 24.1% and Rfree = 28.4%. The full data collection and refinement statistics are shown in Table 1. Each asymmetric unit consists of 10 chains labeled A through J, and these 10 chains are organized in five domain-swapped dimers. The five dimers are packed into a cage-like assembly (Fig. 2). Interestingly, the five dimers are not related to one another by a fivefold axis. This is an unusual observation because protein oligomers almost always possess some point group symmetry. The only non-crystallographic symmetry axis in the decameric assembly of T7EI−NNQQNY is a quasi-twofold axis shown in Figure 2A. Figure 2B shows the surface representation of T7EI−NNQQNY from various angles. A slice view reveals a large cavity inside the decamer (Fig. 2B).
Table 1.
Data collection and refinement statistics
Figure 2.
The crystal structure of decameric T7EI−NNQQNY. (A) A ribbon diagram of the T7EI−NNQQNY structure. The dotted line represents a quasi-twofold non-crystallographic symmetry axis. Chains related by the twofold symmetry axis are colored the same but with different shades. The Cα atoms of residue 109 are shown in spheres to help guide the eye to define the subunit orientation. (B) A surface representation of the T7EI−NNQQNY structure. The top and bottom view are related by the same twofold axis shown in panel A. The slice view shows the cavity inside the decamer of T7EI−NNQQNY.
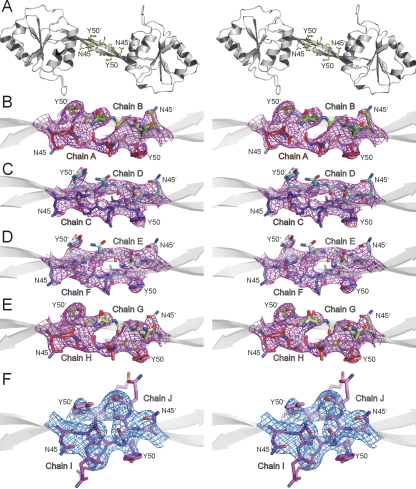
The electron density maps for the NNQQNY residues show that NNQQNY is located in the hinge region of the domain-swapped dimer (Fig. 3A). The hinge region does not make extensive contacts with the rest of the protein, resulting in high flexibility. The flexibility is manifested by the relatively weak electron density of the NNQQNY residues (Fig. 3B–F). However, the electron density for the main chain atoms of NNQQNY in all ten chains is largely resolved. The side chains of the tyrosyl residues of NNQQNY in all 10 chains have relatively strong density.
Figure 3.
Annealed Fo–Fc omit electron density maps for the NNQQNY segment inserted in T7EI, showing that the conformation of NNQQNY differs from that in its steric zipper structure. This figure is shown in stereoview. (A) A ribbon representation of a T7EI−NNQQNY dimer with NNQQNY residues shown as sticks. (B−F) Annealed Fo–Fc omit map for NNQQNY residues contoured at 6.0σ. Residues NNQQNY are shown as sticks. Notice that the main-chain atoms of NNQQNY are largely resolved and the side chain of tyrosine at position 50 in all 10 chains has relatively strong density.
NNQQNY adopts two conformations in the structure of T7EI−NNQQNY: an antiparallel β-ribbon in the dimers AB, CD, EF, and GH (Supplemental Fig. S3A), and an intertwined coil conformation in the dimer IJ (Supplemental Fig. S3B). One notable feature of the β-ribbon structure formed by NNQQNY is that the β-strand has a right-handed twist. The average strand twist angle for NNQQNY in these four dimers is 34 ± 9°, ranging from 21° to 43°. And the inter-strand twist angle is ≈20°. The right-handed twist in β-strands has been commonly observed in protein structures, and antiparallel β-sheets generally have larger twists than parallel ones (Chothia 1973; Ho and Curmi 2002). The degree of twist angle for NNQQNY is comparable to the twist angle for other β-sheet proteins (Ho and Curmi 2002; Zandomeneghi et al. 2004). In dimer IJ, the coil conformation for NNQQNY seems to be a result of crystal packing, which accommodates optimal interactions of the two globular domains connected by the NNQQNY insert with other chains. In amyloid fibrils, the inter-strand twist angle is usually very small, ranging from 1.5° to 2.5°, corresponding to a helical pitch of ≈1200 − 700 Å (Jiménez et al. 2002). Studies also suggest that the twist angle for the β-sheet may be smaller in amyloid fibrils than in native β-sheet proteins (Zandomeneghi et al. 2004). Therefore, the structure of NNQQNY in the context of T7EI is not suitable for a steric zipper.
Structural studies of fibril-forming segments derived from natively unfolded proteins show extended segments in steric zipper structures (Nelson et al. 2005; Sawaya et al. 2007). Is the fibril-forming proclivity of these segments strong enough to ensure that such segments always force an amyloid structure? Here we observe a globular structure for a protein that contains NNQQNY, showing that the segment does not condemn every protein containing it to form fibrils. In fact, the globular structure displays two conformations of the NNQQNY sequence. One is the extended β-conformation; the other is a coil conformation, which accommodates the packing of the domain-swapped dimers into the decamer (Supplemental Fig. S3). That insertion of NNQQNY into T7EI does not create a disordered protein suggests that the presence of such segments in natively disordered proteins is not the sole reason for these proteins to be disordered. Our results suggest that the conformational flexibility may be a common property for the fibril-forming segments, and the conformations they adopt are influenced by the context of the globular fold.
Materials and Methods
T7EI−NNQQNY constructs
The full-length T7EI construct used in this work contains 172 residues, including an N-terminal His-tag sequence of 23 residues (Déclais et al. 2001). The T7EI construct also contains an E65K mutation, which makes the enzyme inactive (Déclais et al. 2001) and allows high level expression in E. coli. The crystal structures of T7EI E65K mutant (Hadden et al. 2001) and wild type T7EI (Hadden et al. 2002) show that E65K mutation does not change the structure of T7EI. To insert NNQQNY sequence into the hinge region of T7EI, restriction sites for endonucleases XhoI and SacI were introduced to T7EI template at amino acid positions 44 and 49, respectively, using QuikChange site-directed mutagenesis kit (Stratagene). The numbering of residues starts from the first residue of the physiological form of T7EI. The T7EI sequence containing XhoI and SacI sites was digested by XhoI and SacI and then ligated with DNA oligos encoding for the NNQQNY insert. The ligation reaction was transformed to XL1-Blue subcloning grade competent cells (Stratagene). This construct is named T7EI−NNQQNY. For crystallization of T7EI−NNQQNY, the His-tag sequence and residues 2 − 17 of the T7EI sequence were removed by a single step of QuikChange mutagenesis. This construct is called Δ2 − 17 T7EI−NNQQNY. All constructs were verified by DNA sequencing.
Protein expression and purification
The T7EI−NNQQNY proteins were expressed and purified as previously described (Guo and Eisenberg 2006). The Δ2 − 17 T7EI−NNQQNY construct was transformed into E. coli BL21(DE3) pLysS cells (Novagen) and protein expression was induced with 1 mM isopropyl β-D-thiogalactoside when cell density reached OD600nm ≈ 0.6 at 37°C. The induction was allowed to proceed at 25°C for 4 h. The cells were harvested by centrifugation and resuspended in buffer A (50 mM Tris, pH 7.5, 40 mM NaCl). The cells were then sonicated, and the cell debris was pelleted by centrifugation. The supernatant was filtered using 0.22 μm filter and loaded onto a 1-mL HiTrap SP HP column (GE Healthcare) equilibrated with buffer A. Proteins were eluted with a linear NaCl gradient (40 − 1000 mM). Protein concentration was determined by UV absorption at 280 nm using an extinction coefficient of 24.75 × 103 M−1 cm−1 for both constructs.
Fibril formation and characterization
Purified T7EI−NNQQNY proteins were buffer-exchanged to buffer B (20 mM Tris, pH 8.0, 50 mM NaCl), concentrated to ≈600 μM, and was incubated at 4°C without agitation. The solution became gel-like after 4–6 wk. For electron microscopy, fibril samples were applied onto glow discharged copper grids covered with 400 mesh formvar/carbon film (Ted Pella) and stained with 2% uranyl acetate. Samples were examined under a Hitachi H-7000 electron microscope with an accelerating voltage of 75 kV. For X-ray fiber diffraction, 7 μL fibrils were placed between the sealed ends of two silanized glass capillaries and allowed to dry slowly at 4°C. Diffraction data were collected at room temperature using a Rigaku FRD generator with an R-AXIS IV++ detector. For ThT binding assay, ThT (Sigma) was dissolved in buffer C (5 mM potassium phosphate, pH 7.4, 150 mM NaCl) and filtered with 0.22 μm filter. Proteins were diluted to a final concentration of 10 μM in buffer C containing 50 μM ThT. Fluorescence scan was performed on a PTI spectrofluorometer. Excitation was at 440 nm (4-nm slit width), and emission was scanned from 460 to 600 nm (8-nm slit width).
Gel electrophoresis
Native PAGE electrophoresis was performed using PhastSystem (GE Healthcare). Native buffer strips for basic proteins (2% agarose, 4.4% β-alanine, and 4% acetic acid) were prepared, and 4% − 15% gradient PhastGel were used. The proteins were loaded directly on the gel without any treatment.
Crystallization of Δ2 − 17 T7EI−NNQQNY
Crystallization was carried out by screening a wide range of crystallization conditions and then by screening additives using the hanging-drop vapor diffusion method. The crystals for the reported structure were obtained by mixing 2 μL of protein solution (450 μM in buffer B) with an equal volume of well buffer (2.6 M ammonium sulfate, 0.1 M sodium citrate, 2% dimethyl formamide, pH 5.5). Crystallization drops were suspended over 1 mL of the same well buffer.
Data collection, structural determination, and refinement
For data collection, crystals were flash-frozen in a cryostream of N2 gas at 100 K using mineral oil as a cryoprotectant. Diffraction data were collected at 100 K at the Advanced Light Source using beamline 8.2.2. Data reduction and scaling were done with the programs DENZO and SCALEPACK (Otwinowski and Minor 1996). The data processing statistics are listed in Table 1.
The structure of T7EI−NNQQNY was determined by the molecular replacement method using the program PHASER (McCoy et al. 2005). The starting model consists of residues 18 − 43 of molecule A and residues 51 − 145 of molecule B of the T7EI structure (PDB ID: 1FZR). The structure was refined using the program CNS (Brunger et al. 1998). The refinement was monitored using the free R-factor. A subset (5%) of the diffraction data was chosen randomly and omitted from refinement for calculation of the free R-factor. The model was rebuilt manually during refinement using programs COOT (Emsley and Cowtan 2004) and O (Jones et al. 1991). There are 10 T7EI−NNQQNY molecules in each asymmetric unit. Residues 44 − 52 for each molecule in the asymmetric unit were built individually through several rounds of refinement. Final refinement statistics are listed in Table 1. To reduce model bias, the annealed Fo–Fc maps of NNQQNY residues were generated by performing a simulated annealing refinement with residues NNQQNY and residues in its 3.5 Å neighborhood omitted from refinement, and these maps are shown in Figure 3. Lys 126 was found to be in a disallowed region of the Ramachandran plot in all chains, but the electron density for our search model (PDB ID: 1FZR) supports this geometry (Hadden et al. 2001; 2002).
The average twist angle for the NNQQNY segment was computed as a function of the backbone dihedral angles (φ, ψ, ω) according to the equations described in Chou and Scheraga (1982). The structures were illustrated using program PyMOL (http://www.pymol.org).
Accession codes
Coordinates and structure factors of T7EI−NNQQNY have been deposited in the Protein Data Bank under accession code 3CAE.
Electronic supplemental material
Supplemental material contains three figures showing the location and sequence of the NNQQNY insert in T7EI; native gel electrophoresis of T7EI and T7EI−NNQQNY; and the two conformations of NNQQNY in decameric T7EI−NNQQNY.
Acknowledgments
We thank David M.J. Lilley at University of Dundee, UK for providing the clone of T7EI E65K mutant, Martin Phillips at UCLA DOE-Biochemistry Instrumentation Facility for assistance, and Duilio Cascio and Michael R. Sawaya for collecting the X-ray diffraction data at Advanced Light Source. This work was supported by the NSF, NIH, and HHMI.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: David Eisenberg, Howard Hughes Medical Institute, UCLA-DOE Institute for Genomics and Proteomics, Molecular Biology Institute, UCLA, Box 951570, Los Angeles, CA 90095-1570, USA; e-mail: david@mbi.ucla.edu; fax: (310) 206-3914.
Abbreviations: T7EI, T7 endonuclease I; T7EI−NNQQNY, T7EI with an insert of peptide NNQQNY; ThT, thioflavin T; EM, electron microscopy.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036368.108.
References
- Balbirnie, M., Grothe, R., Eisenberg, D.S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proc. Natl. Acad. Sci. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Choe, S., Eisenberg, D. Domain swapping: Entangling alliances between proteins. Proc. Natl. Acad. Sci. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard, M., Zurdo, J., Nettleton, E.J., Dobson, C.M., Robinson, C.V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chiti, F., Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chothia, C. Conformation of twisted β-pleated sheets in proteins. J. Mol. Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- Chou, K.C., Scheraga, H.A. Origin of the right-handed twist of β-sheets of poly(L-Val) chains. Proc. Natl. Acad. Sci. 1982;79:7047–7051. doi: 10.1073/pnas.79.22.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déclais, A.C., Hadden, J., Phillips, S.E.V., Lilley, D.M.J. The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol. 2001;307:1145–1158. doi: 10.1006/jmbi.2001.4541. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esteras-Chopo, A., Serrano, L., López de la Paz, M. The amyloid stretch hypothesis: Recruiting proteins toward the dark side. Proc. Natl. Acad. Sci. 2005;102:16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, J.R., Kowal, A.S., Schirmer, E.C., Patino, M.M., Liu, J.J., Lindquist, S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Guo, Z., Eisenberg, D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc. Natl. Acad. Sci. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Eisenberg, D. The mechanism of the amyloidogenic conversion of T7 endonuclease I. J. Biol. Chem. 2007;282:14968–14974. doi: 10.1074/jbc.M609514200. [DOI] [PubMed] [Google Scholar]
- Hadden, J.M., Convery, M.A., Declais, A.C., Lilley, D.M.J., Phillips, S.E.V. Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I. Nat. Struct. Biol. 2001;8:62–67. doi: 10.1038/83067. [DOI] [PubMed] [Google Scholar]
- Hadden, J.M., Declais, A.C., Phillips, S.E.V., Lilley, D.M.J. Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I. EMBO J. 2002;21:3505–3515. doi: 10.1093/emboj/cdf337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson, K., Fraser, P.E., Kirschner, D.A., Lansbury, P.T. Molecular determinants of amyloid deposition in Alzheimer's disease: Conformational studies of synthetic β-protein fragments. Biochemistry. 1990;29:2639–2644. doi: 10.1021/bi00463a003. [DOI] [PubMed] [Google Scholar]
- Ho, B.K., Curmi, P.M.G. Twist and shear in β-sheets and β-ribbons. J. Mol. Biol. 2002;317:291–308. doi: 10.1006/jmbi.2001.5385. [DOI] [PubMed] [Google Scholar]
- Ivanova, M.I., Sawaya, M.R., Gingery, M., Attinger, A., Eisenberg, D. An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc. Natl. Acad. Sci. 2004;101:10584–10589. doi: 10.1073/pnas.0403756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, J.L., Nettleton, E.J., Bouchard, M., Robinson, C.V., Dobson, C.M., Saibil, H.R. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kirschner, D.A., Inouye, H., Duffy, L.K., Sinclair, A., Lind, M., Selkoe, D.J. Synthetic peptide homologous to β protein from Alzheimer disease forms amyloid-like fibrils in vitro. Proc. Natl. Acad. Sci. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Eisenberg, D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat. Struct. Biol. 2003;10:725–730. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C., Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Nelson, R., Sawaya, M.R., Balbirnie, M., Madsen, A.O., Riekel, C., Grothe, R., Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Sambashivan, S., Liu, Y., Sawaya, M.R., Gingery, M., Eisenberg, D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- Sawaya, M.R., Sambashivan, S., Nelson, R., Ivanova, M.I., Sievers, S.A., Apostol, M.I., Thompson, M.J., Balbirnie, M., Wiltzius, J.J.W., McFarlane, H.T., et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Surewicz, W.K., Jones, E.M., Apetri, A.C. The emerging principles of mammalian prion propagation and transmissibility barriers: Insight from studies in vitro. Acc. Chem. Res. 2006;39:654–662. doi: 10.1021/ar050226c. [DOI] [PubMed] [Google Scholar]
- Ventura, S., Zurdo, J., Narayanan, S., Parreno, M., Mangues, R., Reif, B., Chiti, F., Giannoni, E., Dobson, C.M., Aviles, F.X., et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc. Natl. Acad. Sci. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen, M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E.M., Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl. Acad. Sci. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark, P., Benson, M.D., Buxbaum, J.N., Cohen, A.S., Frangione, B., Ikeda, S.I., Masters, C.L., Merlini, G., Saraiva, M.J., Sipeo, J.D. A primer of amyloid nomenclature. Amyloid. 2007;14:179–183. doi: 10.1080/13506120701460923. [DOI] [PubMed] [Google Scholar]
- Zandomeneghi, G., Krebs, M.R.H., Mccammon, M.G., Fandrich, M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]