Abstract
Isocitrate dehydrogenases (IDHs) catalyze oxidative decarboxylation of isocitrate (ICT) into α-ketoglutarate (AKG). We report here the crystal structures of Saccharomyces cerevesiae mitochondrial NADP-IDH Idp1p in binary complexes with coenzyme NADP, or substrate ICT, or product AKG, and in a quaternary complex with NADPH, AKG, and Ca2+, which represent different enzymatic states during the catalytic reaction. Analyses of these structures identify key residues involved in the binding of these ligands. Comparisons among these structures and with the previously reported structures of other NADP-IDHs reveal that eukaryotic NADP-IDHs undergo substantial conformational changes during the catalytic reaction. Binding or release of the ligands can cause significant conformational changes of the structural elements composing the active site, leading to rotation of the large domain relative to the small and clasp domains along two hinge regions (residues 118–124 and residues 284–287) while maintaining the integrity of its secondary structural elements, and thus, formation of at least three distinct overall conformations. Specifically, the enzyme adopts an open conformation when bound to NADP, a quasi-closed conformation when bound to ICT or AKG, and a fully closed conformation when bound to NADP, ICT, and Ca2+ in the pseudo-Michaelis complex or with NADPH, AKG, and Ca2+ in the product state. The conformational changes of eukaryotic NADP-IDHs are quite different from those of Escherichia coli NADP-IDH, for which significant conformational changes are observed only between two forms of the apo enzyme, suggesting that the catalytic mechanism of eukaryotic NADP-IDHs is more complex than that of EcIDH, and involves more fine-tuned conformational changes.
Keywords: catalytic mechanism, conformational change, IDH, Idp1p, induced-fit, isocitrate dehydrogenase, NADP
Isocitrate dehydrogenases (IDHs) are a family of enzymes responsible for catalyzing the oxidative decarboxylation of isocitrate (ICT) into α-ketoglutarate (AKG). Most bacteria and archea contain a single type of NADP-dependent IDH (NADP-IDH) to catalyze this reaction during the Krebs cycle, whereas eukaryotes contain two types of IDHs using either NAD or NADP as a coenzyme. Eukaryotic NAD-dependent IDHs (NAD-IDHs) are localized in the mitochondrial matrix and play an essential role in the Krebs cycle as bacterial NADP-IDHs. Eukaryotic NADP-IDHs are located primarily in cell cytosol and mitochondria, but can also be found in peroxisomes of human, rat, and mouse cells because the cytosolic NADP-IDHs of these species contain a type 1 peroxisomal targeting sequence at the C terminus (Nekrutenko et al. 1998; Geisbrecht and Gould 1999; Yoshihara et al. 2001; Lu et al. 2008). In Saccharomyces cerevesiae, there is an NADP-IDH isozyme specifically targeted to peroxisomes (Henke et al. 1998; van Roermund et al. 1998; Minard and McAlister-Henn 1999). In addition to their potential role in the Krebs cycle, eukaryotic NADP-IDHs have been demonstrated to play important roles in cellular defense against oxidative damage, detoxification of reactive oxygen species, and synthesis of fat and cholesterol through generation of NADPH and AKG (Jo et al. 2001; Lee et al. 2002, 2007; Kim and Park 2003; Koh et al. 2004; Kim et al. 2005, 2007; Valderrama et al. 2006; Frederiks et al. 2007; Mailloux et al. 2007; Marino et al. 2007; Margittai and Banhegyi 2008).
Based on sequence analysis, the NADP-IDHs can be mainly divided into two subfamilies (Steen et al. 2001). Escherichia coli NADP-IDH (EcIDH) is a representative of subfamily I which includes most bacterial and archaeal NADP-IDHs. The structure and function of EcIDH has been studied extensively (Hurley et al. 1989, 1991; Stoddard and Koshland Jr. 1993; Stoddard et al. 1993; Bolduc et al. 1995; Finer-Moore et al. 1997). EcIDH exists as a homodimer in both solution and crystal structure, and each subunit consists of three domains: a large domain, a small domain, and a clasp domain. The latter two domains of one subunit interact with those of the other to form a tight dimer interface. The catalytic active site is formed by the large and small domains of one subunit and the small domain of the other. The apo EcIDH adopts two distinct conformations in two different crystal forms in which the large domain displays a 16° rotation relative to the small and clasp domains, denoted as open and closed conformations, respectively. When it is bound to NADP, or ICT and Mg2+, or ICT, NADP, and Ca2+, or NADPH, AKG, and Ca2+, EcIDH always adopts the closed conformation. The enzymatic activity of EcIDH is regulated through phosphorylation and dephosphorylation of Ser113 at the ICT-binding site by a bifunctional IDH kinase/phosphatase (Hurley et al. 1989, 1990; LaPorte 1993). The structures of several other dimeric bacterial and archaeal NADP-IDHs of subfamily I have also been reported, which exhibit high structural similarities with EcIDH (Singh et al. 2001, 2002; Karlstrom et al. 2005; Stokke et al. 2007). In addition to the dimeric NADP-IDHs, a group of monomeric NADP-IDHs has also been identified in several bacterial species, and the crystal structures of the enzymes from Azotobacter vinelandii (Yasutake et al. 2002, 2003) and Corynebacterium glutamicum (Imabayashi et al. 2006) have also been determined. The monomeric NADP-IDHs are possibly formed by domain duplication and might have a different catalytic mechanism.
Subfamily II NADP-IDHs include mainly eukaryotic NADP-IDHs and a few bacterial NADP-IDHs (Steen et al. 2001). For eukaryotic NADP-IDHs, the crystal structures of porcine mitochondrial NADP-IDH (PmIDH) in complex with ICT and Mn2+ (PmIDH–ICT–Mn) (Ceccarelli et al. 2002), and human cytosolic NADP-IDH (HcIDH) in complexes with NADP (HcIDH–NADP) and with NADP, ICT, and Ca2+ (HcIDH–NADP–ICT–Ca) (Xu et al. 2004) have been reported. The two eukaryotic enzymes share a moderate level of sequence identity (69%) with each other, but low degrees of sequence identity with EcIDH (17%–18%) and other subfamily I NADP-IDHs. The overall structures of PmIDH and HcIDH are very similar to that of EcIDH. Nevertheless, there are some notable differences between these eukaryotic NADP-IDHs and EcIDH. In contrast to EcIDH, the eukaryotic NADP-IDHs in different enzymatic states show significant conformational differences both at the active site and in the overall structure. In the HcIDH–NADP complex, the enzyme adopts a conformation more similar to the open conformation of EcIDH (Xu et al. 2004). In the PmIDH–ICT–Mn complex, the enzyme adopts a conformation more similar to the closed conformation of EcIDH (Ceccarelli et al. 2002). In the HcIDH–NADP–ICT–Ca complex, the enzyme assumes a conformation which is more compact than the closed conformation of EcIDH (Xu et al. 2004). In addition, in both HcIDH and PmIDH, the clasp domains of the two subunits in the homodimer interwind together to form two layers of four-stranded antiparallel β-sheets. On the other hand, in EcIDH the clasp domains form two antiparallel α-helices and a four-stranded antiparallel β-sheet. Furthermore, although the eukaryotic NADP-IDHs also contain a strictly conserved Ser at the position equivalent to Ser113 of EcIDH (corresponding to Ser94 of HcIDH or Ser95 of PmIDH), neither eukaryotic IDH kinase/phosphatase nor its homolog has been identified, nor phosphorylated eukaryotic NADP-IDH has been detected in vivo (Mas and Colman 1984; Ceccarelli et al. 2002; Xu et al. 2004). Recently, the crystal structures of two unique bacterial NADP-IDHs of subfamily II have also been described. Thermotoga maritime NADP-IDH (TmIDH) in apo form assumes an open conformation (Karlstrom et al. 2006) and Desulfotalea psychrophila NADP-IDH (DpIDH) in both apo form and in complex with ICT assumes a closed conformation (Fedøy et al. 2007). These bacterial NADP-IDHs share moderate sequence homology (∼50%) and high structural similarity with eukaryotic NADP-IDHs. Thus, it is conceivable that, along with the functional and structural divergence of NADP-IDHs, there may be some differences in the catalytic mechanism between eukaryotic NADP-IDHs (or subfamily II NADP-IDHs) and EcIDH (or subfamily I NADP-IDHs) and among different eukaryotic isozymes. Due to the lack of a systematic structural study of any eukaryotic NADP-IDH, the catalytic mechanism of eukaryotic NADH-IDHs is thus far still unclear.
S. cerevesiae contains three NADP-IDH isozymes (Idp1p, Idp2p, and Idp3p) which share sequence identity of ∼70% among themselves and ∼55%–65% with PmIDH and HcIDH. The three isozymes are localized in different subcellular compartments and presumably exert some divergent biological functions. Idp1p is constitutively expressed and targeted to mitochondria; Idp2p is located in the cytosol and highly expressed when yeast cells are supplied with non-fermentable carbon sources; and Idp3p is localized specifically to peroxisomes and detected only in cells grown on fatty acids (Contreras-Shannon et al. 2005; Lu et al. 2008). Thus, S. cerevesiae NADP-IDHs are ideal models for studying the catalytic mechanism of eukaryotic NADP-IDHs and the differences among different isozymes and with the bacterial enzymes. We report here the crystal structures of Idp1p in complexes with different ligands which represent several enzymatic states during the catalytic reaction: in binary complexes with NADP, or ICT, or AKG, and in a quaternary complex with NADPH, AKG, and Ca2+. Comparisons among these structures and with the previously reported eukaryotic NADP-IDH structures reveal significant conformational changes of the enzyme along the catalytic reaction and provide insights into the catalytic mechanism of eukaryotic NADP-IDHs.
Results
Overall structure of Idp1p
The structures of Idp1p in binary complexes with NADP, ICT, and AKG, and in a quaternary complex with NADPH, AKG, and Ca2+ were determined at 2.3 Å, 2.6 Å, 2.1 Å, and 2.7 Å resolution, respectively (Table 1). The structure of Idp1p in complex with NADP (Idp1p–NADP) contains four Idp1p molecules forming two homodimers related by a pseudo twofold non-crystallographic symmetry (NCS) in an asymmetric unit. Each Idp1p is bound to an NADP molecule at the active site. The structure of Idp1p in complex with ICT (Idp1p–ICT), or NADPH, AKG, and Ca2+ (Idp1p–NADPH–AKG–Ca), or AKG (Idp1p–AKG) contains six Idp1p molecules that form three homodimers related by a pseudo threefold NCS in an asymmetric unit. Each Idp1p is bound to an active D-stereoisomer of ICT, or an NADPH, an AKG, and a Ca2+ ion, or an AKG at the active site. In all four structures, the protein model is well defined with high-quality electron density except that a few N-terminal and/or C-terminal amino acids are disordered, and the bound NADP/NADPH, ICT, AKG, and Ca2+ are also fitted to high-quality electron density in their respective complexes (Supplemental Fig. S1).
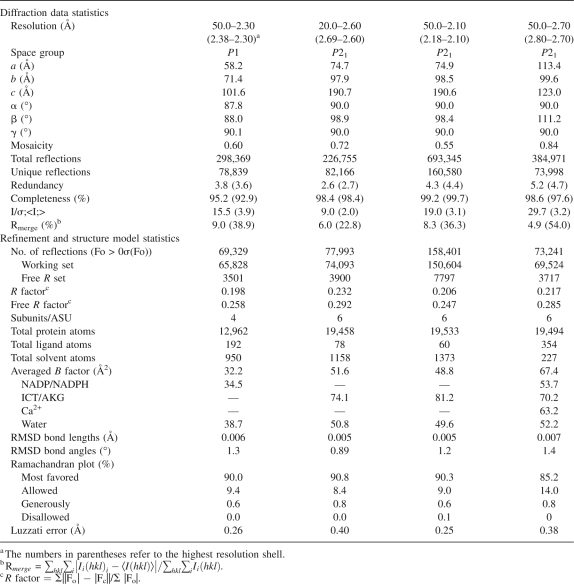
Table 1.
Statistics of X-ray diffraction data and structure refinement
The overall structure of Idp1p is very similar to that of PmIDH and HcIDH and is composed of three domains: a large domain (residues 1–104 and 288–412) assuming a typical Rossmann fold, a small domain (residues 105–137 and 188–287) with an α/β sandwich fold, and a clasp domain (residues 138–187) consisting of two stacked four-stranded antiparallel β-sheets (Fig. 1A,B). Like PmIDH and HcIDH, Idp1p forms a homodimer in both solution (as revealed by dynamic light scattering analysis) and the crystal structures. Dimerization is mediated by extensive hydrophilic and hydrophobic interactions primarily between the small and clasp domains of both subunits. In all four structures, the two subunits in the homodimer adopt very similar conformation with an RMSD of 0.4–0.5 Å for 410 Cα atom pairs. The catalytic active site resides in a large cleft formed by the large and small domains of one subunit and the small domain of the other and consists of the NADP-binding site and the ICT-binding site (Fig. 1A).
Figure 1.
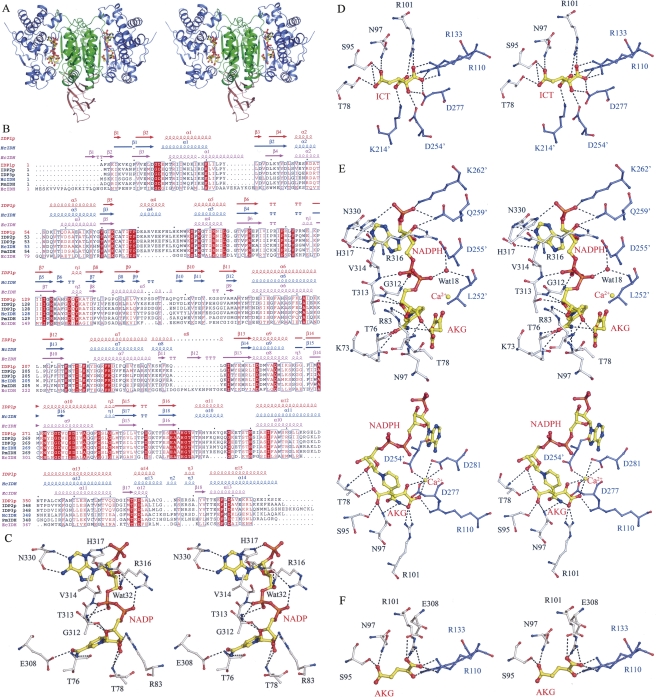
Structure of Idp1p in complexes with different ligands. (A) Overall structure of the Idp1p–NADPH–AKG–Ca complex. Idp1p is colored with the large domain in slate-blue, the small domain in green, and the clasp domain in salmon. The bound NADPH and AKG are shown in ball-and-stick models and the Ca2+ ion as a gold sphere. (B) Structure-based sequence alignment of Idp1p with other NADP-IDHs. The sequences include S. cerevesiae Idp2p and Idp3p, HcIDH, PmIDH, and EcIDH. Invariant residues are highlighted by shaded red boxes and conserved residues by open red boxes. The secondary structures of the Idp1p–NADP (PDB code 2QFV), HcIDH–NADP (PDB code 1T09), and EcIDH–NADP (PDB code 9ICD) complexes are placed above the alignment. (C) A stereoview showing the interactions of NADP with the surrounding residues in the Idp1p–NADP complex. (D) A stereoview showing the interactions of ICT with the surrounding residues in the Idp1p–ICT complex. (E) A stereoview showing the interactions of NADPH (upper panel), AKG, and Ca2+ (lower panel) with the surrounding residues in the Idp1p–NADPH–AKG–Ca complex. (F) A stereoview showing the interactions of AKG with the surrounding residues in the Idp1p–AKG complex. The residues involved in interactions are shown with side chains with those of the large domain in silver and those of the small domain in blue. The residues from the adjacent subunit are labeled with an apostrophe. The hydrogen-bonding interactions are indicated with dashed lines.
Although the overall structures of Idp1p in all four complexes are similar, there are substantial conformational differences at the active site and in the overall structure among them (Fig. 2; Table 2). In the Idp1p–NADP complex, the enzyme assumes an open conformation. In the Idp1p–ICT and Idp1p–AKG complexes, the enzyme assumes a quasi-closed conformation. In the Idp1p–NADPH–AKG–Ca complex, the enzyme assumes a closed conformation. Structural comparisons indicate that, in different complexes, the dimer interface formed between the small and clasp domains of both subunits exhibits very little change (for example, dimerization buries 6872 Å2 or 21.1% of the solvent accessible surface area of each subunit in the Idp1p–ICT complex and 6425 Å2 or 19.6% in the Idp1p–NADP complex); however, the large domain assumes considerably differing conformations. The conformational changes between different complexes can be visualized as a rotation of the large domain relative to the small and clasp domains or the dimer interface (see Discussion) (Fig. 2; Table 2). Crystallographic analyses of these structures indicate that the observed conformational differences are not caused by crystal packing and thus should be biologically relevant.
Figure 2.
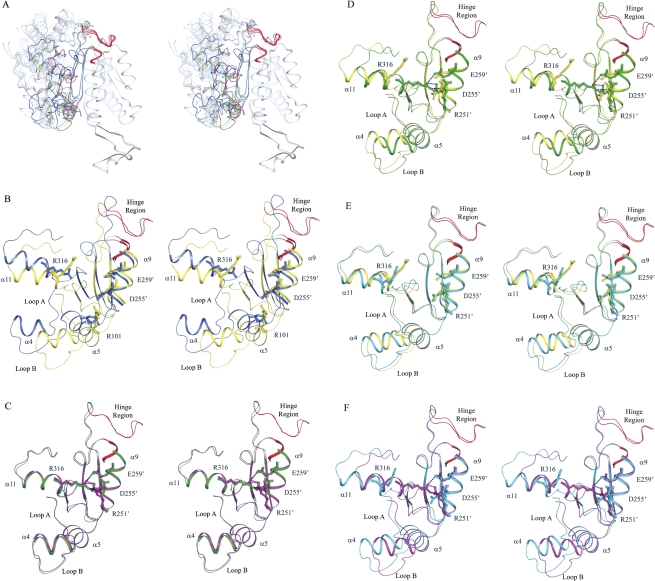
Structural comparison of Idp1p in different enzymatic states. (A) Structural comparison of Idp1p in different enzymatic states showing conformational differences of the overall structure. The bound ligands are omitted for clarity. The comparison is based on superposition of the small and clasp domains relative to the Idp1p–NADP complex. The enzyme assumes an open conformation in the Idp1p–NADP complex (in slate-blue), a quasi-closed conformation in the Idp1p–ICT (in yellow) and Idp1p–AKG (in cyan) complexes, and a closed conformation in the Idp1p–NADPH–AKG–Ca complex (in magenta). (B) Comparison of the active site between the Idp1p–NADP (in slate-blue) and Idp1p–ICT (in yellow) complexes. The large domain in the ICT-bound complex rotates ∼17° toward the small and clasp domains along the hinge regions of residues 118–124 and 284–287 (in red) relative to that in the NADP-bound complex. (C) Comparison of the active site between the Idp1p–NADPH–AKG–Ca (in magenta) and HcIDH–NADP–ICT–Ca (in green) complexes. Both enzymes adopt the closed conformation without notable difference. (D) Comparison of the active site between the Idp1–ICT (in yellow) and HcIDH–NADP–ICT–Ca (in green) complexes. The large domain in the HcIDH–NADP–ICT–Ca complex rotates ∼8.7° further toward the small and clasp domains along the hinge regions (in red) relative to that in the Idp1p–ICT complex. (E) Comparison of the active site between the Idp1p–AKG (in cyan) and Idp1p–ICT (in yellow) complexes. The enzyme in both complexes adopts the quasi-closed conformation. However, the large domain in the AKG-bound complex rotates slightly away from the small and clasp domains relative to that in the ICT-bound complex. (F) Comparison of the active site between the Idp1p–AKG (in cyan) and Idp1p–NADPH–AKG–Ca (in magenta) complexes. The large domain in the binary complex rotates away from the small and clasp domains along the hinge regions (in red) relative to that in the quaternary complex.
Table 2.
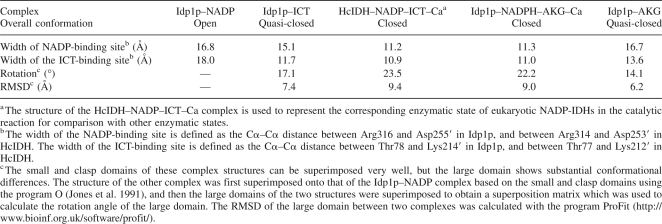
Structural comparison of eukaryotic NADP-IDHs in different enzymatic states
Structural comparisons between Idp1p and EcIDH indicate that the open conformation of Idp1p in the Idp1p–NADP complex is more similar to the open conformation than the closed conformation of EcIDH (an RMSD of 1.8 Å vs. 1.8 Å for the small and clasp domains [145 Cα atom pairs] and 4.3 Å vs. 7.8 Å for the large domain [171 Cα atom pairs]). The quasi-closed conformation of Idp1p in the Idp1p–ICT complex is more similar to the closed conformation than the open conformation of EcIDH (an RMSD of 1.7 Å vs. 1.7 Å for the small and clasp domains and 2.9 Å vs. 5.6 Å for the large domain). The closed conformation of Idp1p in the Idp1p–NADPH–AKG–Ca complex resembles but is more compact than the closed conformation of EcIDH (an RMSD of 1.7 Å for the small and clasp domains and 2.9 Å for the large domain).
Recognition and binding of NADP
The NADP-binding site is located at the upper part of the active site. In the Idp1p–NADP complex, the bound NADP has extensive hydrophilic interactions with the surrounding residues from the large domain, including Thr76, Thr78, Arg83, Glu308, Gly312, Thr313, Val314, Arg316, His317, and Asn330 (Fig. 1C; Supplemental Table S1). Structural comparison between the Idp1p–NADP and HcIDH–NADP complexes indicates that both enzymes assume the open conformation with an RMSD of 1.4 Å for 385 Cα atom pairs. The key residues involved in the recognition and binding of NADP are strictly conserved, and the interactions between NADP and the enzyme are strongly conserved in all eukaryotic NADP-IDHs (Fig. 1B), suggesting that other eukaryotic NADP-IDHs might also assume the open conformation when bound to NADP. However, there are a few notable conformational differences at the NADP-binding site between the two complexes. Particularly, Arg316 of Idp1p helps to stabilize the 3′-OH group of the adenosine ribose moiety and the second phosphate group of NADP, whereas the equivalent residue Arg314 of HcIDH forms a salt bridge with the 2′-phosphomonoester group (Xu et al. 2004). Instead, His317 of Idp1p forms a hydrogen bond with the 2′-phosphomonoester group and appears to play a critical role in the discrimination of NADP against NAD. It is also intriguing to note that in the Idp1p–NADP complex the region equivalent to the structural segment (residues 271–286), which was proposed to play an important role in the regulation of the enzymatic activity of HcIDH through conformational changes (Xu et al. 2004), forms a standard α-helix with well-defined electron density (Supplemental Fig. S1B). This suggests that the self-regulatory mechanism proposed for HcIDH does not apply to Idp1p and possibly other mitochondrial NADP-IDHs.
The structure of the apo Idp1p is unavailable for comparison because Idp1p was co-purified with ICT, and attempts to grow crystals of apo Idp1p were unsuccessful (see Materials and Methods). The structure of the apo HcIDH is also unavailable because HcIDH was co-purified with NADP (Xu et al. 2004). Thus, it is unknown whether the binding of NADP would induce any conformational change of the active site and/or the overall structure. Nevertheless, the apo TmIDH structure provides valuable insight for deciphering this issue. Compared with the open Idp1p–NADP complex, the apo TmIDH assumes a more open conformation (an RMSD of 0.9 Å for the small and clasp domains and 1.9 Å for the large domain) in which the large domain has a 5.8° rotation along the hinge regions away from the small and clasp domains. This results in more open NADP- and ICT-binding sites (the widths are 19.7 Å and 18.5 Å, respectively). Thus, it seems very likely that the binding of NADP can cause moderate conformational changes of the active site and the overall structure of the eukaryotic NADP-IDHs.
Recognition and binding of ICT
The ICT-binding site is located at the lower part of the active site. In the Idp1p–ICT complex, the bound ICT forms extensive hydrogen-bonding interactions with several conserved residues of the small and large domains, including Thr78, Ser95, Asn97, Arg101, Arg110, Arg133, Asp277, Lys214′, and Asp254′ (residues from the second subunit are indicated with an apostrophe hereafter) (Fig. 1D; Supplemental Table S2). Structural comparison between the Idp1p–ICT complex and the PmIDH–ICT–Mn complex shows that both enzymes adopt the quasi-closed conformation with an RMSD of 0.61 Å for 385 Cα atom pairs, and the majority of the interactions between ICT and the enzyme are conserved. However, due to the absence of a metal ion in the Idp1p–ICT complex, ICT assumes a slightly different position, and several residues involved in interactions with the metal ion and the substrate (Asp277, Asp254′, Asp255′, and Tyr140) also adopt different side-chain conformations. For example, the hydroxyl group of ICT forms hydrogen-bonding interactions with the side chains of Asp277 and Asp254′ (equivalent to Asp275 and Asp252′ of PmIDH, respectively) instead of coordinating with the metal ion and, correspondingly, the C2 atom of ICT moves closer to the small domain by ∼0.6 Å. Also, the side chain of Asp255′ has a different orientation whose equivalent residue Asp253 in the PmIDH–ICT–Mn complex serves as a ligand for the metal ion. Reorientation of the side chain of Tyr140 results in the loss of its interaction with the β-carboxylate group of ICT. Overall, these results indicate that the binding of the metal ion can induce subtle but important conformational changes of the residues at the active site.
Idp1p assumes an open conformation in the Idp1p–NADP complex, but a quasi-closed conformation in the Idp1p–ICT complex. Superposition of the two complexes based on the small and clasp domains yields an RMSD of 0.87 Å for the small and clasp domains (178 Cα atom pairs) and 7.4 Å for the large domain (215 Cα atom pairs), indicating a marked conformational change of the large domain (Fig. 2A,B; Table 2). Compared with that in the NADP-bound complex, the large domain in the ICT-bound complex rotates ∼17.1° toward the small and clasp domains along two hinge regions (residues 118–124 and residues 284–287) while maintaining the integrity of its secondary structural elements. Meanwhile, several residues of the large domain move substantially closer toward ICT to have direct interactions with the substrate. In particular, Thr78 and Ser95 on the two loops flanking helix α4 (designated as loop A and loop B, respectively) both move by 6.5 Å, and Asn97 and Arg101 on helix α5 move by 5.8 Å and 5.0 Å, respectively (Fig. 2B). Arg101 rotates its side chain toward the substrate and forms hydrogen bonds with both the α- and β-carboxylates of ICT. These movements result in a partial closure of the active site, which is reflected by a significantly narrowed width of the ICT-binding site (11.7 Å in the Idp1p–ICT complex vs. 18.0 Å in the Idp1p–NADP complex) (Table 2). These results suggest that the binding of ICT can cause conformational changes of the active site and the large domain even though the exact details of these conformational changes are still unclear due to the lack of a structure of apo Idp1p for comparison.
Structure of the Idp1p–NADPH–AKG–Ca complex
The structure of the Idp1p–NADPH–AKG–Ca complex represents the enzymatic state after the catalysis. In contrast with the structure of the EcIDH–NADPH–AKG–Ca complex in which the nicotinamide ring and the phosphate moiety of NADPH were not observed (Stoddard and Koshland Jr.1993), the bound NADPH, AKG, and the metal ion are well defined with high-quality electron density (Supplemental Fig. S1D). The enzymatic activity of NADP-IDHs requires the participation of a divalent metal ion such as Mn2+ or Mg2+ but is inhibited by Ca2+ (Stoddard et al. 1993). In the Idp1p–NADPH–AKG–Ca complex, the Ca2+ ion is bound at the active site and coordinated by one α-carboxylate oxygen and the carbonyl group of AKG, one carboxylate oxygen of Asp277, two carboxylate oxygens of Asp281, and one carboxylate oxygen of Asp254′ in an octahedral geometry similar to those observed in other known structures of NADP-IDHs (Fig. 1E; Hurley et al. 1991; Stoddard et al. 1993; Ceccarelli et al. 2002; Xu et al. 2004).
Compared with the open Idp1p–NADP complex and the quasi-closed Idp1p–ICT complex, in the Idp1p–NADPH–AKG–Ca complex the enzyme assumes a closed overall conformation with a closed active site, and more residues of the small domains of both subunits participate in the binding of NADPH and AKG (Fig. 1E; Supplemental Table S3). Specifically, in addition to its interaction with His317, the 2′-phosphomonoester group of NADPH forms one hydrogen bond each with the side chains of Gln259′ and Lys262′. The 3′-OH group of the adenosine ribose also forms a hydrogen bond with the side chain of Gln259′. The second phosphate group forms indirect hydrogen bonds with the main-chain amide of Leu252′ and the side chain of Asp255′ through a conserved water molecule. The nicotinamide ribose moiety undergoes notable changes due to the presence of AKG: The 2′-OH group of the ribose makes interactions with the main-chain amide of Thr78 and the γ-carboxylate group of AKG instead of Arg316; the amide group of the nicotinamide moiety forms hydrogen bonds with the side chain of Asn97 and the α-carboxylate group of AKG instead of Thr76; and its carbonyl group forms two hydrogen bonds with the main-chain amide of Thr76 and the side chain of Lys73 instead of Glu308. The position of AKG also differs slightly from that of ICT in the Idp1p–ICT complex. The α-carboxylate group forms hydrogen bonds with the amide group of the nicotinamide moiety of NADPH and the side chains of Arg101 and Arg110, and is coordinated with the Ca2+ ion. The carbonyl group is also coordinated with the Ca2+ ion. The γ-carboxylate group forms hydrogen bonds with the side chains of Thr78, Ser95, and Asn97.
Since the structure of the pseudo-Michaelis complex is not available, we utilize the structure of the HcIDH–NADP–ICT–Ca complex as a representative of the corresponding states of eukaryotic NADP-IDHs for comparison with other enzymatic states. Structural comparison between the HcIDH–NADP–ICT–Ca complex and the Idp1p–NADPH–AKG–Ca complex indicates that the two enzymes adopt very similar closed conformations of the overall structure and the active site (an RMSD of 0.65 for 385 Cα atom pairs), and the bound ligands and metal ion maintain comparable interactions with the surrounding residues of the enzyme (Fig. 2C). Nevertheless, there exist several minor differences at the active site. Firstly, the position of AKG differs slightly from that of ICT: Although both the α- and γ-carboxylates of AKG maintain positions similar to those of ICT, loss of the β-carboxylate group and its interactions with the enzyme leads to positional displacements of atoms C3 and C4. Additionally, the coordination geometry of the Ca2+ ion in the two complexes are slightly different (octahedral vs. distorted pentagonal bipyramidal): In the Idp1p–NADPH–AKG–Ca2+ complex the Ca2+ ion forms an extra hydrogen bond with Oδ2 of Asp281 (equivalent to Asp279 of HcIDH) but has no interaction with the main-chain carbonyl oxygen of Asp277 (equivalent to Asp275 of HcIDH) and water molecule. These results suggest that during the catalysis the enzyme does not undergo significant conformational changes except for minor conformational adjustments of the active site.
On the other hand, structural comparison of the HcIDH–NADP–ICT–Ca complex with the Idp1p–NADP and Idp1p–ICT complexes shows substantial conformational differences of the large domain. Compared with the Idp1p–NADP complex, the large domain in the quaternary complex of HcIDH has a 23.5° rotation along the hinge regions toward the small and clasp domains, and both NADP- and ICT-binding sites are significantly narrowed. Consequently, NADP makes more interactions with the surrounding residues. Similarly, compared with the Idp1p–ICT complex, the large domain has a 8.7° rotation toward the small and clasp domains, but only the NADP-binding site is markedly narrowed whereas the ICT-binding site shows no major conformational change (Fig. 2D). Due to the binding of NADP, the active site of HcIDH is covered up with the side chain of Arg314, and ICT adopts a slightly different conformation by rotating toward the clasp domain with the C4 atom moving by 2.4 Å. These results suggest that further binding of ICT to the Idp1p–NADP complex would induce substantial conformational changes of both NADP- and ICT-binding sites, whereas additional binding of NADP to the Idp1p–ICT complex would cause only major conformational changes of the NADP-binding site.
Structure of the Idp1p–AKG complex
The structure of the Idp1p–AKG complex represents the intermediate state of the enzyme after the release of NADPH post catalysis. Structural comparison indicates that the overall structure of the Idp1p–AKG complex assumes the quasi-closed conformation very similar to that of the Idp1p–ICT complex (an RMSD of 0.27 Å for the small and clasp domains and 1.4 Å for the large domain, respectively) (Fig. 2A; Table 2). However, the widths of both NADP- and ICT-binding sites are slightly wider than those in the Idp1p–ICT complex (Fig. 2E), and the positions of AKG and ICT are slightly different. Compared with the Idp1p–NADPH–AKG–Ca complex, several structural elements (including helices α4 and α5) at the active site show moderate movements away from the small and clasp domains, leading to a partial expansion of the active site (in particular the NADP-binding site) (Fig. 2F). Subsequently, Arg316 moves away from the small and clasp domains and loses its interactions with residues of the adjacent subunit as seen in the quaternary complex. In addition, the position of AKG is also notably different in the two complexes and the interactions between AKG and the surrounding residues are changed (Supplemental Fig. S2). Specifically, the α-carboxylate group forms a salt bridge with Arg110 and hydrogen bonds with the side chains of Arg101 and Glu308, but loses interactions with NADPH and Ca2+; the carbonyl group forms a hydrogen bond with the side chain of Arg133 instead of Ca2+; and the γ-carboxylate group forms two hydrogen bonds with the side chains of Ser95 and Asn97, rather than forming interactions with Thr78 (Fig. 1F; Supplemental Table S4). These results indicate that, after the release of NADPH, the enzyme has transformed from the closed conformation back to the quasi-closed conformation and, consequently, the interactions of the product with the enzyme are weakened.
Discussion
Eukaryotic NADP-IDHs can be found in mitochondria, cytoplasm, and peroxisomes and most likely serve distinct biological functions. Sequence comparison shows that eukaryotic NADP-IDHs share a moderate level of sequence identity among themselves (>60%), but a low level of sequence identity from EcIDH (<20%). Previous structural and biochemical studies have shown that, although eukaryotic NADP-IDHs (PmIDH and HcIDH) have overall structures similar to that of EcIDH, there are nonetheless substantial conformational differences in the overall structure and at the active site. Thus, we suspected that eukaryotic NADP-IDHs might adapt these changes in the recognition and binding of the substrate and coenzyme and the catalytic mechanism during the evolution process. To address this issue, we determined the crystal structures of Idp1p in binary complexes with NADP, ICT, and AKG, and in a quaternary complex with NADPH, AKG, and Ca2+, which represent several intermediate states of the enzyme during the catalytic reaction.
Structural analyses of the Idp1p complexes as well as the previously reported PmIDH and HcIDH complexes enable us not only to define the structural elements and key residues involved in the recognition and binding of the coenzyme, substrate, and product, but also to reveal the conformational changes of the enzyme associated with the binding or release of the ligands. In contrast with EcIDH, eukaryotic NADP-IDHs display substantial conformational differences among different enzymatic states of the catalytic reaction. Specifically, several structural elements composing the active site (particularly helices α4 and α5) make marked conformational changes accompanying the binding or release of the coenzyme, substrate, and product, leading to the rotation of the large domain toward or outward the small and clasp domains and thus the formation of at least three distinct overall conformations of the enzyme: an open conformation in the NADP-bound complexes, a quasi-closed conformation in the ICT- and AKG-bound complexes, and a fully closed conformation in the pre- and post-turnover complexes. Due to the unavailability of an apo eukaryotic NADP-IDH structure, it is unknown whether NADP binding would cause any conformational change of the enzyme. However, comparison with the apo TmIDH suggests that the binding of NADP most likely causes moderate conformational changes of the active site and the overall structure. On the other hand, it is clear that the binding of ICT causes marked conformational changes of the active site and the overall structure, and the further binding of NADP to the ICT-bound complex or ICT to the NADP-bound complex induces additional conformational changes of the active site and the overall structure. During the catalysis, no major conformational changes occur either at the active site or in the overall structure. After the catalysis, the enzyme retains the closed conformation when both NADPH and AKG are bound but transforms back to the quasi-closed conformation after NADPH is released. It seems likely that the enzyme assumes an open conformation similar to that of the NADP-bound complex when only NADPH is bound.
Besides our work, thorough analyses of crystal structures representing different enzymatic states of NADP-IDH from a single organism have only been performed for EcIDH. Although, like EcIDH, eukaryotic NADP-IDHs also use a random sequential kinetic scheme to bind the coenzyme NADP and the substrate ICT (Northrop and Cleland 1974; Uhr et al. 1974; Hurley et al. 1991; Dean and Koshland Jr. 1993), the substantial conformational changes observed during the catalysis of isocitrate decarboxylation by eukaryotic NADP-IDHs are quite different from those observed for EcIDH. Specifically, EcIDH can adopt an open conformation and a closed conformation in the apo form but always takes a closed conformation in all ligand-bound complexes, and only subtle conformational changes occur in the vicinity of the active site during the catalytic reaction (Hurley et al. 1989, 1991; Stoddard and Koshland Jr. 1993; Stoddard et al. 1993; Finer-Moore et al. 1997). Upon the binding of NADP, the nicotinamide ring and the phosphate backbone of the bound NADP are disordered, and these moieties appear to be flexible in order to allow further binding of ICT and metal ion in the closed conformation (Stoddard et al. 1993). The binding of ICT also stabilizes the enzyme in the closed conformation in which, however, the NADP-binding site is widely open and accessible by the coenzyme. Upon the binding of both NADP and ICT, no further conformational change is needed because the enzyme is already in the closed conformation and both the coenzyme and the substrate are properly positioned for catalysis. Compared with EcIDH, eukaryotic NADP-IDHs contain two insertions (residues 81–99 and 319–323 of Idp1p or residues 82–90 and 320–324 of HcIDH) forming two extra α-helices (α4 and α11) at the active site. These α-helices cover the top of the NADP- and ICT-binding sites and thus significantly narrow the entrance to the active site. As a result, in the NADP-bound complex, the enzyme has to adopt the open conformation to allow further binding of ICT. Similarly, in the ICT-bound complex, the enzyme has to assume the quasi-closed conformation so that the NADP-binding site can be accessed by the coenzyme. Therefore, upon the binding of both NADP and ICT, profound conformational changes are required for the proper positioning of the coenzyme, substrate, and metal ion at the active site for the catalysis to take place. These results indicate that the catalytic mechanism of eukaryotic NADP-IDHs is more complex than that of EcIDH and involves more fine-tuned conformational changes of the active site and the overall structure. These differences may hold between other subfamily I and subfamily II dimeric NADP-IDHs.
Materials and Methods
Cloning, expression, and purification
The Idp1 gene used for clone construction and protein expression was amplified by PCR from genomic DNA of S. cerevesiae (strain YPH499) with the forward PCR primer 5′-CCCCCGGGGCATATGGCTTTCAGTAAGATTAAGGTC-3′ and the reverse primer 5′-GCACTCGAGTTACTCGATCGACTTGATTTCTTT-3′. The N-terminal 15 residues (MSMLSRRLFSTSRLA) which are suggested to be the signal peptide for the enzyme's translocation into mitochondria (Haselbeck and McAlister-Henn 1991) were purposely removed during construction to facilitate crystallization. The gene fragment was inserted into the SmaI and XhoI restriction sites of the pET-3E-His expression plasmid. The plasmid was transformed into E. coli BL21 (DE3) pLysS strain (Novagen). When the culture of the transformed cells reached an OD600 of 0.6 ∼0.8, protein expression was induced by 0.1 mM IPTG for 18 h at 16°C. The bacterial cells were harvested by centrifugation and lysed by sonication in the lysis buffer (20 mM Tris-HCl, pH 7.4, 500 mM NaCl, 10% glycerol, and 7.2 mM β-ME) supplemented with 1 mM PMSF.
The target protein Idp1p was purified by affinity chromatography using a Ni-NTA column (Qiagen) with the lysis buffer supplemented with 20, 40, and 200 mM imidazole serving as washing buffer 1, washing buffer 2, and elution buffer, respectively. The elution fractions containing the target protein were dialyzed against the storage buffer (20 mM Tris-HCl, pH 7.0, 100 mM NaCl, and 7.2 mM β-ME) for 5 h, and then concentrated to ∼40 mg/mL for crystallization. The purification and dialysis processes were carried out at 4°C to reduce potential proteolysis and denaturation of the target protein. SDS-PAGE analysis of the purified protein shows a single band with molecular mass of ∼48 kDa. Dynamic light scattering analysis confirms the purity and homogeneity of the protein and reveals the dimeric state of the protein in solution.
Crystallization and diffraction data collection
Crystallization was carried out at 20°C using the hanging-drop vapor diffusion method. Attempts to grow crystals of apo Idp1p were unsuccessful because later structure determination shows that the crystals grown from the crystallization solution containing the purified protein but without addition of the substrate ICT yielded a structure with an ICT molecule bound at the active site, indicating that the enzyme was copurified with ICT using the expression and purification procedures described above. However, the electron density for the bound ICT in this structure was not very good probably due to a partial occupancy. Thus, to obtain high-quality structure of the Idp1p–ICT complex, 10 mM DL-isocitrate was added into the storage buffer during dialysis. Similarly, to obtain structures of the other Idp1p complexes, the purification procedure was modified to facilitate formation of the respective complexes. For the Idp1p–NADP complex, 5 mM β-NADP sodium salt was added into the lysis and storage buffers. For the Idp1p–AKG and the Idp1p–NADPH–AKG–Ca2+ complexes, 5 mM α-ketoglutaric acid disodium salt was added into the lysis and storage buffers to replace the bound ICT.
Crystals of the Idp1p–ICT and Idp1p–AKG complexes were grown in drops with equal volumes of the protein solution (10 mg/mL) and the reservoir solution (0.1 M HEPES-Na, pH 7.2, 0.2 M Li2SO4, 0.1 M NaF, and 20% PEG 4000). Crystals of the Idp1p–NADP complex were grown in drops with equal volumes of the protein solution (5 mg/mL) and the reservoir solution (100 mM MES, pH 6.1, and 25% PEG 1500). Crystals of the Idp1p–NADPH–AKG–Ca complex were grown in drops with equal volumes of the protein solution (20 mg/mL) supplemented with 10 mM NADPH, 150 mM AKG, and 10 mM CaCl2, and the reservoir solution (100 mM Tris-HCl, pH 8.2, 25% PEG 4000, 0.2 M Na-acetate, and 4% 1,3-propanediol).
Diffraction data of the Idp1p–NADP, Idp1p–AKG, and Idp1p–NADPH–AKG–Ca complexes were collected from flash-cooled crystals at beamline NW12 or BL17A of Photon Factory, Japan, and processed using the HKL2000 program (Otwinowski and Minor 1997). Diffraction data of the Idp1p–ICT complex were collected from a flash-cooled crystal with a Rigaku R-AXIS IV++ diffractometer and processed using the CrystalClear program (Pflugrath 1999). Statistics of the diffraction data are summarized in Table 1.
Structure determination and refinement
The structure of the Idp1p–NADP complex was solved with the molecular replacement (MR) method implemented in the program CNS (Brunger et al. 1998) using the structure of the HcIDH–NADP complex (PDB code 1T09) (Xu et al. 2004) as the search model. The structure of the Idp1p–ICT complex was solved using the structure of the PmIDH–ICT–Mn complex (PDB code 1LWD) (Ceccarelli et al. 2002) as the search model. The structures of the Idp1p–AKG and Idp1p–NADPH–AKG–Ca complexes were solved using the structure of the Idp1p–ICT complex as the search model. The initial structure refinement was carried out with the program CNS using standard protocols and the final structure refinement was performed with the maximum likelihood algorithm implemented in the program REFMAC5 (Murshudov et al. 1997). A free R factor monitor calculated with 5% of randomly chosen reflections and a bulk solvent correction were applied throughout the refinement. The monomers in the asymmetric unit were refined with strict NCS constraints in the early stage of refinement, but refined independently in the later stage. Model building was performed with the program O (Jones et al. 1991) and guided by SIGMAA-weighted 2Fo − Fc and Fo − Fc maps and composite-omit maps. In the initial difference Fourier maps, there was strong electron density at the active site that matches well the bound NADP/NADPH, ICT, AKG, and metal ion in their respective complexes (Supplemental Fig. S1). Electron density peaks in difference Fourier maps at a height of >2.5σ were assigned as water molecules if they had reasonable geometry in relation to hydrogen bond donors or acceptors and their B factors did not rise above 70 Å2 during subsequent refinements. A summary of the structure refinement statistics is given in Table 1.
Electronic supplemental material
The Supplemental material provides additional details on the following: (1) possible salt bridges and hydrogen bonds between the bound ligands and the enzyme at the active site of the respective complexes of Idp1p (Supplemental Tables S1–4); (2) composite-omit electron density maps of the bound ligands (Supplemental Fig. S1); and (3) structural differences at the AKG-binding site between the Idp1p–AKG and Idp1p–NADPH–AKG–Ca complexes (Supplemental Fig. S2).
Protein Data Bank deposition
The atomic coordinates and structure factors of the Idp1p–NADP, Idp1p–ICT, Idp1p–NADPH–AKG–Ca, and Idp1p–AKG complexes have been deposited with the Protein Data Bank under accession codes 2QFV, 2QFW, 2QFX, and 2QFY, respectively.
Acknowledgments
We thank Dr. Jinqiu Zhou for providing us with the genomic DNA of S. cerevesiae. We thank the staff members at Photon Factory, Japan, for technical support in diffraction data collection. This work was supported by grants from the Ministry of Science and Technology of China (2006AA02A313, 2006AA02Z112, and 2007CB914302), the Natural Science Foundation of China (30570379, 30770480, and 30730028), and the Science and Technology Commission of Shanghai Municipality (07XD14032 and 07ZR14131).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Jianping Ding, Institute of Biochemistry and Cell Biology, Shanghai Insitutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China; e-mail: jpding@sibs.ac.cn; fax: 086-21-5492-1116.
Abbreviations: IDH, isocitrate dehydrogenase; ICT, isocitrate; AKG, α-ketoglutarate; NADP-IDH, NADP-dependent IDH; DpIDH, Desulfotalea psychrophila NADP-IDH; EcIDH, Escherichia coli NADP-IDH; PmIDH, porcine mitochondrial NADP-IDH; HcIDH, human cytosolic NADP-IDH; TmIDH, Thermotoga maritima NADP-IDH.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035675.108.
References
- Bolduc, J.M., Dyer, D.H., Scott, W.G., Singer, P., Sweet, R.M., Koshland D.E., Jr, Stoddard, B.L. Mutagenesis and Laue structures of enzyme intermediates: Isocitrate dehydrogenase. Science. 1995;268:1312–1318. doi: 10.1126/science.7761851. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ceccarelli, C., Grodsky, N.B., Ariyaratne, N., Colman, R.F., Bahnson, B.J. Crystal structure of porcine mitochondrial NADP+-dependent isocitrate dehydrogenase complexed with Mn2+ and isocitrate. Insights into the enzyme mechanism. J. Biol. Chem. 2002;277:43454–43462. doi: 10.1074/jbc.M207306200. [DOI] [PubMed] [Google Scholar]
- Contreras-Shannon, V., Lin, A.P., McCammon, M.T., McAlister-Henn, L. Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+-specific isocitrate dehydrogenases. J. Biol. Chem. 2005;280:4469–4475. doi: 10.1074/jbc.M410140200. [DOI] [PubMed] [Google Scholar]
- Dean, A.M., Koshland D.E., Jr Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry. 1993;32:9302–9309. doi: 10.1021/bi00087a007. [DOI] [PubMed] [Google Scholar]
- Fedøy, A.E., Yang, N., Martinez, A., Leiros, H.K., Steen, I.H. Structural and functional properties of isocitrate dehydrogenase from the psychrophilic bacterium Desulfotalea psychrophila reveal a cold-active enzyme with an unusual high thermal stability. J. Mol. Biol. 2007;372:130–149. doi: 10.1016/j.jmb.2007.06.040. [DOI] [PubMed] [Google Scholar]
- Finer-Moore, J., Tsutakawa, S.E., Cherbavaz, D.R., LaPorte, D.C., Koshland D.E., Jr, Stroud, R.M. Access to phosphorylation in isocitrate dehydrogenase may occur by domain shifting. Biochemistry. 1997;36:13890–13896. doi: 10.1021/bi9711691. [DOI] [PubMed] [Google Scholar]
- Frederiks, W.M., Kummerlin, I.P., Bosch, K.S., Vreeling-Sindelarova, H., Jonker, A., Van Noorden, C.J. NADPH production by the pentose phosphate pathway in the zona fasciculata of rat adrenal gland. J. Histochem. Cytochem. 2007;55:975–980. doi: 10.1369/jhc.7A7222.2007. [DOI] [PubMed] [Google Scholar]
- Geisbrecht, B.V., Gould, S.J. The human PICD gene encodes a cytoplasmic and peroxisomal NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- Haselbeck, R.J., McAlister-Henn, L. Isolation, nucleotide sequence, and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J. Biol. Chem. 1991;266:2339–2345. [PubMed] [Google Scholar]
- Henke, B., Girzalsky, W., Berteaux-Lecellier, V., Erdmann, R. IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the β-oxidation of unsaturated fatty acids. J. Biol. Chem. 1998;273:3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., Thorsness, P.E., Ramalingam, V., Helmers, N.H., Koshland D.E., Jr, Stroud, R.M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl. Acad. Sci. 1989;86:8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J.H., Dean, A.M., Sohl, J.L., Koshland D.E., Jr, Stroud, R.M. Regulation of an enzyme by phosphorylation at the active site. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., Dean, A.M., Koshland D.E., Jr, Stroud, R.M. Catalytic mechanism of NADP+-dependent isocitrate dehydrogenase: Implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry. 1991;30:8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- Imabayashi, F., Aich, S., Prasad, L., Delbaere, L.T. Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase. Proteins. 2006;63:100–112. doi: 10.1002/prot.20867. [DOI] [PubMed] [Google Scholar]
- Jo, S.H., Son, M.K., Koh, H.J., Lee, S.M., Song, I.H., Kim, Y.O., Lee, Y.S., Jeong, K.S., Kim, W.B., Park, J.W., et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Karlstrom, M., Stokke, R., Steen, I.H., Birkeland, N.K., Ladenstein, R. Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability. J. Mol. Biol. 2005;345:559–577. doi: 10.1016/j.jmb.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Karlstrom, M., Steen, I.H., Madern, D., Fedoy, A.E., Birkeland, N.K., Ladenstein, R. The crystal structure of a hyperthermostable subfamily II isocitrate dehydrogenase from Thermotoga maritima . FEBS J. 2006;273:2851–2868. doi: 10.1111/j.1742-4658.2006.05298.x. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Park, J.W. Cellular defense against singlet oxygen-induced oxidative damage by cytosolic NADP+-dependent isocitrate dehydrogenase. Free Radic. Res. 2003;37:309–316. doi: 10.1080/1071576021000050429. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Kang, B.S., Park, J.W. Cellular defense against heat shock-induced oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. Free Radic. Res. 2005;39:441–448. doi: 10.1080/10715760500066265. [DOI] [PubMed] [Google Scholar]
- Kim, S.J., Yune, T.Y., Han, C.T., Kim, Y.C., Oh, Y.J., Markelonis, G.J., Oh, T.H. Mitochondrial isocitrate dehydrogenase protects human neuroblastoma SH-SY5Y cells against oxidative stress. J. Neurosci. Res. 2007;85:139–152. doi: 10.1002/jnr.21106. [DOI] [PubMed] [Google Scholar]
- Koh, H.J., Lee, S.M., Son, B.G., Lee, S.H., Ryoo, Z.Y., Chang, K.T., Park, J.W., Park, D.C., Song, B.J., Veech, R.L., et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J. Biol. Chem. 2004;279:39968–39974. doi: 10.1074/jbc.M402260200. [DOI] [PubMed] [Google Scholar]
- LaPorte, D.C. The isocitrate dehydrogenase phosphorylation cycle: Regulation and enzymology. J. Cell. Biochem. 1993;51:14–18. doi: 10.1002/jcb.240510104. [DOI] [PubMed] [Google Scholar]
- Lee, S.M., Koh, H.J., Park, D.C., Song, B.J., Huh, T.L., Park, J.W. Cytosolic NADP+-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic. Biol. Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Kim, S.Y., Kil, I.S., Park, J.W. Regulation of ionizing radiation-induced apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2007;282:13385–13394. doi: 10.1074/jbc.M700303200. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Minard, K.I., McAlister-Henn, L. Dual compartmental localization and function of mammalian NADP+-specific isocitrate dehydrogenase in yeast. Arch. Biochem. Biophys. 2008;472:17–25. doi: 10.1016/j.abb.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux, R.J., Beriault, R., Lemire, J., Singh, R., Chenier, D.R., Hamel, R.D., Appanna, V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE. 2007;2:e690. doi: 10.1371/journal.pone.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai, E., Banhegyi, G. Isocitrate dehydrogenase: A NADPH-generating enzyme in the lumen of the endoplasmic reticulum. Arch. Biochem. Biophys. 2008;471:184–190. doi: 10.1016/j.abb.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Marino, D., Gonzalez, E.M., Frendo, P., Puppo, A., Arrese-Igor, C. NADPH recycling systems in oxidative stressed pea nodules: A key role for the NADP+-dependent isocitrate dehydrogenase. Planta. 2007;225:413–421. doi: 10.1007/s00425-006-0354-5. [DOI] [PubMed] [Google Scholar]
- Mas, M.T., Colman, R.F. Phosphorus-31 nuclear magnetic resonance studies of the binding of nucleotides to NADP+-specific isocitrate dehydrogenase. Biochemistry. 1984;23:1675–1683. doi: 10.1021/bi00303a015. [DOI] [PubMed] [Google Scholar]
- Minard, K.I., McAlister-Henn, L. Dependence of peroxisomal β-oxidation on cytosolic sources of NADPH. J. Biol. Chem. 1999;274:3402–3406. doi: 10.1074/jbc.274.6.3402. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A., Hillis, D.M., Patton, J.C., Bradley, R.D., Baker, R.J. Cytosolic isocitrate dehydrogenase in humans, mice, and voles and phylogenetic analysis of the enzyme family. Mol. Biol. Evol. 1998;15:1674–1684. doi: 10.1093/oxfordjournals.molbev.a025894. [DOI] [PubMed] [Google Scholar]
- Northrop, D.B., Cleland, W.W. The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. II. Dead-end and multiple inhibition studies. J. Biol. Chem. 1974;249:2928–2931. [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Singh, S.K., Matsuno, K., LaPorte, D.C., Banaszak, L.J. Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 Å. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 2001;276:26154–26163. doi: 10.1074/jbc.M101191200. [DOI] [PubMed] [Google Scholar]
- Singh, S.K., Miller, S.P., Dean, A., Banaszak, L.J., LaPorte, D.C. Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 2002;277:7567–7573. doi: 10.1074/jbc.M107908200. [DOI] [PubMed] [Google Scholar]
- Steen, I.H., Madern, D., Karlstrom, M., Lien, T., Ladenstein, R., Birkeland, N.-K. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 2001;276:43924–43931. doi: 10.1074/jbc.M105999200. [DOI] [PubMed] [Google Scholar]
- Stoddard, B.L., Koshland D.E., Jr Structure of isocitrate dehydrogenase with α-ketoglutarate at 2.7 Å resolution: Conformational changes induced by decarboxylation of isocitrate. Biochemistry. 1993;32:9317–9322. doi: 10.1021/bi00087a009. [DOI] [PubMed] [Google Scholar]
- Stoddard, B.L., Dean, A., Koshland D.E., Jr Structure of isocitrate dehydrogenase with isocitrate, nicotinamide adenine dinucleotide phosphate, and calcium at 2.5 Å resolution: A pseudo-Michaelis ternary complex. Biochemistry. 1993;32:9310–9316. doi: 10.1021/bi00087a008. [DOI] [PubMed] [Google Scholar]
- Stokke, R., Karlstrom, M., Yang, N., Leiros, I., Ladenstein, R., Birkeland, N.K., Steen, I.H. Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers. Extremophiles. 2007;11:481–493. doi: 10.1007/s00792-006-0060-z. [DOI] [PubMed] [Google Scholar]
- Uhr, M.L., Thompson, V.W., Cleland, W.W. The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. I. Initial velocity, substrate and product inhibition, and isotope exchange studies. J. Biol. Chem. 1974;249:2920–2927. [PubMed] [Google Scholar]
- Valderrama, R., Corpas, F.J., Carreras, A., Gomez-Rodriguez, M.V., Chaki, M., Pedrajas, J.R., Fernandez-Ocana, A., Del Rio, L.A., Barroso, J.B. The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant Cell Environ. 2006;29:1449–1459. doi: 10.1111/j.1365-3040.2006.01530.x. [DOI] [PubMed] [Google Scholar]
- van Roermund, C.W., Hettema, E.H., Kal, A.J., van den Berg, M., Tabak, H.F., Wanders, R.J. Peroxisomal β-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: Isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 1998;17:677–687. doi: 10.1093/emboj/17.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Zhao, J., Xu, Z., Peng, B., Huang, Q., Arnold, E., Ding, J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- Yasutake, Y., Watanabe, S., Yao, M., Takada, Y., Fukunaga, N., Tanaka, I. Structure of the monomeric isocitrate dehydrogenase: Evidence of a protein monomerization by a domain duplication. Structure. 2002;10:1637–1648. doi: 10.1016/s0969-2126(02)00904-8. [DOI] [PubMed] [Google Scholar]
- Yasutake, Y., Watanabe, S., Yao, M., Takada, Y., Fukunaga, N., Tanaka, I. Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: Insight into the cofactor recognition, catalysis, and evolution. J. Biol. Chem. 2003;278:36897–36904. doi: 10.1074/jbc.M304091200. [DOI] [PubMed] [Google Scholar]
- Yoshihara, T., Hamamoto, T., Munakata, R., Tajiri, R., Ohsumi, M., Yokota, S. Localization of cytosolic NADP-dependent isocitrate dehydrogenase in the peroxisomes of rat liver cells: Biochemical and immunocytochemical studies. J. Histochem. Cytochem. 2001;49:1123–1131. doi: 10.1177/002215540104900906. [DOI] [PubMed] [Google Scholar]