Abstract
Bacillus anthracis is well known in connection with biological warfare. The search for new drug targets and antibiotics is highly motivated because of upcoming multiresistant strains. Thymidylate kinase is an ideal target since this enzyme is at the junction of the de novo and salvage synthesis of dTTP, an essential precursor for DNA synthesis. Here the expression and characterization of thymidylate kinase from B. anthracis (Ba-TMPK) is presented. The enzyme phosphorylated deoxythymidine-5′-monophosphate (dTMP) efficiently with K m and V max values of 33 μM and 48 μmol mg−1 min−1, respectively. The efficiency of deoxyuridine-5′-monophosphate phosphorylation was ∼10% of that of dTMP. Several dTMP analogs were tested, and D-FMAUMP (2′-fluoroarabinosyl-5-methyldeoxyuridine-5′-monophosphate) was selectively phosphorylated with an efficiency of 172% of that of D-dTMP, but l-FMAUMP was a poor substrate as were 5-fluorodeoxyuridine-5′-monophosphate (5FdUMP) and 2′,3′-dideoxy-2′,3′-didehydrothymidine-5′-monophosphate (d4TMP). No activity could be detected with 3′-azidothymidine-5′-monophosphate (AZTMP). The corresponding nucleosides known as efficient anticancer and antiviral compounds were also tested, and d-FMAU was a strong inhibitor with an IC50 value of 10 μM, while other nucleosides—l-FMAU, dThd, 5-FdUrd, d4T, and AZT, and 2′-arabinosylthymidine—were poor inhibitors. A structure model was built for Ba-TMPK based on the Staphylococcus aureus TMPK structure. Docking with various substrates suggested mechanisms explaining the differences in substrate selectivity of the human and the bacterial TMPKs. These results may serve as a start point for development of new antibacterial agents.
Keywords: Bacillus anthracis, thymidylate kinase, TMPK, structure model, nucleoside analogs, drug design, FMAU
Bacillus anthracis, a Gram-positive, spore-forming bacterium that causes anthrax, is primarily an animal pathogen but can infect humans and be fatal (Baillie and Read 2001). B. anthracis has been used as a potential biological weapon recently in connection with the events of 11 September 2001 (Spencer 2003). At least 17 nations and some autonomous terrorist groups are thought to have or have had offensive biological weapons programs (Inglesby et al. 1999). As such, anthrax is considered a substantial threat.
The regular treatment and prophylaxis of inhalation anthrax rely on antibiotics such as ciprofloxacin, fluoroquinolones, and doxycycline. There are also vaccines available, but they suffer from problems such as relatively high costs of production and associated transient side effects (Spencer 2003). There is also a danger for upcoming antibiotic resistance and thus a need for new antimicrobial agents with novel targets and mechanisms, which may successfully treat people exposed to or infected with drug-resistant strains of B. anthracis.
Most organisms synthesize their DNA precursors by either the de novo or the salvage pathways. The de novo synthesis involves initial synthesis of ribonucleotides, while the salvage pathway reuses nucleosides from DNA degradation. The two pathways vary in activity in different types of cells and during different stages of the cell cycle.
Thymidine 5′-monophosphate kinase (TMPK), expressed in all organisms, catalyzes the transfer of gamma phosphate from ATP to thymidine 5′-monophosphate (dTMP), yielding thymidine 5′-diphosphate (dTDP) and ADP. TMPK is the site where the de novo and salvage pathways meet in dTTP synthesis and, hence, is a good target for drug design.
Most nucleoside analogs used in antiviral and anticancer therapy require stepwise phosphorylation by cellular nucleoside/tide kinases to the respective triphosphates to exert their therapeutic effects. The poor recognition of 3′-azido-dTMP by human TMPK has been a bottleneck for anti-HIV therapy with the therapeutic nucleoside analog 3′-azidothymidine (AZT) (Lavie et al. 1998b). β-l-Nucleosides, such as β-l-(−)-2′,3′-dideoxy-3′-thiacytidine (3TC) and 1-(2′-deoxy-2′-fluoro-β-l-arabinofuranosyl)-5-methyluracil (l-FMAU), have been recognized as potent and selective inhibitors of HIV and hepatitis B virus replication (Balakrishna Pai et al. 1996; Buti and Esteban 2003). Human TMPK selectively phosphorylates the d-enantiomer of dTMP and its analogs, for example, the efficiency for d-FMAUMP phosphorylation was 70-fold higher than that of l-FMAUMP (Hu et al. 2005; Alexandre et al. 2007). The enantioselectivity of nucleoside-activating enzymes most likely have a strong impact on the efficacy and specificity of new antimicrobial agents.
TMPK from B. anthracis Sterne strain (AAT52367) (Ba-TMPK) shares only 16% sequence identity with human TMPK (hTMPK), while the sequence identity to TMPK of Staphylococcus aureus (Sa-TMPK) is ∼50%. Nevertheless, all known TMPKs showed a highly conserved 3D fold (Lavie et al. 1998a,b; Ostermann et al. 2000; Li de la Sierra et al. 2001; Kotaka et al. 2006). Still, there are (structural) differences that could be used in drug design. For instance, studies with Mycobacterium tuberculosis TMPK have led to the identification of potent TMPK inhibitors, which also inhibited the growth of the organism (Vanheusden et al. 2002, 2003; Haouz et al. 2003; Pochet et al. 2003; Fioravanti et al. 2005; Van Daele et al. 2006, 2007).
Here, we describe the expression and characterization of TMPK from B. anthracis. Several dTMP analogs used as anticancer and antiviral drugs were tested as substrates. The nucleoside forms of these analogs, that is, 5-FdUrd, AZT, FMAU, and d4T, are used against cancer and viral infections (Furman et al. 1986; Balzarini et al. 1989; Balakrishna Pai et al. 1996; Chu et al. 1998; Marsh and McLeod 2001; Eriksson et al. 2002; Dawson and Lawrence 2004). The specificity of Ba-TMPK was investigated using analogs with substitutions at the 5-, 3′-, and 2′-positions, respectively. Structural models of Ba-TMPK in complex with several analogs were built based on the 3D structure of Sa-TMPK. These results were used to explain the substrate specificity and catalytic rates observed with the enzymes of human and bacterial origin. Specifically, the mechanism involved in stereoisomer selectivity has been addressed by a computational approach. These studies provide a basis for the future design of new agents against B. anthracis and related pathogenic bacteria.
Results
Expression and purification
Recombinant Ba-TMPK was expressed as an N-terminally His-tagged protein and purified in one step by metal affinity chromatography. Based on SDS-PAGE analysis, the final protein preparation was more than 95% pure (Fig. 1). The amount of recombinant Ba-TMPK was estimated to be ∼40% of the total protein, and the yield was ≈180 mg of pure protein per liter of culture. Purified Ba-TMPK was stable upon repeated freezing and thawing but started to lose activity after 2 d when stored at +4°C. The addition of detergents, for example, Triton X-100, 3-cyclohexylamino-1-propanesulfonic acid, did not affect Ba-TMPK activity.
Figure 1.
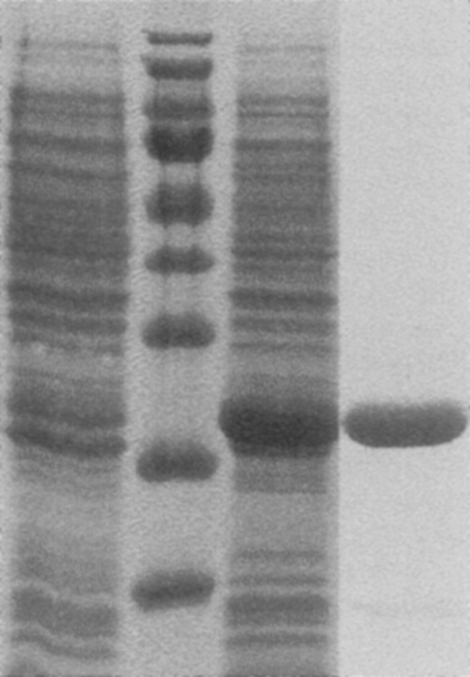
SDS-PAGE of Ba-TMPK. (From the left) Uninduced cell extract, marker, induced cell extract, purified Ba-TMPK. The bands in the marker correspond to ∼170, 130, 100, 70, 55, 40, 35, 25, and 15 kDa.
Purified Ba-TMPK was analyzed by size exclusion chromatography using a Superdex 200 column. The enzyme eluted as a single peak at a molecular size corresponding to 54 kDa, which suggested that Ba-TMPK is a dimer since the calculated molecular weight of the monomer is 26 kDa, including the His-tag.
Substrate specificity
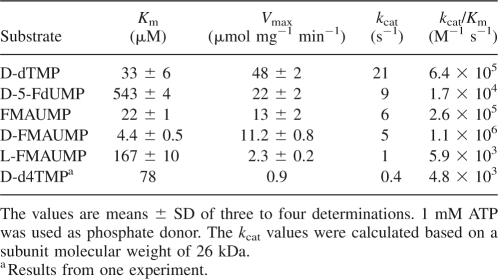
Ba-TMPK was thymidylate-specific and phosphorylated dTMP with high efficiency and to some extent dUMP. All other deoxyribonucleotides and ribonucleotides showed <4% activity (Table 1). dTMP phosphorylation followed the Michaelis-Menten kinetics with a K m value of 33 μM and a V max value of 48 μmol mg−1 min−1 of dTDP formed. This resulted in a high k cat value (21 s−1) and catalytic efficiency (k cat/K m = 6.4 × 105 M−1 s−1) (Table 2).
Table 1.
Phosphate acceptor specificity of purified recombinant Ba-TMPK
Table 2.
Kinetic parameters of purified recombinant Ba-TMPK
In addition to natural nucleoside monophosphates, several dTMP analogs were also tested. FMAUMP, as a racemic mixture, had a slightly lower K m value (22 μM) and a lower V max value (13 μmol mg−1 min−1), as compared with those of dTMP. The catalytic efficiency (k cat/K m) of this mixture was threefold lower than that of dTMP. When the individual enantiomers, that is, l- and d-FMAUMP were tested, l-FMAUMP had a K m value of 167 μM and V max value of 2.3 μmol mg−1 min−1, while d-FMAUMP had a K m value of 4.4 μM and V max value of 11 μmol mg−1 min−1, demonstrating that d-FMAUMP was a much better substrate for Ba-TMPK with an efficiency of 172% of that of dTMP. 5-FdUMP and d4TMP were poor substrates with high K m and low V max values. 5-FdUMP had a K m value about 16 times higher than that of dTMP. The rate-limiting step for d4TMP appeared instead to be the turnover number/rate (k cat), which was more than 50 times lower than that of dTMP (Table 2). AZTMP was also tested, but no activity could be detected (Table 1).
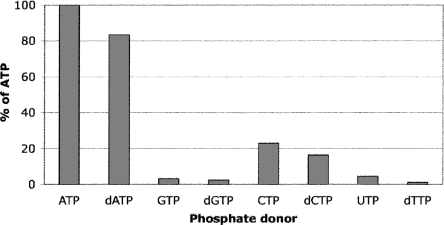
While ATP was used as phosphate donor to mimic physiological conditions, Ba-TMPK also accepted dATP as phosphate donor (83% relative activity of ATP), and other natural ribonucleotide and deoxyribonucleotide triphosphates were poor donors. The order of base preference was adenine > cytosine > guanine > uracil > thymine (Fig. 2). Interestingly, the end-product dTTP could be used as phosphate donor, ∼3% as compared with ATP.
Figure 2.
Phosphate acceptor specificity of purified recombinant Ba-TMPK. The phosphorylation rate with ATP was set to 100%. Phosphate donors and dTMP were 2 mM and 100 μM, respectively.
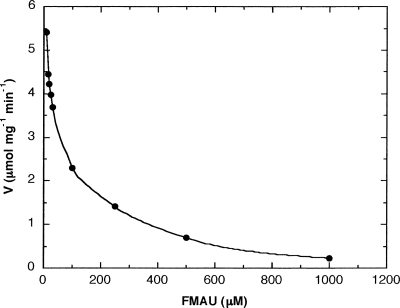
Some anticancer and antiviral pyrimidine nucleoside analogs were tested for their inhibitory effect on dTMP phosphorylation. Initially, a FMAU preparation, which is a mixture of d- and l-FMAU, was used and found to be the only nucleoside analog that gave a pronounced inhibition with an IC50 value of 22 ± 4 μM (Table 3). At 500 μM FMAU, <10% of the activity was found using 10 μM dTMP as substrate (Fig. 3). The individual enantiomers, that is, l- and d-FMAU, were also tested and d-FMAU strongly inhibited Ba-TMPK activity with an IC50 value of 10 μM, while l-FMAU did not produce any detectable inhibition (Table 3). All the other tested nucleoside analogs were poor inhibitors in the order of d4T > dThd > AZT > AraT > 5-FdUrd. The IC50 values for d4T, dThd, and AZT were between 200 and 700 μM, whereas the IC50 value of AraT was >1.5 mM. No inhibition was detected with 5-FdUrd (IC50 > 4 mM) (Table 3).
Table 3.
IC50 values of nucleosides as inhibitors with recombinant Ba-TMPK
Figure 3.
Activity of Ba-TMPK with dTMP in the presence of FMAU, 10 μM dTMP, and 1 mM ATP were used together with different concentrations of FMAU ranging from 10 to 1000 μM. The experiment was done three times, and the IC50 value was calculated to be 22 ± 4 μM.
The inhibitory effect of d- and l-FMAU toward human TMPK was also examined. l-FMAU did not inhibit hTMPK activity; instead, an unexpected ∼30% stimulation was observed. d-FMAU inhibited hTMPK activity only at concentrations >100 μM, and the IC50 value was >400 μM.
Structural model of Ba-TMPK
We have attempted to crystallize the Ba-TMPK but so far without success. Therefore, a structural model was built based on the Sa-TMPK structure (pdb code: 2CCJ) (Kotaka et al. 2006), since the two enzymes share ∼50% sequence identity. d-dTMP as well as l- and d- FMAUMP were docked in the active site of the Ba-TMPK model using the ArgusLab software (www.arguslab.com/arguslab40.htm). l- and d-FMAUMP were also docked into the hTMPK active site with this procedure.
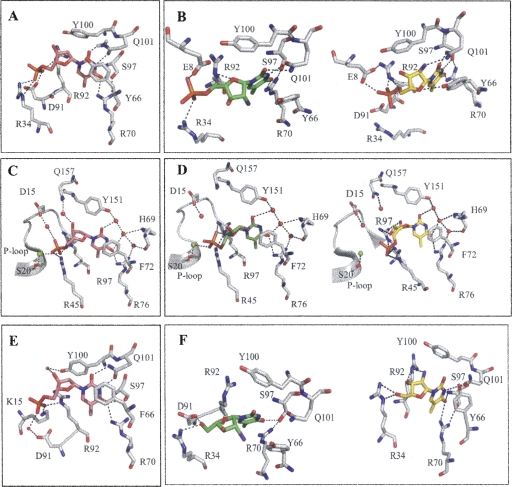
Similar to Sa-TMPK, the binding of d-dTMP to Ba-TMPK is through a network of hydrogen bonds to the pyrimidine ring involving residues R70, S97, and Q101 (Fig. 4A,E), and the base is stacked by an aromatic residue (Y66), which is also observed in all known TMPK structures (in hTMPK, it corresponds to F72) (Fig. 4C). Q101 interacts with O2 and N3 of the base, and R70 interacts with O4. The 3′-OH group interacts with Y100 via a water molecule. The phosphate group is tightly coordinated by R34, D91, and R92.
Figure 4.
Molecular docking of D-dTMP (in pink), L-FMAUMP (in green), and D-FMAUMP (in yellow) in the active site of Ba-TMPK and hTMPK. (A) D-dTMP complexed with Ba-TMPK; (B) L-FMAUMP and D-FMAUMP in complex with Ba-TMPK. Fluorine atom is colored purple. (C) D-dTMP in hTMPK active site. (D) L-FMAUMP and D-FMAUMP in complex with hTMPK. Magnesium ion is represented by a green sphere and water molecules are in red. (E) D-dTMP in SaTMPK active site. (F) L-FMAU (green) and D-FMAU (yellow) in the active site of Ba-TMPK.
The binding of l-FMAUMP and d-FMAUMP to Ba-TMPK is shown in Figure 4B. The pyrimidine ring is bound in the same fashion as for d-dTMP. Two additional hydrogen bonds are predicted between the 2′-fluorine of d-FMAUMP and the amino group of Q101, and between the O1′ from the arabinosyl moiety and the hydroxyl group from Y66. Binding of the 2′-fluorine and O1′ with Q101 and Y66, respectively, may be responsible for the increased binding affinity of d-FMAUMP to Ba-TMPKs. The stacking interaction is also enhanced because the base is closer to Y66. Altogether, this may account for the increased binding affinity for d-FMAUMP (K M (d-FMAUMP) = 4.4 μM as compared with the K M for d-dTMP = 33 μM). The binding of the phosphate groups is different as a hydrogen bond appears between E8 and the arabinose derivatives. The interaction between D91 and the phosphate moiety is missing as compared with d-dTMP (Fig. 4A). In the case of d-FMAUMP, R92 provides most likely two H-bonds with the phosphate group, whereas this interaction is missing in l-FMAUMP (Fig. 4B). This may explain why d-FMAUMP is a better substrate (K M (d-FMAUMP) = 4.4 μM and k cat = 6 s−1) than its l-enantiomer (K M (d-FMAUMP) = 167 μM and k cat = 1.0 s−1).
In the case of hTMPK, Y151, H69, and R76 are the main residues involved in the H-bond network to the pyrimidine ring. The 3′-OH of d-dTMP is bound to residue Q157 via a water molecule in the LID domain, and D15 and S20 from the P-loop play a key role in d-dTMP phosphate binding (Alexandre et al. 2007). The binding of the pyrimidine ring of d- and l-FMAUMP is similar to that of d-dTMP. However, R97 is able to bind both the 2′-F and 3′-OH of the arabinose of d-FMAUMP, which enhances its binding affinity to hTMPK. The phosphate moiety is closely bound to R45 instead of D15 and S20, explaining the reduced catalytic activity of d-FMAUMP as compared with d-dTMP. In case of l-FMAUMP, the 3′-OH is bound to the core domain via R45 but not to the LID domain (Q157 via a water molecule) as observed with the d-dTMP (Fig. 4C,D).
l- and d-FMAU were also docked onto Ba-TMPK active site using the same conditions as for the monophosphate forms (Fig. 4F). l-FMAU bound to the enzyme in a different fashion from the monophosphate counterpart. The pyrimidine ring is weakly bound to the active site. Only the O4 atom interacts with S97, while O2 and N3 are not involved in H-bond interactions. O1′ is maintained by R92, and the 5′-OH is interacting via H-bonds to R34 and D91. This weak binding may explain the high IC50 value observed (>5 mM). d-FMAU, on the other hand, is tightly bound to the active site since the base is stacked to Y66 very well, and N3 and O4 participate with H-bond interacting with R70, S97, and Q101. In addition, R92 interacts with the 2′-fluorine group and 3′-OH via two hydrogen bonds, and R34 is close enough to 5′-OH to establish two H-bonds. These interactions may explain why d-FMAU is an efficient inhibitor of Ba-TMPK (IC50 = 10 μM), although it lacks a 5′-phosphate group.
Discussion
Targeting thymidylate biosynthesis is an efficient way to block DNA synthesis, eventually leading to cell death. Because of its unique position in both the de novo and the salvage synthesis of dTTP, Ba-TMPK is a promising drug target for antibiotic development. Therefore, we have characterized Ba-TMPK with focus on its substrate specificity to get a deeper understanding of the enzyme and substrate interactions. The acquired knowledge concerning Ba-TMPK will also help in understanding the functional differences and similarities between TMPKs from other species.
All known TMPKs are dimers, and a classification of TMPKs has been proposed with respect to the location of specific residues in the active site (Lavie et al. 1998b). Class I TMPKs, for example, human and yeast TMPK, have an arginine in the P-loop interacting with the phosphate donor, while class II TMPKs, for example, Escherichia coli and Staphylococcus aureus, instead have a glycine in the same position. Additionally, class II enzymes have interacting basic residues, mostly arginines, in the LID region. Ba-TMPK was shown to be a dimer in this study and had the sequence characteristics of a class II TMPK.
The phosphate donor study showed that ATP/dATP and to a lower extent CTP/dCTP were accepted by Ba-TMPK, similar to Streptococcus pneumoniae TMPK (Sp-TMPK) (Petit and Koretke 2002), whereas other TMPKs use a broader set of phosphate donors. The Ba- and Sp-TMPK sequences do not show any special feature or motif at the amino acid level that could explain their narrow phosphate donor specificity. An alignment of TMPK sequences from eukaryotes, bacteria, and archaea demonstrates large sequence diversity between different organisms (Li de la Sierra et al. 2001).
Ba-TMPK was strictly thymidylate-specific and had low affinity for dUMP, in line with earlier publications with other TMPKs (Jong and Campbell 1984; Petit and Koretke 2002; Vanheusden et al. 2002). An exception is Vaccinia virus TMPK (vv-TMPK), which also phosphorylates dGMP, although at a fourfold lower rate (Topalis et al. 2005). The poor activity with dUMP for TMPK of B. anthracis and other organisms could be a safety mechanism to prevent a buildup of dUTP, which may cause replication mistakes. The very high K m value and almost 40 times lower efficiency for 5-FdUMP (compared to dTMP) further showed the sensitivity of Ba-TMPK for modifications at the 5 position of the base. In the modeled Ba-TMPK structure, the pyrimidine ring is located in the middle of a hydrophobic pocket delineated by five nonpolar residues (Y66, P36, H73, F93, and Y100) (Fig. 4A). The 5-methyl group contributes to the stability of the substrate/enzyme complex. Substitutions such as H (dUMP) or F (5FdUMP) are not likely to fit in this hydrophobic pocket. Therefore, dUMP and 5FdUMP are poor substrates.
The base moiety plays a key role in substrate recognition of Ba-TMPK, but the 2′- and 3′-positions on the sugar are also very important as demonstrated in this study. UMP is 10 times less efficient than dUMP. Thus, the 2′-position of the ribose does not tolerate any substitutions larger than a hydrogen atom. Concerning the arabinose derivatives, that is, FMAUMP, a 2′-arabinosyl fluoro substitution is advantageous since FMAUMP had a lower K m value (22 μM) as compared with dTMP (33 μM) and a catalytic efficiency ∼40% of that of dTMP. When the individual enantiomers were tested, it turned out that d-FMAUMP was the selective substrate but not the corresponding l-form. d-FMAUMP was the most efficient substrate and a highly promising substrate analog. As observed in the modeled d- and l-FMAUMP/Ba-TMPK binary complexes, the binding of the phosphate group to the enzyme is similar with E8 involved in both cases. It is the interactions with the sugar moiety in the two compounds that differ. The hydrogen bonding of the 2′-F of the arabinosyl moiety in d-FMAUMP to residue Q101 and O′1 to the hydroxyl group of residue Y66 contributed to its high affinity for the enzyme.
The 3′-position modifications are, in comparison with the 2′-position, more restricted since the tested 3′-analogs, d4TMP and AZTMP, were poor substrates for Ba-TMPK, as well as the corresponding nucleoside analogs d4T and AZT were poor inhibitors. The efficiency for d4TMP was <1% of that of dTMP, and AZTMP had no detectable activity. These results are in accordance with previous studies with hTMPK, Mt-TMPK, and vv-TMPK (Munier-Lehmann et al. 2001; Ostermann et al. 2003; Hu et al. 2005; Topalis et al. 2005). The lack of phosphorylation of AZTMP by Ba-TMPK could be an explanation for the poor growth inhibition of B. anthracis with AZT. AZT is readily phosphorylated by Ba-TK (Carnrot et al. 2006), but if AZTMP cannot be further phosphorylated by Ba-TMPK, for example, it will not reach its active form—AZTTP. Therefore, with AZT in the growth media, the effective dose (ED50) is high, >1 mM (Carnrot et al. 2006).
The enantioselectivity of hTMPK also favors the d-configuration of dTMP and its analogs as shown in recent studies with d- and l-dTMP as well as with d- and l-FMAUMP (Hu et al. 2005; Alexandre et al. 2007). The kinetic data show that d-FMAUMP (K m = 6 μM and V max = 2.6 μmol min−1 mg−1) is a much better substrate than l-FMAUMP (K m = 21.7 μM and V max = 0.12 μmol min−1 mg−1) (Hu et al. 2005). The selectivity of d-FMAUMP over l-FMAUMP is explained here in the modeled d- and l-FMAUMP/hTMPK complex. Both the 2′-F and 3′-OH of the d-FMAUMP interact with residue R97, but this is not the case with the l-configuration.
When nucleosides were tested as inhibitors toward Ba-TMPK, FMAU had an IC50 value that was almost identical to the K m value of FMAUMP, and of the two enantiomers, it was d-FMAU that inhibited the enzyme but not l-FMAU. These results strongly indicate that the substrate binding affinity is determined to a large extent by the nucleoside moiety and that the 5′-phosphate played a minor role in the case of d-FMAU and Ba-TMPK. This hypothesis is strengthened by docking of the two nucleosides in the active site of the Ba-TMPK model structure (Fig. 4F). However, as shown here, this was not the case with hTMPK since d-FMAU was not an inhibitor and l-FMAU instead apparently stimulated the hTMPK activity. These significant differences between human and bacterial TMPKs, which was also observed with the M. tuberculosis enzyme (Pochet et al. 2003), should be investigated further.
The phosphate group of dTMP provides much of the binding energy in catalysis carried out by TMPKs. An ideal drug candidate should not carry a charged group because of its difficulties in uptake through cell membranes. The differences observed in the inhibitory effect of d- and l-FMAU on Ba-TMPK and human TMPK suggest that there are differences in the binding mode of these nucleoside analogs to the active site of the enzymes. This difference may be advantageous in designing specific nonsubstrate inhibitors, for example, nucleoside analogs for Ba-TMPK with little or no effect on hTMPK. The development of narrow-spectrum rather than broad-spectrum antibiotics may reduce the risk of transfer of drug resistance between different bacterial species. This study provides a good starting point for future development of antibiotics against B. anthracis and other pathogenic bacteria such as S. aureus.
Materials and Methods
Materials
[2′,3′-3H]Thymidine-5′-monophosphate ([3H]dTMP; 34.6 Ci/mmol), (1-β-D)-2′-fluoroarabinosyl-5-methyldeoxyuridine (d-FMAU), and 2′,3′-dideoxy-2′,3′-didehydrothymidine-5′-monophosphate (d4TMP) were purchased from Moravek Biochemicals Inc. 5-Fluorodeoxyuridine (5-FdUrd), 5-fluorodeoxyuridine-5′-monophosphate (5-FdUMP), 3′-azidothymidine (AZT), 2′,3′-didehydrothymidine (d4T), 2′-arabinosylthymidine (AraT), deoxythymidine (dThd), and dTMP were from Sigma-Aldrich. 2′-Fluoroarabinosyl-5-methyldeoxyuridine (FMAU), a mixture of the d- and l-form, was a gift from J. Fox (Memorial Sloan Kettering Cancer Institute, New York). 3′-Azidothymidine-5′-monophosphate (AZTMP) was a gift from W. Tjarks (College of Pharmacy and Center for Microbial Interface Biology, Ohio State University). l-FMAU was kindly provided by Hee-Won Yoo (Bukwang Pharm. Co., Ltd). The FMAU 5′-monophosphates were synthesized enzymatically as described previously (Jacobsson et al. 1998).
Cloning and expression of the B. anthracis tmpk gene
The plasmid DNA containing the B. anthracis Sterne strain (34F2) tmpk gene in the pCR4-TOPO vector (Invitrogen) was kindly provided by A.J. Phipps (Department of Veterinary Biosciences, The Center for Microbial Interface Biology, Ohio State University, Columbus) (Byun et al. 2007). The tmpk gene was then subcloned into the pET-14b expression vector and transformed into the chemical competent E. coli strain, BL21(DE3) pLysS (Novagen), as previously described (Carnrot et al. 2006). The bacteria were cultured as previously described at 37°C to an OD600 of 0.6. Recombinant Ba-TMPK was expressed by induction for 6 h with 0.4 mM IPTG at 25°C. Purification of recombinant Ba-TMPK was done as previously described (Carnrot et al. 2003), and the enzyme was analyzed with SDS-PAGE. Human TMPK was expressed and purified as previously described (Alexandre et al. 2007).
Molecular weight estimation
Purified recombinant Ba-TMPK was applied to a Superdex 200 column (Amersham Biosciences, GE Healthcare) attached to an FPLC instrument (Amersham Biosciences, GE Healthcare). The column was equilibrated with 20 mM Tris-HCl pH 7.6, 0.2 M NaCl, 1 mM DTT, and 2 mM MgCl2. The column was initially calibrated with blue dextran (2000 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (42 kDa), and cytochrome c (12.4 kDa; Sigma-Aldrich). The molecular size of active Ba-TMPK was estimated by using the calibration curve.
Enzyme assays
TMPK activity was followed by the ADP production in a coupled enzyme system with pyruvate kinase and lactate dehydrogenase (Blondin et al. 1994). The standard reaction mixture contained 50 mM Tris-HCl pH 7.6, 2 mM MgCl2, 1 mM ATP, 5 mM DTT, 1 mM phosphoenolpyruvate, 2 units/mL pyruvate kinase, 2 units/mL lactate dehydrogenase, 100 μM NADH, and a final concentration of 0.5 μg/mL Ba-TMPK in a total volume of 1 mL. The reaction was performed at 37°C with a Cary 3 spectrophotometer (Varian Techtron). Phosphate donor specificity was determined by using 100 μM [3H]dTMP as substrate as described previously (Wang 2007). The reaction products were separated by thin layer chromatography and quantified by liquid scintillation counting (Beckman Coulter LS 6500).
The mean and the standard deviation (SD) were calculated, and the IC50 values were defined as the concentration of an inhibitor that decreases the enzyme maximum velocity with 50%. Kinetic parameters were calculated by using the Michaelis-Menten equation in KALEIDAGRAPH (Synergy Software).
Modeling studies
d- and l-FMAUMP as well as the corresponding nucleosides were drawn with Molsoft (http://www.molsoft, 2D to 3D converter) and Smiles Translator (http://cactus.nci.nih.gov/services/translate/) to produce the pdb file. The conformation of the sugar ring was checked by visualizing the molecules within the PyMOL graphic system (DeLano Scientific) and, if necessary, modified to the β-l configuration, as desired. Docking of the l- and d-FMAUMP and l- and d-FMAU and d-dTMP was performed using ArgusLab software (www.arguslab.com/arguslab40.htm). The binding site was defined from the coordinates of the ligand in the original PDB files 1E2D for hTMPK and a model of Ba-TMPk based on Sa-TMPK (pdb code: 2CCJ). Docking precision was set to “high,” and the “flexible ligand docking” mode was used for each docking run. Resulting complexes were visualized with the PyMOL graphic system.
Acknowledgments
This work was supported by grants from the Swedish Research Council for the Environment, Agricultural Sciences and Spatial Planning (FORMAS), and the Swedish Research Council. We thank Dominique Devill-Bonne for providing us with the human TMPK plasmid.
Footnotes
Reprint requests to: Staffan Eriksson, Department of Anatomy, Physiology and Biochemistry, The Swedish University of Agricultural Sciences, The Biomedical Centre, PO Box 575, S-751 23 Uppsala, Sweden; e-mail: staffan.eriksson@afb.slu.se; fax: 46-18-55-07-62.
Abbreviations: AraT, 2′-arabinosylthymidine; AZT, 3′-azidothymidine; AZTMP, 3′-azidothymidine-5′-monophosphate; d4T, 2′,3′-dideoxy-2′,3′-didehydrothymidine; dThd, deoxythymidine; d4TMP, 2′,3′-dideoxy-2′,3′-didehydrothymidine-5′-monophosphate; dTMP, thymidylate, thymidine-5′-monophosphate; dUMP, deoxyuridine-5′-monophosphate; 5-FdUMP, 5-fluorodeoxyuridine-5′-monophosphate; 5-FdUrd, 5-fluorodeoxyuridine; FMAU, 2′-fluoroarabinosyl-5-methyldeoxyuridine; FMAUMP, 2′-fluoroarabinosyl-5-methyldeoxyuridine-5′-monophosphate; TMPK, thymidylate kinase.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034199.107.
References
- Alexandre, J.A.C., Roy, B., Topalis, D., Pochet, S., Périgaud, C., Deville-Bonne, D. Enantioselectivity of human AMP, dTMP and UMP-CMP kinases. Nucleic Acids Res. 2007;35:4895–4904. doi: 10.1093/nar/gkm479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie, L., Read, T. Bacillus anthracis, a bug with attitude! Curr. Opin. Microbiol. 2001;4:78–81. doi: 10.1016/s1369-5274(00)00168-5. [DOI] [PubMed] [Google Scholar]
- Balakrishna Pai, S., Liu, S.H., Zhu, Y.L., Chu, C.K., Cheng, Y.C. Inhibition of hepatitis B virus by a novel L-nucleoside, 2′-fluoro-5-methyl-β-L-arabinofuranosyl uracil. Antimicrob. Agents Chemother. 1996;40:380–386. doi: 10.1128/aac.40.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini, J., Herdewijn, P., De Clercq, E. Differential patterns of intracellular metabolism of 2′,3′-didehydro2′,3′-dideoxythymidine and 3′-azido-2′,3′-dideoxythymidine, two potential anti-human immunodeficiency virus compounds. J. Biol. Chem. 1989;264:6127–6133. [PubMed] [Google Scholar]
- Blondin, C., Serina, L., Wiesmuller, L., Gilles, A., Barzu, O. Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal. Biochem. 1994;220:219–221. doi: 10.1006/abio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Buti, M., Esteban, R. Entecavir, FTC, L-FMAU, LdT and others. J. Hepatol. 2003;39:S139–S142. doi: 10.1016/s0168-8278(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Byun, Y., Carnrot, C., Vogel, S., Usova, E., Phipps, A., Eriksson, S., Tjarks, W. Synthesis and biological evaluation of inhibitors of thymidine kinase and thymidine monophosphate kinase from Bacillus anthracis . Nucleosides Nucleotides Nucleic Acids. 2007;27:244–260. doi: 10.1080/15257770701845238. [DOI] [PubMed] [Google Scholar]
- Carnrot, C., Wehelie, R., Eriksson, S., Bölske, G., Wang, L. Molecular characterization of thymidine kinase from Ureaplasma urealyticum: Nucleoside analogues as potent inhibitors of mycoplasma growth. Mol. Microbiol. 2003;50:771–780. doi: 10.1046/j.1365-2958.2003.03717.x. [DOI] [PubMed] [Google Scholar]
- Carnrot, C., Vogel, S., Byun, Y., Wang, L., Tjarks, W., Eriksson, S., Phipps, A. Evaluation of thymidine kinase as a potential target for development of nucleoside analogs against Bacillus anthracis . Biol. Chem. 2006;387:1575–1581. doi: 10.1515/BC.2006.196. [DOI] [PubMed] [Google Scholar]
- Chu, C.K., Boudinot, F.D., Peek, S.F., Hong, J.H., Choi, Y., Korba, B.E., Gerin, J.L., Cote, P.J., Tennant, B.C., Cheng, Y.C. Preclinical investigation of L-FMAU as an anti-hepatitis B virus agent. Antivir. Ther. 1998;3(Suppl 3):113–121. [PubMed] [Google Scholar]
- Dawson, L., Lawrence, T. The role of radiotherapy in the treatment of liver metastases. Cancer J. 2004;10:139–144. doi: 10.1097/00130404-200403000-00009. [DOI] [PubMed] [Google Scholar]
- Eriksson, S., Munch-Petersen, B., Johansson, K., Eklund, H. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti, E., Adam, V., Munier-Lehmann, H., Bourgeois, D. The crystal structure of Mycobacterium tuberculosis thymidylate kinase in complex with 3′-azidodeoxythymidine monophosphate suggests a mechanism for competitive inhibition. Biochemistry. 2005;44:130–137. doi: 10.1021/bi0484163. [DOI] [PubMed] [Google Scholar]
- Furman, P., Fyfe, J., St. Clair, M., Weinhold, K., Rideout, J., Freeman, G., Nusinoff Lehrmann, S., Bolognesi, D., Broder, S., Mitsuya, H., et al. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouz, A., Vanheusden, V., Munier-Lehmann, H., Froeyen, M., Herdewijn, P., Van Calenbergh, S., Delarue, M. Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. J. Biol. Chem. 2003;278:4963–4971. doi: 10.1074/jbc.M209630200. [DOI] [PubMed] [Google Scholar]
- Hu, R., Li, L., Degrève, B., Dutschman, G., Lam, W., Cheng, Y.-C. Behavior of thymidylate kinase toward monophosphate metabolites and its role in the metabolism of 1-(2′-deoxy-2′-fluoro-b-L-arabinofuranosyl)-5-methyluracil (Clevudine) and 2′,3′-didehydro-2′,3′-dideoxythymidine in cells. Antimicrob. Agents Chemother. 2005;49:2044–2049. doi: 10.1128/AAC.49.5.2044-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby, T., Henderson, D., Bartlett, J., Ascher, M., Eitzen, E., Friedlander, A., Hauer, J., McDade, J., Osterholm, M., O'Toole, T., et al. Anthrax as a biological weapon: Medical and public health management. Working group on civilian Biodefense. JAMA. 1999;281:1735–1745. doi: 10.1001/jama.281.18.1735. [DOI] [PubMed] [Google Scholar]
- Jacobsson, B., Britton, S., Tornevik, Y., Eriksson, S. Decrease in thymidylate kinase activity in peripheral blood mononuclear cells from HIV-infected individuals. Biochem. Pharmacol. 1998;56:389–395. doi: 10.1016/s0006-2952(98)00032-x. [DOI] [PubMed] [Google Scholar]
- Jong, A., Campbell, J. Characterization of Saccharomyces cerevisiae thymidylate kinase, the CDC8 gene product. J. Biol. Chem. 1984;259:14394–14398. [PubMed] [Google Scholar]
- Kotaka, M., Dhaliwal, B., Ren, J., Nichols, C.E., Angell, R., Lockyer, M., Hawkins, A.R., Stammers, D.K. Structures of S. aureus thymidylate kinase reveal an atypical active site configuration and an intermediate conformational state upon substrate binding. Protein Sci. 2006;15:774–784. doi: 10.1110/ps.052002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, A., Konrad, M., Brundiers, R., Goody, R., Schlichting, I., Reinstein, J. Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5′-adenosyl) P5-(5′-thymidyl) pentaphosphate (TP5A) at 2.0 Å resolution: Implications for catalysis and AZT activation. Biochemistry. 1998a;37:3677–3686. doi: 10.1021/bi9720787. [DOI] [PubMed] [Google Scholar]
- Lavie, A., Ostermann, N., Brundiers, R., Goody, R.S., Reinstein, J., Konrad, M., Schlichting, I. Structural basis for efficient phosphorylation of 3′-azidothymidine monophosphate by Escherichia coli thymidylate kinase. Proc. Natl. Acad. Sci. 1998b;95:14045–14050. doi: 10.1073/pnas.95.24.14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li de la Sierra, I., Munier-Lehmann, H., Gilles, A.M., Barzu, O., Delarue, M. X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 Å resolution. J. Mol. Biol. 2001;311:87–100. doi: 10.1006/jmbi.2001.4843. [DOI] [PubMed] [Google Scholar]
- Marsh, S., McLeod, H. Thymidylate synthase pharmacogenetics in colorectal cancer. Clin. Colorectal Cancer. 2001;1:175–178. doi: 10.3816/CCC.2001.n.018. [DOI] [PubMed] [Google Scholar]
- Munier-Lehmann, H., Chaffotte, A., Pochet, S., Labesse, G. Thymidylate kinase of Mycobacterium tuberculosis: A chimera sharing properties common to eukaryotic and bacterial enzymes. Protein Sci. 2001;10:1195–1205. doi: 10.1110/ps.45701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann, N., Schlichting, I., Brundiers, R., Konrad, M., Reinstein, J., Veit, T., Goody, R., Lavie, A. Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structures of enzyme complexes along the reaction coordinate. Structure. 2000;8:629–642. doi: 10.1016/s0969-2126(00)00149-0. [DOI] [PubMed] [Google Scholar]
- Ostermann, N., Segura-Pena, D., Meier, C., Veit, T., Monnerjahn, C., Konrad, M., Lavie, A. Structures of human thymidylate kinase in complex with prodrugs: Implications for the structure-based design of novel compounds. Biochemistry. 2003;42:2568–2577. doi: 10.1021/bi027302t. [DOI] [PubMed] [Google Scholar]
- Petit, C., Koretke, K. Characterization of Streptococcus pneumoniae thymidylate kinase: Steady-state kinetics of the forward reaction and isothermal titration calorimetry. Biochem. J. 2002;363:825–831. doi: 10.1042/0264-6021:3630825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochet, S., Dugué, L., Labesse, G., Delepierre, M., Munier-Lehmann, H. Comparative study of purine and pyrimidine nucleoside analogues acting on the thymidylate kinases of Mycobacterium tuberculosis and of humans. ChemBioChem. 2003;4:742–747. doi: 10.1002/cbic.200300608. [DOI] [PubMed] [Google Scholar]
- Spencer, R. Bacillus anthracis . J. Clin. Pathol. 2003;56:182–187. doi: 10.1136/jcp.56.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalis, D., Collinet, B., Gasse, C., Dugué, L., Balzarini, J., Pochet, S., Deville-Bonne, D. Substrate specificity of vaccinia virus thymidylate kinase. FEBS J. 2005;272:6254–6265. doi: 10.1111/j.1742-4658.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- Van Daele, I., Munier-Lehmann, H., Hendrickx, P.M.S., Marchal, G., Chavarot, P., Froeyen, M., Qing, L., Martins, J.C., Van Calenbergh, S. Synthesis and biological evaluation of bicyclic nucleosides as inhibitors of M. tuberculosis thymidylate kinase. ChemMedChem. 2006;1:1081–1090. doi: 10.1002/cmdc.200600028. [DOI] [PubMed] [Google Scholar]
- Van Daele, I., Munier-Lehmann, H., Froeyen, M., Balzarini, J., Van Calenbergh, S. Rational design of 5′-thiourea-substituted α-thymidine analogues as thymidine monophosphate kinase inhibitors capable of inhibiting mycobacterial growth. J. Med. Chem. 2007;50:5281–5292. doi: 10.1021/jm0706158. [DOI] [PubMed] [Google Scholar]
- Vanheusden, V., Munier-Lehmann, H., Pochet, S., Herdewijn, P., Van Calenbergh, S. Synthesis and evaluation of thymidine-5′-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg. Med. Chem. Lett. 2002;12:2695–2698. doi: 10.1016/s0960-894x(02)00551-6. [DOI] [PubMed] [Google Scholar]
- Vanheusden, V., Van Rompay, A., Munier-Lehmann, H., Pochet, S., Herdewijn, P., Van Calenbergh, S. Thymidine and thymidine-5′-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg. Med. Chem. Lett. 2003;13:3045–3048. doi: 10.1016/s0960-894x(03)00643-7. [DOI] [PubMed] [Google Scholar]
- Wang, L. The role of Ureaplasma nucleoside monophosphate kinases in the synthesis of nucleoside triphosphates. FEBS J. 2007;274:1983–1990. doi: 10.1111/j.1742-4658.2007.05742.x. [DOI] [PubMed] [Google Scholar]