Abstract
SUMOylation, the covalent attachment of SUMO (small ubiquitin-like modifier), is a eukaryotic post-translational event that has been demonstrated to play a critical role in several biological processes. When used as an N-terminal tag or fusion partner, SUMO has been shown to enhance functional protein production significantly by improving folding, solubility, and stability. We have engineered several SUMOs and, through their fusion, developed a system for enhancing the expression and secretion of complex proteins. To demonstrate the fidelity of this fusion technology, secreted phospholipase A2 proteins (sPLA2) were produced using HEK-293T and CHO-K1 cells. Five mouse sPLA2 homologs were expressed and secreted in mammalian cell cultures using SUMO or SUMO-derived, N-terminal fusion partners. Mean and median increases of 43- and 18-fold, respectively, were obtained using novel SUMO mutants that are resistant to digestion by endogenous deSUMOylases.
Keywords: SUMO, SUMO fusion, phospholipase A2, mammalian expression, deSUMOylase
SUMO (Small Ubiquitin-like MOdifier) and the SUMO pathway are highly conserved throughout eukaryotes. SUMOylation of proteins in humans was discovered recently by Blobel and colleagues in studies of protein compartmentalization (Matunis et al. 1996). Although SUMO has low (18%) sequence identity with ubiquitin, the two proteins are very similar structurally. The SUMO and ubiquitin pathways also share many features, for instance, both proteins are synthesized as precursors requiring a proteolytic processing step prior to target protein conjugation, and each has a series of activating, conjugating, and ligating enzymes that attach the C termini of the respective proteins to the ε-amino group of lysines in the target proteins (Johnson 2004). SUMO and ubiquitin hydrolases both play important regulatory roles by their removal of SUMO or ubiquitin from target proteins; however, the mechanistic similarity ends at the fate of the modified protein. Unlike ubiquitin, SUMO is not a signal for proteolysis by the 26S proteasome. Although SUMOylation is recognized as an important regulatory modification, its precise role is still the subject of active study. Several papers implicate SUMOylation as a translocation signal for nuclear-cytosolic trafficking. Paradoxically, lysine residues in target proteins that are SUMOylated are protected from ubiquitylation, while SUMOylation of proteins can also serve as a signal for ubiquitylation (Prudden et al. 2007). Thus, nature has evolved two pathways that have remarkably similar mechanical and biochemical features, yet have very distinct roles in cellular physiology (Johnson 2004).
Fusion to SUMO has been shown to enhance the expression of recombinant proteins in Escherichia coli (Malakhov et al. 2004; Butt et al. 2005; Zuo et al. 2005; Marblestone et al. 2006; Panavas et al. 2008), while fusion to hSUMO3 led to similar enhancement in E. coli (data not shown) and Pichia pastoris (R.J. Peroutka, D. Roy, J. Strickler, and T.R. Butt, in prep.). Based on the original observation of split-ubiquitin (Johnsson and Varshavsky 1994), C-terminal half SUMO (CTHS) was developed for baculovirus/insect cell expression, such that it could be removed only in the presence of its N-terminal half (NTHS). We found that CTHS fusion enhanced the production of fusion partners in insect cells while avoiding endogenous cleavage (data not shown). SUMOstar was developed with the same goal—creating a SUMO fusion, which in a eukaryotic host would not be cleaved in vivo, but would undergo enhanced expression similar to the enhancement demonstrated in prokaryotes. Following crystal structure analysis of SUMO bound to its natural protease Ulp1 (PDB: 1EUV), a rational mutagenesis screening campaign resulted in the modification of two interfacial amino acids. These modifications, R64T and R71E, resulted in a SUMO that was resistant to cleavage by Ulp1 regardless of enzyme concentration. This novel SUMO, termed SUMOstar, displayed an enhancement in the expression of its fusion partner in prokaryotes equivalent to that obtained with wild-type SUMO (data not shown); however, it was not clear whether SUMOstar would be cleaved by mammalian deSUMOylases.
To determine (1) if the novel SUMO was equally resistant to mammalian deSUMOylases and (2) whether a similar enhancement could be accomplished in mammalian cells, we examined the effect of various SUMOs on the expression of several members of the secreted phospholipase A2 family (sPLA2) in human embryonic kidney (HEK-293T) and Chinese hamster ovary (CHO-K1) cells.
The sPLA2 family was selected for expression studies because these enzymes, like many complex proteins, have traditionally been expressed in E. coli as inclusion bodies and refolded for characterization. In addition, a native N terminus is required for accurate biological activity, which can be quantified by any of a number of facile assays. Secretory PLA2 was first discovered in cobra venom in the 1890s, but most sPLA2s have been discovered only within the past two decades (for review, see Six and Dennis 2000; Schaloske and Dennis 2006). PLA2s are named for their specific cleavage of phospholipids at the sn-2 acyl bond yielding free fatty acids, such as arachidonic acid (AA), and lysophospholipids (lyso PL). Both products of PLA2 digestion are precursors of a variety of signaling molecules involved in diverse biological pathways. To date, five principal PLA2 families have been identified, with enzyme assignment based on specific function, method of function, and structure (Schaloske and Dennis 2006). The sPLA2 family is comprised of enzymes that are relatively small in size (12–18 kDa), possess five to eight disulfide bonds, contain active sites with a His/Asp dyad, and require μM Ca2+ for activity. To date, 10 sPLA2s have been identified in humans and 11 in mice, exhibiting various functions and tissue distribution. Mammalian sPLA2s have been implicated in a wide range of diseases such as asthma (Henderson Jr. et al. 2007), atherosclerosis (Webb 2005), and cancer (Murakami et al. 2005). Biological relevance is not limited to enzymatic activity, as sPLA2s have been shown to act as ligands themselves, interacting with at least two receptors in mammals (Lambeau and Lazdunski 1999). Of the 11 mouse sPLA2s, groups IIC, IIE, III, V, and X were selected for the present study because they cover the widest range of expression levels in prokaryotes, ∼0.8 to 20 mg/L (Rouault et al. 2007).
Our results show that: (1) Native SUMO fusions can be processed in mammalian cell culture; (2) when overexpressed, and released from their fusion partner, some sPLA2s exhibit moderate toxicity; and (3) fusion to SUMO mutants, not recognized by endogenous deSUMOylases, can lead to significantly enhanced expression of sPLA2s while avoiding cell toxicity. We believe that the development of these mutant SUMOs and their related enzymes will provide valuable tools for enhancing the expression and secretion of difficult to express recombinant proteins in eukaryotic cells.
Results
Development of novel SUMO tags
To evaluate the potential utility of expressing SUMO-fusion proteins in the mammalian secretory pathway, mouse sPLA2-X was used as a model protein. Initially, the following four N-terminal fusions were tested: SUMO (SUMO), the C-terminal half of SUMO comprising amino acids 55–98 (CTHS), a R64T R71E double mutant SUMO (SUMOstar), and human SUMO-3 (hSUMO3). All tags were created with a hexahistidine (6xHis) N terminus and directed for secretion using the IgG kappa secretory signal from mouse. For control purposes, a vector was created with only the signal sequence and 6xHis tag, creating a total of five vectors differing only in their SUMO-based tag.
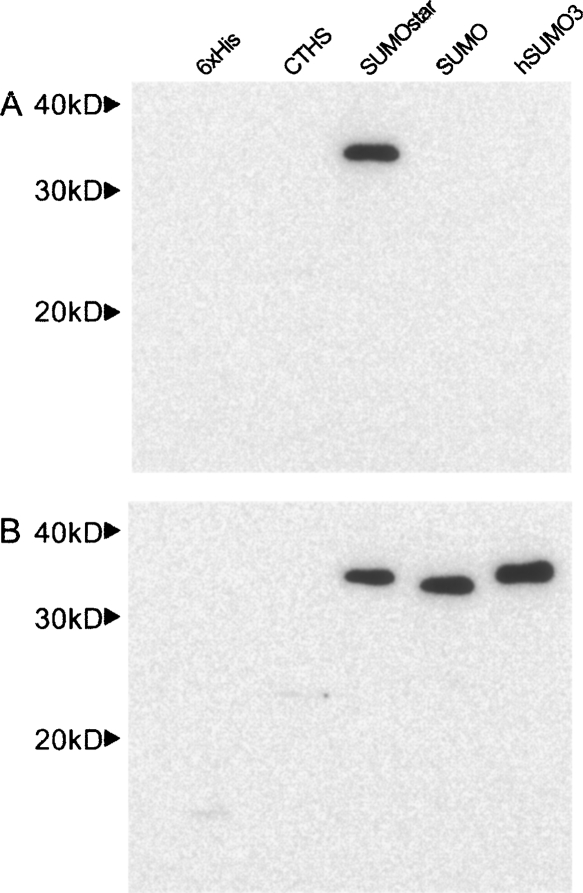
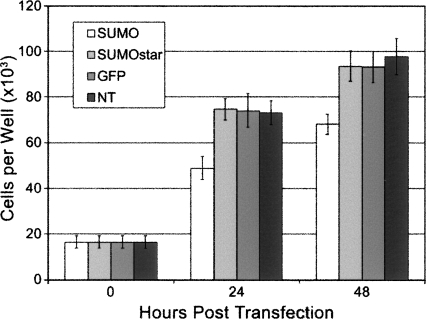
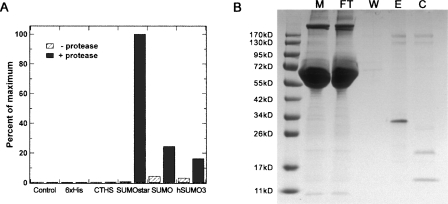
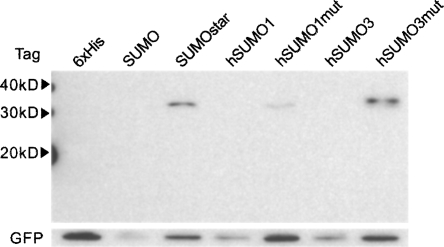
Expression and secretion of sPLA2-X in monolayer HEK-293T cells were examined by Western blot of cell culture media after 48 h (Fig. 1A). SUMOstar showed a significant enhancement in the production of sPLA2-X compared to expression of the 6xHis or CTHS tag fusions, while no SUMO or hSUMO3 fusion was detected. The SUMO and hSUMO3 cultures appeared less confluent, compared to the others, 48 h post-transfection. These differences in growth rate were quantified by determining viable HEK-293T cell counts at 0, 24, and 48 h post-transfection. Cell counts were compared among SUMO, SUMOstar, and GFP transfections. In addition to growth of GFP-transfected cells as a control, growth of nontransformed cells was measured. Cultures transfected with SUMOstar-sPLA2-X or GFP displayed growth equivalent to that of nontransfected cells, while SUMO-sPLA2-X-transfected cultures contained significantly reduced numbers of viable cells (Fig. 2). On average, the cultures transfected with SUMO-sPLA2-X contained 34% and 28% fewer cells after 24 and 48 h, respectively. To investigate whether any SUMO or hSUMO3 fused sPLA2-X was produced, all fusions were purified from medium on a small scale using Ni+-NTA resin in batch mode, and the eluates were digested with their respective enzymes. Although SUMO and hSUMO3 fusions were not detected in Western blots, some SUMO-protease-dependent and -independent activity was detected after Ni+-NTA purification and assay (Fig. 3A). Presumably, the SUMO-protease-independent activity is the result of release of active sPLA2 by endogenous deSUMOylases. The lack of 6xHis sPLA2-X activity in Figure 3A is not indicative of its expression level (∼15-fold less than SUMOstar), as the tag did not contain a protease cleavage site. The transfection, purification, and assay of the sPLA2-X constructs were repeated several times with the same results. To confirm that inactive sPLA2-X was produced and could be activated by removal of the SUMOstar moiety, we purified the fusion using a Ni+-loaded IMAC Sepharose resin and digested the partially purified, imidazole eluted protein with SUMOstar protease (Fig. 3B). The imidazole eluate (E) clearly shows a band with the expected molecular weight of the fusion protein (∼34 kD), which is lost following digestion by SUMOstar protease (C) to yield bands with the expected molecular weights of SUMO (∼20 kD)1 and sPLA2-X (∼14 kD). The eluate fraction demonstrated no PLA2 activity prior to digestion with SUMOstar protease (data not shown).
Figure 1.
Western blot analyses of sPLA2-X expression. Samples of media (15 μL) from mouse sPLA2-X constructs, 48 h post-transfection (HEK-293T), were separated by SDS-PAGE and transferred to nitrocellulose as described. The blots were probed with a monoclonal anti-6xHis antibody. The following five N-terminal fusion tags were tested: 6xHis, 6xHis-CTHS, 6xHis-SUMOstar, 6xHis-SUMO, and 6xHis-hSUMO3. All constructs were directed for secretion via the mouse IgG kappa secretory signal. Results are representative of at least three independent experiments. (A) Expression of wild-type sPLA2-X. (B) Expression of ΔGly-sPLA2-X.
Figure 2.
The effect of sPLA2-X expression on cell growth and viability. HEK-293T cells were seeded in 24-well dishes, and the number of viable cells was determined at 0, 24, and 48 h post-transfection with SUMO-PLA2, SUMOstar-PLA2, GFP, or no DNA. Data points represent the average number of viable cells from three independent transfections.
Figure 3.
Purification and determination of PLA2 enzymatic activity. (A) Small-scale Ni+-NTA purifications of the initial fusions were performed in batch mode using equal volumes of media, 48 h post-transfection. The eluted samples were digested with the proteases specific for each tag and assayed for sPLA2 activity, which is represented as a percent of the most active sample. (B) Purification of SUMOstar-sPLA2-X using a Ni+-Sepharose column and subsequent cleavage the fusion with SUMOstar protease. The SUMOstar-sPLA2 fusion can be seen in the media (M) just below 34 kDa, while it is absent from the flow-through (FT). The column was washed (W), and the fusion (∼34 kD) was eluted (E) with imidazole and cleaved (C) with protease, yielding SUMOstar (∼20 kD) and free sPLA2-X (∼14 kD).
Mouse sPLA2-X is naturally produced as a zymogen; its mature form preceded by an 11-amino-acid N-terminal propeptide, which has been observed to reduce activity to 8% (Morioka et al. 2000). The mature form, beginning with GLLEL, was cloned behind the various tags such that fully active PLA2 would be produced only after in vitro release with protease. The recombinant production of active mouse and human sPLA2-X in mammalian cells has been described, with no mention of toxicity (Cupillard et al. 1997; Morioka et al. 2000), so we did not anticipate the behavior of the SUMO-sPLA2-X- and hSUMO3-sPLA2-X-transfected cells. To investigate whether endogenous SUMO cleavage and the resulting phospholipase activity contributed to the differential expression of SUMO and hSUMO3 fusions, compared to SUMOstar, or if the difference was a result of the SUMO proteins themselves, a series of mutant sPLA2-X fusions was generated. Based on the crystal structure of human sPLA2-X (PDB: 1LE6), which shares 90% similarity with mouse, we theorized that omitting the N-terminal glycine of sPLA2-X (ΔGly-sPLA2-X) would result in a diglycine that would be less accessible to proteolytic cleavage. Expression of the various fusion tags with ΔGly-sPLA2-X in HEK-293T cells can be seen in Figure 1B. By densitometry analysis of 6xHis- and SUMOstar-tagged ΔGly-sPLA2-X constructs, SUMOstar led to ∼15-fold higher sPLA2-X production. The results demonstrate that sPLA2-X activity and the susceptibility of its N-terminal pro-peptide to cleavage clearly play a role in overexpression.
Expanded expression study
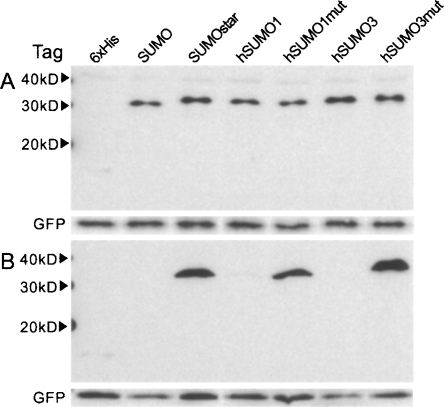
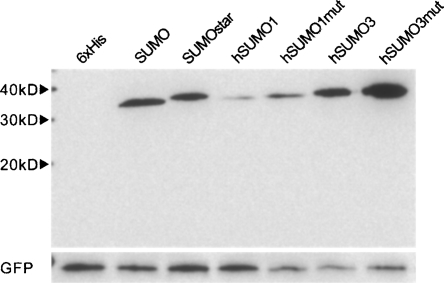
From the initial expression data, we re-evaluated which fusion tags should be tested. We decided that CTHS would not be tested further and that the expression levels of hSUMO-3 with inactive sPLA2-X warranted further investigation of the human SUMOs. We noticed a strong conservation among SUMOs while evaluating the crystal structures of human SUMO-1, 2, 3, and SUMO; the nearly identical location of two interfacial arginine residues. Since hSUMO-2 and 3 share 97% identity, we elected to investigate only hSUMO-1 and 3. The following modifications were made: for hSUMO1, R63T and R70E; for hSUMO3, R58T and R60E. Wild-type sPLA2-X and ΔGly-sPLA2-X fusions were made with the mutant and wild-type versions of SUMO, hSUMO1, and 3. In addition to generating these constructs, a vector containing GFP (without tag or secretory signal) was created for co-transfection purposes. The results of expressing ΔGly-sPLA2-X and sPLA2-X fusions for 48 h can be seen in Figure 4, A and B, respectively. Equal amounts of intracellular GFP can be seen in an anti-GFP Western blot below the anti-6xHis blot in Figure 4A, indicating equal transfection efficiencies. The cultures expressing fusions to wild-type hSUMO1, like those expressing SUMO and hSUMO3, displayed limited growth, and therefore less GFP coexpression per culture. Expression of the wild-type forms of SUMO and hSUMO1 could occasionally be detected in replicate culture Western blots, as in Figure 4B, where a faint band of hSUMO1-sPLA2-X can be seen. Both human SUMO mutants displayed expression comparable to or greater than the original SUMOstar when fused to sPLA2-X, as seen in Figure 4B. On average, by densitometry analysis of at least three independent transfections, fusions of the three SUMO mutants expressed 15–20 times more sPLA2-X than fusions of the 6xHis tag alone. Expression of sPLA2-X in cultured mammalian cells was further analyzed by the transfection of monolayer CHO-K1 cells. Under the transfection and culture conditions used for HEK-293T cells, CHO-K1 cells displayed very similar expression and secretion after 48 h by Western blot (Fig. 5). It is interesting to note that the CHO-K1 cells appeared to be more sensitive than HEK-293T cells to expression of the natural SUMO fusions, as judged by the confluence of cultures after 48 h and the levels of intracellular GFP.
Figure 4.
Expression analysis of seven N-terminal fusion tags in HEK-293T cells. (A) Comparison of the ΔGly-sPLA2-X fusions. All SUMO tags led to an enhancement as compared to the 6xHis tag alone. (B) Expression of wild-type sPLA2-X fusions. GFP was co-transfected to demonstrate equal transfection. Media samples (15 μL) from the revised mouse sPLA2-X constructs, 48 h post-transfection (HEK-293T), were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with monoclonal anti-6xHis. Intracellular GFP was detected using polyclonal anti-GFP. Results are representative of at least three independent experiments.
Figure 5.
Expression of wild-type sPLA2-X fusions by CHO-K1 cells. As described in Figure 4, seven N-terminal fusion tags were tested with wild-type sPLA2-X. Media samples (15 μL) from the revised mouse sPLA2-X constructs, 48 h post-transfection (CHO-K1), were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with monoclonal anti-6xHis. Intracellular GFP was detected using polyclonal anti-GFP. Results are representative of three independent experiments.
Given the expression data using mouse sPLA2-X, we were curious to see whether the enhancements seen in HEK-293T and CHO-K1 production would be comparable using other sPLA2 groups. We chose four additional mouse sPLA2 genes based on their varied levels of recombinant expression previously reported (Rouault et al. 2007). Mouse sPLA2-IIC and III have been produced in insect cells with yields of 150 and 70 μg/L, respectively. There are currently no refolding protocols for either sPLA2, and both enzymes are naturally glycosylated, making eukaryotic production a necessity. Mouse sPLA2-IIE represents the lowest reported yield in bacterial production at 800 μg/L, while sPLA2-V represents the highest yield at 20 mg/L. Mouse sPLA2-X reportedly has been expressed at 10 mg/L in E. coli.
We proceeded with the fusion tag comparisons using mouse sPLA2-IIC, IIE, III, and V in HEK-293T cells. In the interest of time, only the wild-type versions of these enzymes were tested. All transfections were performed with GFP to monitor efficiencies. The intracellular expression of sPLA2-IIC after 48 h can be seen in Figure 6. His-tagged protein could not be detected in the media. Despite an apparently large increase in expression with all the SUMO tags, secretion was somehow inhibited. The expression and secretion of sPLA2-IIE can be seen in Figure 7A. After 48 h, significantly more sPLA2-IIE is visible in most of the SUMO fusions, with 140 times more SUMOstar and 190 times more hSUMO3mut than His-tag alone by densitometry analysis. Mouse and human sPLA2-III is expressed as a 55-kDa protein but often matures via post-translational and cell-specific proteolytic processing to a 28-kDa active domain (Murakami et al. 2003, 2005). The active or S domain is preceded by an N domain and followed by a C domain. We generated fusions with the full-length sPLA2-III, only replacing its native secretory signal with SUMO and the kappa signal. In HEK-293T cells, all sPLA2-III fusions were apparently processed at their first cleavage point, dividing the N and S domains as seen in Figure 7B, where the His-tagged protein is 12 kDa and the various SUMO fusions are ∼32 kDa. Intracellular blotting demonstrated the production of a 55-kDa protein with no additional forms visible (data not shown). The expression and secretion of sPLA2-V can be seen in Figure 7C. Similar to group X, there is a strong preference for the SUMOstar and mutant hSUMO fusions in the expression of sPLA2-V. Although there was clearly a lack of expression in the fusions to wild-type SUMO, similar cell culture problems were not seen in the case of sPLA2-V, where, with the exception of hSUMO1, there was little difference in the level of coexpressed GFP. Transfections were performed several times for each sPLA2 group, using all fusion tags, with comparable results, as analyzed by Western blotting.
Figure 6.
Intracellular expression of sPLA2-IIC. All constructs being directed for secretion via the mouse IgG kappa secretory signal; however, sPLA2-IIC was not detected in the medium. Cell lysates, 48 h post-transfection (HEK-293T), were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with monoclonal anti-6xHis. Results are representative of two independent experiments. GFP was co-transfected and detected with polyclonal anti-GFP.
Figure 7.
Analysis of N-terminal fusion tags with sPLA2-IIE, sPLA2-III, and sPLA2-V. (A) sPLA2-IIE samples. (B) sPLA2-III samples. (C) sPLA2-V samples. Media samples (15 μL), 48 h post-transfection (HEK-293T), were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with monoclonal anti-6xHis. Results are representative of two to three independent experiments.
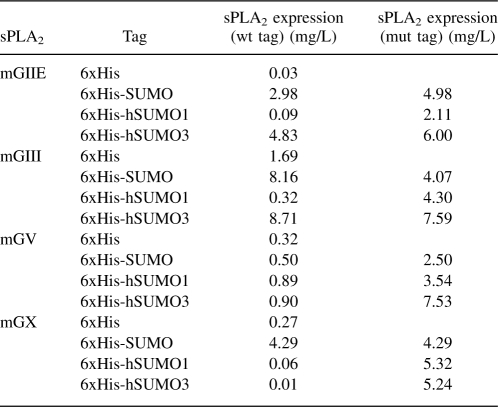
Expression levels of the various mouse sPLA2 fusions were quantified by blotting representative SUMOstar-sPLA2 samples from each sPLA2 group, from several transfections with serial dilutions of bacterially expressed SUMO-sPLA2-X (see Materials and Methods). Although this method is only semiquantitative, it was our best option with such small culture volumes. We found that, on average, SUMOstar-sPLA2-IIE was produced at 5 ± 0.4 mg/L, while SUMOstar-sPLA2-III was produced at 4.1 ± 0.1 mg/L. SUMOstar-sPLA2-V and SUMOstar-sPLA2-X were, on average, produced at 2.5 ± 1.0 mg/L and 4.3 ± 1.5 mg/L, respectively. The values for the other fusion constructs were extrapolated from the densitometry data of the SUMOstar fusions (Table 1).
Table 1.
Comparison of sPLA2 (groups IIE, III, V, and X) production from monolayer HEK-293T cells 48 h post-transfection
Discussion
Secreted phospholipases were used as model proteins to determine the impact of wild-type and mutant SUMOs on the expression, secretion, and stability of biologically active enzyme. Although mammalian cells contain highly active SUMO proteases, it was not clear whether these enzymes would affect secreted SUMO fusions, as deSUMOylases have not been identified within the secretory pathway. Some, but not all, of the various sPLA2 fusions were cleaved, indicating an absence of deSUMOylases in the secretory pathway. However, in constructs containing sPLA2-V and sPLA2-X, a small amount of presumably extracellular digestion and PLA2 release was sufficient to elicit a toxic cellular response. This study has shown that, by minor modification, mutant human and yeast SUMO fusions were able to circumvent cleavage in mammalian cell culture while maintaining and/or improving recombinant production levels obtained by wild-type SUMO fusion. Furthermore, phospholipases expressed in large amounts in mammalian cells can be toxic and lead to decreased cell viability. This study has allowed us to make three main observations. Attachment of a SUMO moiety to the N terminus of target proteins dramatically enhances their expression in cells. Wild-type yeast and human SUMO tags are susceptible to release from their fusion partner, presumably in cell culture media. Toxic proteins such as sPLA2-X and sPLA2-V can be produced as dormant fusions, while cleavage of mutant SUMO fusions with mutant SUMO protease in vitro generates an active enzyme.
Since they contain large numbers of disulfide bonds, sPLA2s are difficult to express in E. coli and refold following solubilization, typically resulting in poor yields. The recombinant production of secreted mammalian phospholipase A2s has been established in various hosts including insect cells and larvae (Singer et al. 2002; Zhang et al. 2006), yeast (Lefkowitz et al. 1999; Zhang et al. 2006), chemical synthesis (Hackeng et al. 1997; Dong et al. 2002), mammalian cells (Murakami et al. 1998; Morioka et al. 2000), and E. coli (Othman et al. 1996; Singer et al. 2002; Rouault et al. 2006, 2007). Despite the requirement for refolding, the cost of production in E. coli is still lower than that in any other system. Many sPLA2s require a unique refolding procedure, and some of the PLA2 cannot be produced in active form in E. coli (Singer et al. 2002). Therefore, new systems that enable the recombinant production and purification of correctly folded proteins having stringent requirements for defined N termini, and that maintain potentially toxic proteins, such as the secreted phospholipase A2s, in an inactive state prior to tag removal, would be very useful. We have demonstrated that recombinant fusion of the N terminus of sPLA2s to the C terminus of yeast and human SUMOs significantly enhances expression and secretion of sPLA2 proteins in mammalian cell culture. Furthermore, we have developed novel SUMO derivatives that are not processed in vivo, and, when fused to sPLA2 permit overexpression of even the most enzymatically active sPLA2 without producing toxic effects.
The results of the initial expression comparison using mouse sPLA2-X were surprising. Despite our efforts to develop a SUMO derivative that was resistant to natural deSUMOylases, we did not expect to see such a difference in the levels of expression among our initial constructs. We theorized, based on previous work with yeast and insect cells, that wild-type yeast SUMO and hSUMO3 fusions would express at levels comparable to those of SUMOstar, but would be at least partially cleaved. In repeated experiments, however, we observed an almost complete lack of recombinant sPLA2-X expression when fused to SUMO or hSUMO3. Group X is naturally produced as a zymogen with an 11-amino-acid pro-peptide that reduces its enzymatic activity by 92% (Morioka et al. 2000). Since mature sPLA2-X has been described as possessing a “powerful potency for releasing arachidonic acid” (Morioka et al. 2000), mutant sPLA2-X fusions were constructed to investigate whether enzyme function was playing a deleterious role in expression. Once sPLA2-X was mutated, all SUMO-sPLA2-X fusions displayed significant enhancements in expression (Figs. 1B and 4A).
These observations suggest that uncleaved SUMO fusions can be used as tools to both improve expression of potentially toxic proteins and to study the role of SUMO in cell physiology. Given the high degree of conservation of sequence and structure between SUMOs from different species, it is not surprising that mutation of analogous residues in yeast and human SUMOs generate versions of the proteins that are not recognized or processed by the cognate deSUMOylases. These observations suggest that the SUMO hydrolases recognize similar surfaces of SUMOs in yeast and human cells.
Materials and Methods
Construction of fusion tag vectors
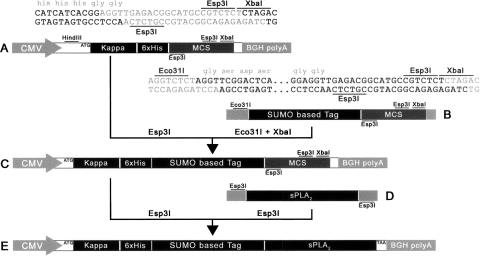
For all vector constructs, pcDNA3.1/V5-His (Invitrogen) was used as a backbone. Platinum Taq DNA Polymerase High Fidelity (Invitrogen) was used for all PCR reactions, while all restriction enzymes and T4 DNA ligase were from Fermentas. Cloning was performed according to standard techniques. All clones were verified by sequencing. All primers used can be found in Table 2. Initially, a kappa S.S. and 6xHis tag were generated via overlapping primers with a region of homology between the two (primers 1 + 2 and 3 + 4, respectively). The kappa S.S and His tag were joined in a secondary PCR reaction using primers 1 + 4. The kappa-6xHis fusion was inserted into pcDNA3.1 via HindIII and XbaI restriction sites, generating pcDNA3.1-kappa-6xHis (Fig. 8A). Primers 3 and 4 were designed so that the His tag was followed by two glycines and an Esp3I/BsmBI restriction site on the non-coding strand, upstream of the XbaI site. Between the Esp3I/BsmBI and XbaI sites was an additional Esp3I/BsmBI site on the coding strand. Digestion with Esp3I was designed to generate a 4-base overhang on the non-coding strand consisting of TCCA from the di-glycine GGAGGT coding sequence and an XbaI-compatible CTAG overhang on the opposite end of the vector. A single-enzyme, directional double digest was therefore accomplished. CTHS, SUMO, SUMOstar, and hSUMO3 were amplified with primers 5 + 6, 7 + 6, 7 + 6, and 8 + 9, respectively. All reverse primers were designed to recreate the Esp3I recognition sites downstream from the various SUMO terminal di-glycine codons (Fig. 8B). SUMO tags were inserted into pcDNA3.1-kappa-6xHis via Eco31I and XbaI restriction sites generating the following vectors: pcDNA3.1-kappa-6xHis-CTHS, pcDNA3.1-kappa-6xHis-SUMO, pcDNA3.1-kappa-6xHis-SUMOstar, and pcDNA3.1-kappa-6xHis-hSUMO3 (Fig. 8C).
Table 2.
Primers used for generating the SUMO vectors and sPLA2 fusions
Figure 8.
Representative detail of vector construction and sPLA2 fusion. Initially, pcDNA3.1 was modified by inserting a kappa secretory signal followed by a 6xHis tag and a modified multiple cloning site via a HindIII/XbaI double digestion. The resulting vector (A) was used directly to create 6xHis-tagged sPLA2 constructs. In order to generate the various SUMO fusions, vector A was digested with Esp3I and ligated to Eco31I/XbaI digested, PCR-amplified, SUMO derivatives containing a new multiple cloning site (B) to give vector C. SUMO vectors (C) were digested with Esp3I and fused with sPLA2 PCR products (D) digested with Eco31I, Esp3I, or BpiI. This strategy resulted in the creation of in-frame fusions (E) between the various SUMOs and sPLA2s.
Initial mouse sPLA2-X construct creation
Active sPLA2-X was PCR amplified from a clone kindly provided by Dr. Michael Gelb's laboratory (University of Washington, Seattle) using primers 10 + 11 (Fig. 8D). ΔGly-sPLA2-X was PCR-amplified from the same clone using primers 12 + 11. Both active and inactive sPLA2-X constructs were created by digesting both PCR product and vectors with Esp3I and ligating digestion products (Fig. 8E).
Expansion of fusion tag vectors
Human SUMO-1 was PCR-amplified from cDNA obtained from Open Biosystems using primers 13 + 14, and cloned into pcDNA3.1-kappa-6xHis via Esp3I and XbaI restriction sites generating pcDNA3.1-kappa-6xHis-hSUMO1. Mutant human SUMO-1 and 3 were generated using PCR site-directed mutagenesis in which the N-terminal and C-terminal halves were produced in separate reactions, gel isolated, and joined in a subsequent PCR reaction. Human SUMO-1 primary PCR used primers 13 + 15 and 16 + 14 for the N- and C-terminal reactions, respectively. Human SUMO-3 primary PCR used primers 8 + 17 and 18 + 9 for the N- and C-terminal reactions, respectively. In the secondary PCR-purified primary products were mixed for each human SUMO, and primers 13 + 14 were used for hSUMO1mut, while primers 8 + 9 were used for hSUMO3mut. Products were inserted into pcDNA3.1-kappa-6xHis generating pcDNA3.1-kappa-6xHis-hSUMO1mut and pcDNA3.1-kappa-6xHis-hSUMO3mut.
Expansion of mouse sPLA2 constructs
cDNAs for mouse sPLA2-IIC, IIE, III, and V were purchased from Open Biosystems. PLA2 primers were designed with the goal of generating mature proteins subsequent to purification and tag removal. Secretory signals and propeptides were therefore omitted in primer design, based on literature review and SignalP analysis (Bendtsen et al. 2004). Mouse sPLA2-IIC was cloned from cDNA, corresponding to GenBank entry BC029347, with primers 19 + 20. Mouse sPLA2-IIE was cloned from cDNA, corresponding to GenBank entry BC027524, with primers 21 + 22. Full-length mouse sPLA2-III was cloned from cDNA, corresponding to GenBank entry BC079556, with primers 23 + 24. Mouse sPLA2-V was cloned from cDNA, corresponding to GenBank entry BC030899, with primers 25 + 26. All sPLA2 genes including sPLA2-X active and inactive were subcloned into pcDNA3.1-kappa-6xHis, pcDNA3.1-kappa-6xHis-SUMO, pcDNA3.1-kappa-6xHis-SUMOstar, pcDNA3.1-kappa-6xHis-hSUMO1, pcDNA3.1-kappa-6xHis-hSUMO1mut, pcDNA3.1-kappa-6xHis-hSUMO3, and pcDNA3.1-kappa-6xHis-hSUMO3mut.
Transient transfection in HEK-293T and CHO-K1 cells
HEK-293T or CHO-K1 cells were seeded into six-well plates (Becton Dickinson) at a density of 500,000 cells per well in either DMEM or F12, respectively, containing 10% fetal bovine serum media, and incubated overnight at 37°C with 95% air/CO2. Cells were transiently transfected with various sPLA2 constructs in pcDNA3.1 vector (2.5 μg/well) using Lipofectamine-LTX as described by the manufacturer (Invitrogen). After transfection, cells were incubated for an additional 48 h at 37°C before being analyzed for PLA2 expression.
Expression analysis
After 48 h of incubation, following transfection, media and cells were collected for analysis. Culture media was removed from each well (∼1.5 mL), and debris was separated by centrifugation. For SDS-PAGE/Western blotting, media was mixed with 6× SDS loading buffer and boiled for 5 min. The remaining media was stored at −80°C for later assay. Cells were washed from each well of the plate, separated by centrifugation, re-suspended in 180 μL of cold RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 1% Na deoxycholate), sonicated briefly, mixed with 6× SDS loading buffer, and boiled for 5 min. All samples were resolved on denaturing 15% acrylamide gels with a 4% acrylamide stacking layer. Gels were transferred to Immoblin nitrocellulose (Millipore) using a Trans-Blot SD semidry transfer cell (Bio-Rad). After transfer, blots were blocked with 5% non-fat milk in PBS (pH 7.5) + 0.05% Tween 20 (PBST) for 1 h. Following blocking, the blots were incubated in 1:1000 monoclonal Anti-His Antibody (Sigma) in PBST + milk for at least 1 h. Blots were washed with PBST three times and incubated with 1:2500 anti-mouse HRP conjugated antibody (Sigma) in PBST + milk for 1 h. Blots were again washed three times with PBST. HRP conjugates were detected with SuperSignal West Pico chemiluminescent substrate (Pierce). Blots were imaged using an LAS-3000 (Fujifilm Life Science). Densitometry analysis was performed using Multi Gauge 3.0 (Fujifilm Life Science). Density was calculated by subtracting background and dividing by area. All areas compared were of equal size, while exposures were set to ensure that none of the bands compared had reached saturation. Standard curves were created for quantification by fitting known sample values to the function Y = aX 2 + bX + c in Multi Gauge. The software was then used to determine the relative amounts of the various SUMOstar fusions (loaded on the same gel). Quantities of the other constructs were determined by comparing density values and adjusting for differences relative to SUMOstar. For example: The average value of SUMOstar-PLA2-IIE expression, found by a standard curve fit, was 4.98 ng/μL, and the ratio in densities between SUMO-sPLA2-IIE and SUMOstar-sPLA2-IIE expression in Figure 7A was 3:5, respectively; so SUMO-sPLA2-IIE expression was calculated to be 2.98 ng/μL.
Purification of sPLA2 group X
Small-scale purification was performed in batch mode using Ni+-NTA beads (QIAGEN). Briefly, 50 μL of Ni+-NTA beads was equilibrated with wash buffer (0.05 M Tris-HCl, pH 7.5, 0.5 M NaCl, 0.2 M imidazole). Clarified medium (1 mL) was adjusted to 0.02 M imidazole by addition of 1 M imidazole in 0.05 M Tris-HCl, pH 7.5, 0.5 M NaCl. Samples were gently mixed with beads for at least 1 h at 4°C, washed three times with wash buffer, and eluted with wash buffer containing 0.5 M imidazole.
Larger-scale purification was accomplished on a 5-mL HisTrap FF Ni+ Sepharose column (GE Healthcare). Conditioned medium was filtered through a 0.2-μm filter, adjusted to 20 mM imidazole, and loaded onto the column at a flow rate of 1 mL/min. The column was washed with 95% Buffer A (0.05 M Tris-HCl, pH 8.0, 0.5 M NaCl) and 5% Buffer B (Buffer A + 0.5 M imidazole). Elution was performed in two steps: first with 90% A/10% B, and then with 100% B. Fractions were analyzed for sPLA2 activity following digestion with SUMOstar protease (LifeSensors). Eluted fractions with sPLA2 activity were pooled, concentrated, and exchanged into 0.05 M Tris-HCl pH 7.5 and 2 mM CaCl2.
Assay
Enzymatic activity was analyzed according to the method of Nicholson (Nicholson et al. 2008). Briefly, SUMOstar-sPLA2 was mixed with 20 nM SUMOstar protease and 20 μM NBD C6-HPC (Invitrogen). Release of NBD-fluorescence was measured at 1-min intervals in a Fluorscan plate reader (ThermoScientific) using an excitation wavelength of 460 nm and emission wavelength of 538 nm. Following an initial lag period reflecting the activation of sPLA2 through removal of the SUMOstar fusion partner, steady-state PLA2 activity was determined.
Acknowledgments
We thank Dr. Michael Gelb for his generous gift of the group X sPLA2 clone and Drs. James Strickler and Michael Mattern for helpful discussions and critical reading of the manuscript. Part of this work was funded by NIH/NIGMS grants, GM067271-02 and GM068404-3.
Footnotes
SUMO migrates with an apparent molecular weight of 18–20 kDa on SDS-PAGE.
Reprint requests to: Tauseef R. Butt, LifeSensors, Inc., 271 Great Valley Parkway, Malvern, PA 19355, USA; e-mail: butt@lifesensors.com; fax: (610) 644-8616.
Abbreviations: CHO-K1, Chinese hamster ovary; HEK-293T, human embryonic kidney cells; sPLA2, secretory phospholipase A2; SUMO, small ubiquitin-like modifier; CTHS, C-terminal half SUMO; GFP, green fluorescent protein; NBD C6-HPC, 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035576.108.
References
- Bendtsen, J.D., Nielsen, H., von Heijne, G., Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Butt, T.R., Edavettal, S.C., Hall, J.P., Mattern, M.R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupillard, L., Koumanov, K., Mattei, M.G., Lazdunski, M., Lambeau, G. Cloning, chromosomal mapping, and expression of a novel human secretory phospholipase A2 . J. Biol. Chem. 1997;272:15745–15752. doi: 10.1074/jbc.272.25.15745. [DOI] [PubMed] [Google Scholar]
- Dong, C.Z., Romieu, A., Mounier, C.M., Heymans, F., Roques, B.P., Godfroid, J.J. Total direct chemical synthesis and biological activities of human group IIA secretory phospholipase A2 . Biochem. J. 2002;365:505–511. doi: 10.1042/BJ20011648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackeng, T.M., Mounier, C.M., Bon, C., Dawson, P.E., Griffin, J.H., Kent, S.B. Total chemical synthesis of enzymatically active human type II secretory phospholipase A2 . Proc. Natl. Acad. Sci. 1997;94:7845–7850. doi: 10.1073/pnas.94.15.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson W.R., Jr, Chi, E.Y., Bollinger, J.G., Tien, Y.T., Ye, X., Castelli, L., Rubtsov, Y.P., Singer, A.G., Chiang, G.K., Nevalainen, T., et al. Importance of group X-secreted phospholipase A2 in allergen-induced airway inflammation and remodeling in a mouse asthma model. J. Exp. Med. 2007;204:865–877. doi: 10.1084/jem.20070029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnsson, N., Varshavsky, A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeau, G., Lazdunski, M. Receptors for a growing family of secreted phospholipases A2 . Trends Pharmacol. Sci. 1999;20:162–170. doi: 10.1016/s0165-6147(99)01300-0. [DOI] [PubMed] [Google Scholar]
- Lefkowitz, L.J., Deems, R.A., Dennis, E.A. Expression of group IA phospholipase A2 in Pichia pastoris: Identification of a phosphatidylcholine activator site using site-directed mutagenesis. Biochemistry. 1999;38:14174–14184. doi: 10.1021/bi991432t. [DOI] [PubMed] [Google Scholar]
- Malakhov, M.P., Mattern, M.R., Malakhova, O.A., Drinker, M., Weeks, S.D., Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- Marblestone, J.G., Edavettal, S.C., Lim, Y., Lim, P., Zuo, X., Butt, T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas, E., Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka, Y., Saiga, A., Yokota, Y., Suzuki, N., Ikeda, M., Ono, T., Nakano, K., Fujii, N., Ishizaki, J., Arita, H., et al. Mouse group X secretory phospholipase A2 induces a potent release of arachidonic acid from spleen cells and acts as a ligand for the phospholipase A2 receptor. Arch. Biochem. Biophys. 2000;381:31–42. doi: 10.1006/abbi.2000.1977. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Shimbara, S., Kambe, T., Kuwata, H., Winstead, M.V., Tischfield, J.A., Kudo, I. The functions of five distinct mammalian phospholipase A2S in regulating arachidonic acid release. Type IIa and type V secretory phospholipase A2S are functionally redundant and act in concert with cytosolic phospholipase A2 . J. Biol. Chem. 1998;273:14411–14423. doi: 10.1074/jbc.273.23.14411. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Masuda, S., Shimbara, S., Bezzine, S., Lazdunski, M., Lambeau, G., Gelb, M.H., Matsukura, S., Kokubu, F., Adachi, M., et al. Cellular arachidonate-releasing function of novel classes of secretory phospholipase A2s (groups III and XII) J. Biol. Chem. 2003;278:10657–10667. doi: 10.1074/jbc.M211325200. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Masuda, S., Shimbara, S., Ishikawa, Y., Ishii, T., Kudo, I. Cellular distribution, post-translational modification, and tumorigenic potential of human group III secreted phospholipase A2 . J. Biol. Chem. 2005;280:24987–24998. doi: 10.1074/jbc.M502088200. [DOI] [PubMed] [Google Scholar]
- Nicholson, B., Leach, C.A., Goldenberg, S.J., Francis, D.M., Kodrasov, M.P., Tian, X., Shanks, J., Sterner, D.E., Bernal, A., Mattern, M.R., et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman, R., Baker, S., Li, Y., Worrall, A.F., Wilton, D.C. Human non-pancreatic (group II) secreted phospholipase A2 expressed from a synthetic gene in Escherichia coli: Characterisation of N-terminal mutants. Biochim. Biophys. Acta. 1996;1303:92–102. doi: 10.1016/0005-2760(96)00083-5. [DOI] [PubMed] [Google Scholar]
- Panavas, T., Sanders, C.F., Butt, T.R. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. In: Ullrich H., editor. Methods in molecular biology. Humana Press; Totowa, NJ: 2008. (in press). [DOI] [PubMed] [Google Scholar]
- Prudden, J., Pebernard, S., Raffa, G., Slavin, D.A., Perry, J.J., Tainer, J.A., McGowan, C.H., Boddy, M.N. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, M., Rash, L.D., Escoubas, P., Boilard, E., Bollinger, J., Lomonte, B., Maurin, T., Guillaume, C., Canaan, S., Deregnaucourt, C., et al. Neurotoxicity and other pharmacological activities of the snake venom phospholipase A2 OS2: The N-terminal region is more important than enzymatic activity. Biochemistry. 2006;45:5800–5816. doi: 10.1021/bi060217r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, M., Le Calvez, C., Boilard, E., Surrel, F., Singer, A., Ghomashchi, F., Bezzine, S., Scarzello, S., Bollinger, J., Gelb, M.H., et al. Recombinant production and properties of binding of the full set of mouse secreted phospholipases A2 to the mouse M-type receptor. Biochemistry. 2007;46:1647–1662. doi: 10.1021/bi062119b. [DOI] [PubMed] [Google Scholar]
- Schaloske, R.H., Dennis, E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 2006;1761:1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Singer, A.G., Ghomashchi, F., Le Calvez, C., Bollinger, J., Bezzine, S., Rouault, M., Sadilek, M., Nguyen, E., Lazdunski, M., Lambeau, G., et al. Interfacial kinetic and binding properties of the complete set of human and mouse groups I, II, V, X, and XII secreted phospholipases A2 . J. Biol. Chem. 2002;277:48535–48549. doi: 10.1074/jbc.M205855200. [DOI] [PubMed] [Google Scholar]
- Six, D.A., Dennis, E.A. The expanding superfamily of phospholipase A2 enzymes: Classification and characterization. Biochim. Biophys. Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- Webb, N.R. Secretory phospholipase A2 enzymes in atherogenesis. Curr. Opin. Lipidol. 2005;16:341–344. doi: 10.1097/01.mol.0000169355.20395.55. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Dong, L., Cai, M., Shen, J., Wang, Y. Heterologous expression of lipoprotein-associated phospholipase A2 in different expression systems. Protein Expr. Purif. 2006;48:300–306. doi: 10.1016/j.pep.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Zuo, X., Li, S., Hall, J., Mattern, M.R., Tran, H., Shoo, J., Tan, R., Weiss, S.R., Butt, T.R. Enhanced expression and purification of membrane proteins by SUMO fusion in Escherichia coli . J. Struct. Funct. Genomics. 2005;6:103–111. doi: 10.1007/s10969-005-2664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]