Abstract
Screening of transfer DNA (tDNA) tagged lines of Arabidopsis thaliana for mutants defective in systemic acquired resistance led to the characterization of dir1-1 (defective in induced resistance [systemic acquired resistance, SAR]) mutant. It has been suggested that the protein encoded by the dir1 gene, i.e., DIR1, is involved in the long distance signaling associated with SAR. DIR1 displays the cysteine signature of lipid transfer proteins, suggesting that the systemic signal could be lipid molecules. However, previous studies have shown that this signature is not sufficient to define a lipid transfer protein, i.e., a protein capable of binding lipids. In this context, the lipid binding properties and the structure of a DIR1–lipid complex were both determined by fluorescence and X-ray diffraction. DIR1 is able to bind with high affinity two monoacylated phospholipids (dissociation constant in the nanomolar range), mainly lysophosphatidyl cholines, side-by-side in a large internal tunnel. Although DIR1 shares some structural and lipid binding properties with plant LTP2, it displays some specific features that define DIR1 as a new type of plant lipid transfer protein. The signaling function associated with DIR1 may be related to a specific lipid transport that needs to be characterized and to an additional mechanism of recognition by a putative receptor, as the structure displays on the surface the characteristic PxxP structural motif reminiscent of SH3 domain signaling pathways.
Keywords: DIR1, Arabidopsis thaliana, systemic induced resistance, lipid transfer protein, SH3 signaling pathway, polyproline
Upon interactions with pathogens (virus, fungi, bacteria, etc.) a succession of events takes place in the plant cell. Hypersensitive response (HR) is a widespread mechanism used by hosts to prevent pathogen invasion. HR is mainly a localized plant cell death (necrosis) at the site of infection. The uninfected tissues of the plant undergoing HR are frequently more resistant to infections associated with stronger pathogens (Sticher et al. 1997). This increased tolerance to pathogens (or systemic acquired resistance, SAR) results, in planta, from a complex stimulation of defense mechanisms. SAR is long lasting and requires a signal transmission from the infected tissues toward healthy cells whose chemical nature remains to be characterized. By screening transfer DNA (tDNA) tagged lines of Arabidopsis thaliana for mutants defective in SAR, the dir1-1 mutant (defective in induced resistance) was isolated (Maldonado et al. 2002). Dir1-1 exhibits wild-type local resistance to avirulent Pseudomonas syringae but was unable to develop SAR. This suggested that the protein encoded by the dir1 gene, i.e., DIR1, was involved in the long distance signaling associated with SAR. Interestingly, DIR1 displays the cysteine signature of the lipid transfer protein (LTP) family. LTPs are ubiquitous lipid-binding proteins from plants. LTPs can be divided into two multigenic families, LTP1 (∼90 amino acids long) and LTP2 (∼70 amino acids long). LTPs have been associated in many developmental and stress response processes, but their precise function is still unknown. They have been involved in the mobilization of storage lipids (Edqvist and Farbos 2002), the formation of cuticles (Sterk et al. 1991; Hollenbach et al. 1997), pollen tube adhesion (Chae et al. 2007), cell wall loosening (Nieuwland et al. 2005), antimicrobial activity and response to cold and wounding stress (Pearce et al. 1998; Yubero-Serrano et al. 2003), as well as to microbial invasions (Molina et al. 1993). Besides, LTPs have been described as ubiquitous allergens (Marion et al. 2004). LTPs were also demonstrated to share a number of structural similarities with oomycetous elicitins and to compete for the same plasma membrane receptors (Buhot et al. 2001; Blein et al. 2002).
LTP1 fold is characterized by a bundle of four to five helices stabilized by four disulfide bridges, partly wrapped by a long C-terminal segment. The consensus motif of eight cysteine residues engaged in four disulfide bridges is usually considered as the typical LTP signature. The overall structure delimits a large central hydrophobic cavity, which is able to bind long fatty acid chains. Most of the putative functions of LTPs are related to their ability to bind lipids in their hydrophobic cavity (Maldonado et al. 2002; Buhot et al. 2004; Nieuwland et al. 2005). The folding of LTP2 proteins is related to that of LTP1 with, however, drastic changes in their central hydrophobic cavity. These modifications are induced by a move at the C-terminal segment, revealing the plasticity of the lipid-binding site. The three-dimensional structures of several LTP1 from grains of wheat (Gincel et al. 1994; Charvolin et al. 1999; Tassin-Moindrot et al. 2000), rice (Lee et al. 1998), maize (Shin et al. 1995; Gomar et al. 1996; Han et al. 2001), barley (Heinemann et al. 1996; Lerche et al. 1997), from flower organs of tobacco (Da Silva et al. 2005), and from peach fruit (Pasquato et al. 2006) have been determined in their uncomplexed and lipid-complexed forms by either solution NMR or X-ray diffraction methods. In contrast, only two structures of LTP2 are available, the first one from wheat in complex with a lipid (NMR, [Pons et al. 2003]; X-ray diffraction, [Hoh et al. 2005]), the second one from an unliganded structure from rice (Samuel et al. 2002).
DIR1, which comprises 77 amino acids and displays the characteristic cysteine signature, has a low sequence identity with the two LTP2, the structure of which was already determined (vide infra). DIR1 exhibits an acidic pI of 4.25 whereas most of the LTPs are basic proteins. However, the presence of both the LTP-type cysteine signature and a LTP-like fold is also shared by proteins that are unable to bind lipids as revealed by three-dimensional studies and in vitro lipid-binding analyses (Baud et al. 1993; Tassin et al. 1998).
It has been suggested that DIR1 interacts with a lipid derived molecule to promote long distance signaling in A. thaliana (Maldonado et al. 2002). To understand how DIR1 could play its role at the molecular level in long distance signaling, we have analyzed the affinity of DIR1 for lipids by fluorescence spectroscopy and determined its three-dimensional structure complexed to lipids.
Results and Discussion
Lipid binding
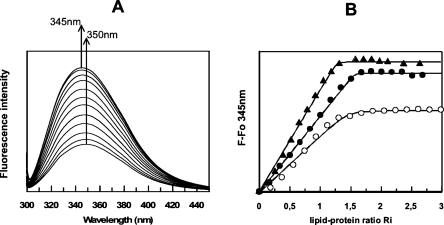
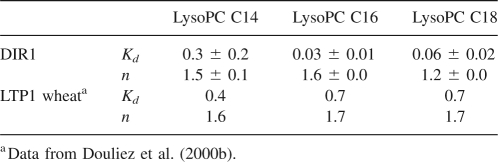
To probe the lipid–protein interaction, lysophosphatidyl cholines (LPC) were used with various fatty acid chain lengths. Lysophosphatidyl cholines, as model lipids, were compared with previous data obtained on plant LTP1s (Douliez et al. 2000a). Actually, these amphiphilic lipids, forming micelles in an aqueous environment, are also efficient to initiate crystallization of LTPs as they mimic natural phospholipids (Charvolin et al. 1999). In an aqueous environment, fluorescence emission of the DIR1 tryptophan 51 residue (Fig. 1) displays a maximum wavelength centered at 350 nm. When the protein binds lysophospholipids, this maximum is only slightly shifted to 345 nm. Therefore, the Trp 51 environment is not expected to change drastically. However, the fluorescence intensity of this residue is highly sensitive to the lipid binding with an increase of up to 200%–250% at saturation that was attributed to a decrease of the fluorescence quenching, itself formerly related to a decrease in mobility of the fluorescent probe. Longer fatty acid chain lengths are more efficient to alleviate tryptophan quenching than short ones with LPC-18>LPC-16>LPC-14, (Fig. 2). This implies that mobility of Trp 51 becomes constrained by the presence of lipids. A similar effect was previously observed for the fluorescence of tyrosine in LTP1s (Douliez et al. 2000b). Fitting of the titration curves, i.e., fluorescence intensity at 345 nm versus the lipid-protein molar ratio, using a noncooperative binding model with identical and independent binding sites provides the dissociation constant (Kd) and the number of lipid binding sites (Table 1). From these results, DIR1 displays higher affinities (nanomolar range) than LTP1s (micromolar range) for lysophospholipids with a chain length >14 carbon atoms and could bind up to two lipids. No lipid-binding data are available for LTP2s, but a study on the complex between rice LTP2 and sterol provided a Kd ∼ 70 μM (Cheng et al. 2004). The lipid-binding properties of DIR1 led us to select lysoPC C18 as the best ligand for crystallization, since it was the most efficient to constrain tryptophan mobility, and, by extension, the protein dynamics.
Figure 1.
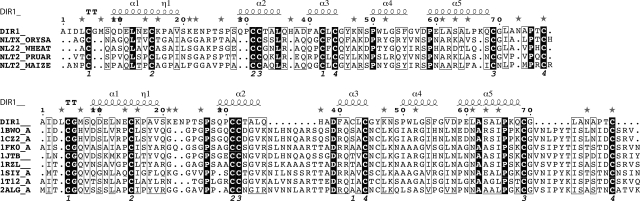
Amino acid sequence alignment of DIR1 and plant ns-LTP1 (top) and LTP2 (bottom) of known three-dimensional structure. The different sequences are labeled after their PDB identification code for LTP1s: 1BWO, wheat; 1JTB, barley; 1FK0, maize; 1RZL, rice; 1T12, tobacco; 2ALG, peach; 1SIY, mung bean. For LTP2s, codes are: NLTX_ORYSA (swissprot|P83210) or 1L6H (PDB) for rice; NL22_WHEAT (swissprot|P82901) or 1TUK (PDB) for wheat; NLT2_PRUAR for Prunus armeniaca (swissprot|P82353), and NLT2_MAIZE for maize (swissprot|P83506). The amino acids are numbered according to the sequence of DIR1. The numbers of the SS bonds are reported below alignments. In the case of LTP1s, two loops must be deleted to fit the sequences correctly.
Figure 2.
(A) Fluorescence emission spectra of DIR1 with increasing amounts of lysoPC c218. (B) Increase of fluorescence intensity (F − Fo) at 345 nm vs. lipid-protein molar ratio: ▲, lysoPC C18; ●, lysoPC C16; ○, lysoPC C14.
Table 1.
Dissociation constants (Kd, μM) and number of sites (n) for binding of lysophosphatidyl cholines to DIR1 and wheat LTP1
From a functional standpoint, Maldonado and colleagues showed that overexpression of DIR1 could not induce SAR without a pathogen infection (Maldonado et al. 2002). Based on homology of the amino acid sequence of DIR1 with those of LTPs, these authors suggested that SAR would require the formation of a complex between DIR1 and a lipid molecule generated from pathogen infection. Herein, we demonstrate that DIR1 is actually able to bind lipids as LTP1s do, but with a higher affinity in the case of monoacylated lipids. Interestingly, such lipids are released on the action of secreted lipases following a pathogen attack, and as such could play a role in defense signaling (Jirage et al. 1999).
X-ray structure of the (1:2) DIR1 lysostearoylphosphatidyl choline complex
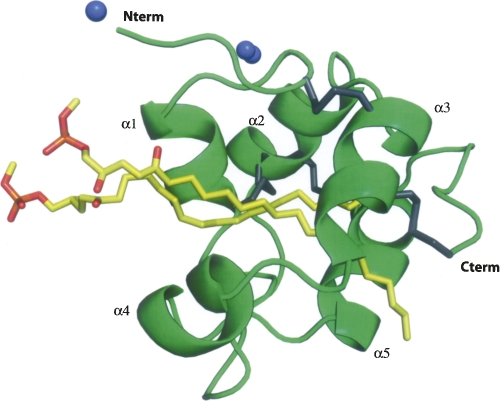
The final model of the lipid–protein complex contains the complete 77 amino acid protein chain, two lipid molecules, two zinc atoms, and 124 water molecules (Fig. 3). The Cter end presents a loop (residues 71–76) less ordered than the core of the structure with larger B factors than the rest of the protein. Alternate conformations could be clearly modeled for 17 residues. One of the two zinc atoms (Zn 2) has also been modeled in two alternate positions. The two C18 (stearoyl) chains of the lipid molecules are clearly defined in the density map. They run parallel to each other with their polar choline heads being located into the solvent area.
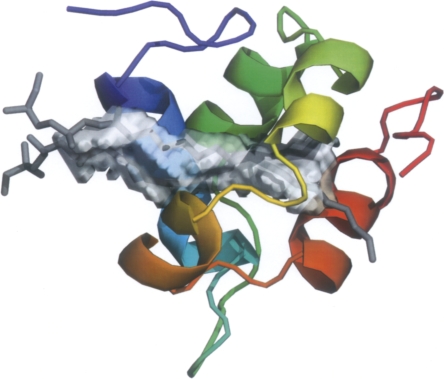
Figure 3.
Overall structure of DIR1. The five helices are labeled, the four disulfide bridges are represented as gray junctions, the zinc ions are denoted as blue spheres, and the two lipids (lyso stearoylphosphatidyl choline) located inside the central cavity are in yellow.
The three-dimensional model of DIR1 follows the general LTP fold with five α-helices connected by four disulfide bonds arranged in a super helical pattern around a central tunnel-shaped cavity. After an elongated N-terminal segment followed by a turn, the DIR1 structure begins with a long α-helix (α1). Three residues in 3/10-helix conformation complete this first α-helix. Between the α1 and α2 helices, the chain is folded in a β-turn followed by a 5-amino acid long poly-proline type II helix (PPII) left-handed helix (Stapley and Creamer 1999). The core of the molecule forms a left-handed super helical arrangement of four α-helices (α2, α3, α4, α5) building the hydrophobic central cavity. The C-terminal segment has no defined secondary structure, except for the last residue, Cys 77, which is involved in a disulfide bond. The four disulfide bridges (Cys 5–Cys 42, Cys 15–Cys 31, Cys 32–Cys 69, Cys 44–Cys 77), with the first and the last bridges being side by side, are reminiscent of a LTP2-type fold (Douliez et al. 2000a).
The two Zn++ ions, the presence of which was compulsory in the crystallization process, act as cross-linkers between two symmetry-related molecules. Each of the two zinc ions exhibits a tetrahedral coordination, involving residues from two different molecules in the packing. Zn 1 is coordinated with N and O from Ala 1, a water molecule and Oδ1 from Asp 58 of a symmetry-related protein. The second zinc (Zn 2) is found in two alternate positions 50:50 and coordinated with Glu 11, Asp 39*, and His 37.
The volume of the large central cavity is 242 Å3, as calculated with VOIDOO (Fig. 4; Kleywegt and Jones 1994) after removing the two lipids. The internal cavity is fully lined by hydrophobic residues: Ile 2, Leu 4, Leu 12, Val 19, Leu 35, Phe 40, Leu 43, Tyr 46 (the hydrophobic part of the aromatic ring), Leu 52, Phe 55, Val 57, Phe 62, Leu 65, and to some extent Trp 51. Some polar residues are located around the large tunnel entrance (Gln 9, Asn 13, Lys 16), guiding the two lipid heads toward the external solvent area. At the other end, the cavity is closed by three proline residues (Pro 59, Pro 66, and Pro 75) and by the Cys 44–Cys 77 bridge. The nonpolar ends (C18) of the two LPC molecules are completely buried within the protein.
Figure 4.
Delineation of the central lipid cavity in DIR1. The two lipid molecules are drawn as gray sticks. The cavity runs at 45° with respect to the five lining helices.
DIR1: LTP1 or LTP2 fold?
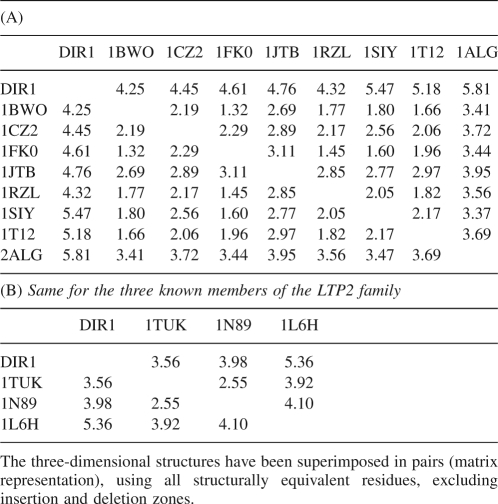
As mentioned previously, the two families of lipid transfer proteins LTP1 and LTP2 differ by their molecular weight (9 kDa and 7 kDa, respectively), and by their disulfide bond patterns (Douliez et al. 2001). In DIR1, the four cysteine pairs, Cys 5–Cys 42, Cys 15–Cys 31, Cys 32–Cys 69, Cys 44–Cys 77, reveal a side-by-side connectivity for the central motif C 42–X–C 44, which is typical of the LTP2 family. In addition, with 77 amino acids, the length of DIR1 is also closer to LTP2s (70 aa) than to LTP1s (90 aa). The DIR1 three-dimensional structure has been further compared with all available structures of LTP2 proteins (Table 2A,B; Samuel et al. 2002; Pons et al. 2003; Hoh et al. 2005). Despite the small number of known structures, a large discrepancy between similar proteins is noticed, e.g., the difference between the solution (1N98) and crystal structure (1TUK) of the wheat LTP2 is unusually large (RMSD 2.55 Å). In this context, the differences between the DIR1 and the LTP2 structures are not surprising. When the structure of DIR1 is compared with plant LTP1 proteins (Lerche et al. 1997; Lee et al. 1998; Charvolin et al. 1999; Han et al. 2001; Da Silva et al. 2005; Lin et al. 2005; Pasquato et al. 2006), even though LTP1 structures show smaller RMS differences within the family than LTP2s—particularly when considering cereal LTP1 proteins—the RMS differences between main-chain atoms of DIR1 and LTP1s still remain >4 Å. DIR1 seems thus farther from LTP1s than LTP2s. Therefore, DIR1 belongs to the LTP2 family rather than LTP1, although being somewhat atypical by its longer sequence and an unusual acidic pI of 4.25.
Table 2.
RMS differences (Å) between the coordinates of equivalent main-chain atoms of DIR1 and LTP1 protein structures, labeled with their PDB code
Structural comparison of DIR1 cavity with known structures of LTP2 highlights sharp contrasts. First, the position of the two octadecyl chains of LPCs is fully extended and, running side-by-side and parallel to each other from the bottom of the cavity to the entrance, has essentially van der Waals contact to the walls (the shortest distance being 3.3 Å between Asn 13-Nδ2 and LPC 1-O1A, at the entrance of the cavity) of DIR1. In the X-ray structure of wheat LTP2 (Hoh et al. 2005) the two L-α-palmitoyl-phosphatidyl-glycerol (LPG) moieties are located in two different compartments of the cavity built by a hydrophobic lining of the three residues Leu 7, Ile 14, and Leu 28. The two lipids are oriented tail-to-tail and only partially buried into the core of the protein. Besides, in the NMR structure of the same wheat LTP2 protein (PDB code 1N89; [Pons et al. 2003]), only one molecule of LPG occupies the long continuous tunnel-shaped central part of the cavity. In DIR1, the central tunnel is large enough to accommodate two parallel lipid molecules. When superimposing the structures of DIR1 and wheat LTP2 structures, one of the two LPCs of DIR1 can be superimposed to the two partial LPC moieties of 1TUK (Hoh et al. 2005). In the DIR1 sequence, the three corresponding residues of the inner wall are Leu 12, Val 19, and Leu 35, respectively. When superimposing DIR1 and wheat LTP2 X-ray structures, Leu 35 (wheat LTP2) exactly superimposes on Leu 28 (DIR1); this is also the case for the pair Ile 14/Val 19. The main difference comes from Leu 7/Leu 12 residues, which belong to the first helix in both 1TUK and DIR1 structures. In 1TUK, the 3/10 helix forms an angle of ∼90° with the α1 helix, while in DIR1 the first α1 helix and the second 3/10 helix are almost collinear. This opens widely the central channel, allowing entry and room for two lipid molecules. The displacement of the α4 helix of DIR1 toward the outside, with respect to the wheat LTP2 structure, also contributes to enlarge the cavity of DIR1.
A putative recognition motif
It has been shown that LTPs bind to high affinity sites of tobacco plasmalemma, which also interact with elicitin (Buhot et al. 2001, 2004). These small cysteine-rich proteins secreted by Phytophtora and Pythium Oomycetes share some structural properties with LTP and trigger a hypersensitive response and SAR in tobacco plants (Ponchet et al. 1999). The initial step of the plant response is the formation of a sterol–elicitin complex (Osman et al. 2001). A number of biophysical methods have shown that rice nsLTP2 can accommodate a planar sterol molecule whereas rice nsLTP1 can only bind linear fatty acids (Samuel et al. 2002; Cheng et al. 2004). The authors hypothesized that such a sterol/LTP2 complex could interact with the plant membrane receptor to induce plant defense response. However, lipid-binding studies indicated that DIR1 was unable to bind dihydroergosterol in vitro (data not shown). In addition, whatever the method used (soaking or co-crystallization), no crystallization of a sterol–DIR1 complex was obtained, unlike elicitin (Lascombe et al. 2002; Rodrigues et al. 2006). Nevertheless, a functional loading of DIR1 by another lipid molecule cannot be ruled out as observed in tobacco where a LTP1/jasmonate complex is required to induce plant protection (Buhot et al. 2004).
It is worth noting that two regions of DIR1 are folded in PPIIs: the five residue segment Pro 24 to Ser 28, located in the long loop between helices α1 and α2, and the four residues Leu 71 to Ala 74 in the C-terminal end after the α5 helix. The left-handed PPIIs are composed of residues whose φ and ψ angle values are about −75° and 145°, respectively (Stapley and Creamer 1999). Short PxxP proline motifs can be found in LTPs structures, in particular in wheat LTP2 (1TUK), where one motif is located in the loop between the two α1 and α2 helices and the other in the C-terminal region, in a similar way to DIR1. Nevertheless, the PxxPxxP (Pro 24 to Pro 30) recognition motif is unique in DIR1 and is not observed in any other LTP2 sequence.
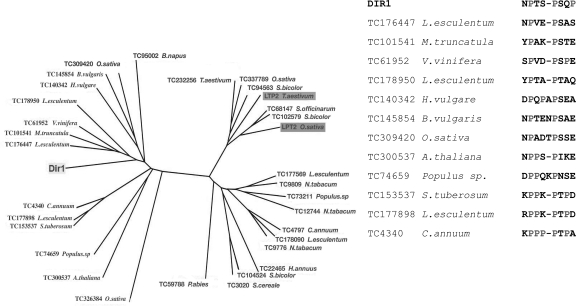
Comparison of the structure and lipid-binding properties of DIR1 with those of LTPs shows that DIR1 differs from LTP2 and represents a new type of lipid transfer protein. This was confirmed by a BLAST in the EST data banks of different plant species that returned DIR1-like putative proteins or putative LTP2 by starting from the DIR1 and from cereal (wheat and rice) seed LTP2, respectively. The corresponding phylogenetic tree constructed from the sequence alignments reveals that DIR1 and seed LTP2 fall in two different protein families (Fig. 5). The putative DIR1-like proteins from other plants that are recovered from BLAST in EST data banks do not highlight a similar PxxPxxP motif but still display high proline content that is not highlighted in proteins homologous to seed LTP2 (Fig. 5). It is noteworthy that proline-rich regions are involved in protein–protein interactions (Williamson 1994). They can interact with various protein domains and, among them, the Src homology 3 domain (SH3), frequently involved in the cell signaling pathway (Li 2005). The two PPII regions in DIR1 are located at the surface of the protein and fully accessible to the solvent (see Supplemental material). Thus, these two regions can be considered as putative candidates for the docking of a partner protein in a signaling function.
Figure 5.
(Left panel) Phylogenetic tree constructed from CLUSTAL alignment (PHYLIP software) of DIR1-like and seed LTP2-like proteins recovered from BLAST in EST data banks of different plant species (http://compbio.dfci.harvard.edu/tgi/). (Right panel) Alignment of the proline-rich domains of DIR1 and DIR1-like plant proteins recovered from BLAST in EST data banks of different plant species.
In summary, the complexation of DIR1 with stearoyl derivatives has been analyzed, and the structure of the complex determined by X-ray diffraction. The polypeptide chain folds in a structure reminiscent of the LTP2 family but displays a number of unique features, such as a large flexible cavity that can bind with high affinity two molecules of long-chain fatty acid derivatives, an acidic pI that makes DIR1 unique in the LTP family, a characteristic PxxPxxP motif on the surface that could be, upon lipid binding, a major structural determinant in the signaling functions of this new type of lipid transfer protein.
Materials and Methods
Expression and purification of DIR1
Cloning and expressing the DIR1 protein in yeast Pichia pastoris have been reported (Lascombe et al. 2006). DIR1 was purified using the following method: The Pichia pastoris culture supernatant was first concentrated and desalted on an adsorption resin (XAD7 resin, Amberlite). The protein was then eluted using an acetonitrile gradient. It was finally purified by reverse phase chromatography and freeze-dried for storage. The homogeneity and the purity were checked by mass spectroscopy.
Lipid binding by intrinsic tryptophan fluorescence spectroscopy
Tryptophan fluorescence emission spectra of DIR1 were recorded at 25°C with a Fluoromax steady-state spectrofluorometer (Spex-Jobin-Yvon). Excitation and emission wavelengths were set at 295 and 335 nm (slits 5 nm). Lysophosphatidyl cholines (from Sigma-Aldrich) were used without further purification. A concentrated lipid solution was prepared in absolute ethanol in order to have an ethanol concentration <5% at the end of titration. Titration curves were fitted using a noncooperative binding model assuming independent and identical sites as detailed in Dubreil et al. (1997).
Crystallization of DIR1
After being incubated with LPC as a putative ligand, in 1.5/1.0 (mol ligand/mol protein) ratio, the mixture was set to crystallize using the vapor diffusion technique in hanging drops. The best crystals were obtained by mixing 1 μL of the protein solution (20 mg/mL in 0.08 M sodium acetate buffer, pH 6.0) with 1 μL of the reservoir solution (20%–25% PEG 550 monomethyl ether, 0.02 M ZnSO4, and 0.1 M MES buffer, pH 6.5). Bipyramidal crystals of ∼0.2 mm were obtained within a month (Lascombe et al. 2006). In the absence of LPC, no crystal can be produced.
X-ray data collection and processing
Diffraction data were collected at the ESRF synchrotron facility, Grenoble (France). The crystals were flash-cooled in liquid nitrogen without any additional cryoprotection and were recorded at 100 K in a stream of gaseous nitrogen. As the presence of zinc sulfate was compulsory in the crystal formation, we expected the Zn++ ions in the crystal lattice, and data were recorded at the zinc absorption edge in order to benefit the anomalous scattering signal. Three data sets were collected using the same crystal up to 1.9 Å resolution on the ID29 beamline operating an ADSC Q315R detector, at the three energies corresponding to the peak (f′ max), the inflexion (f′ max) of the Zn K-edge, and at a remote wavelength (Lascombe et al. 2006). These data sets were used for phase determination. A second crystal allowed us to collect a native data set at a higher resolution (1.6 Å), on the ID14-EH1 beamline with an ADSC Q210 detector, which was used for refinement purposes. All data sets were reduced using the integrating program MOSFLM (Leslie 1999), merged with SCALA and put on an absolute scale with TRUNCATE, all from the CCP4 program suite (Collaborative Computational Project, Number 4 1994). The crystals belong to the orthorhombic P212121 space group with cell parameters: a = 28.65 Å, b = 48.22 Å, c = 54.41 Å. The unit cell volume of 75,171 Å3 suggests the presence of one molecule per asymmetric unit, with a Matthews coefficient of 2.3 Å3/Dalton and a solvent content of 46.9%.
Structure determination, model building, and refinements
The structure was solved using the multiple anomalous dispersion of the zinc atom near its K absorption edge (Zn-MAD). After formatting the data collected at the f′ peak with XPREP (Bruker 1997), the programs SHELXD and SHELXE (Schneider and Sheldrick 2002) were used to determine and refine the substructure of the zinc atoms. Three positions were clearly found, one fully occupied and the two other positions partially occupied. This protocol also allowed us to choose the correct hand of the structure. The SHARP package (de La Fortelle and Bricogne 1997; Bricogne et al. 2003) was then run, following the MAD protocol using the three data sets collected at 1.9 Å resolution. The ARP/Warp (Perrakis et al. 1999) procedure allowed tracing 69 out of the 77 amino acids of the polypeptide chain. The first electron density map revealed the presence of two lipid (LPC) molecules in the central cavity of the protein.
The final completion and adjustments of the model were performed in O (Jones et al. 1991) based on |2Fo − Fc| and |Fo − Fc| maps, at 1.9 Å resolution. Least squared restrained refinements of the structure were performed with REFMAC5 (Murshudov et al. 1997; Winn et al. 2001) in conjunction with the graphic program COOT (Emsley and Cowtan 2004) using the high-resolution data at 1.6 Å resolution. After the last cycle of refinement, the R-factor was 18.9% for the 9863 unique reflections in the 15.0–1.6 Å resolution range. The free R-factor was 25.2% for 4.8% of the data. The distribution of the 62 non-proline and non-glycine residues in a (ϕ, Ψ) Ramachandran plot reveals that 95.2% of the residues fall in the most favorable regions and the rest in the additional allowed regions. The final refinement statistics and stereochemical parameters of the model are reported in Table 3.
Table 3.
DIR1/LPC18 complex: Refinement statistics
Cavity calculations were performed using the VOIDOO program (Kleywegt and Jones 1994) with a 1.4 Å diameter for the accessibility probe, and the figures were drawn using PyMOL from DeLano Software. The structure alignments (Fig. 1) were generated with ESPript (Gouet et al. 2003) using the DIR1 structure as a reference for secondary structure elements. The coordinates and structure factors are deposited with the PDB ID code 2RKN.
Electronic supplemental material
Three additional figures include: initial electron density (X-ray determination), the zinc atom disordered in the structure, and a van der Waals surface of the protein showing the location of the polyproline motifs.
Acknowledgments
We thank the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities. We also thank Joanne McCarthy for assistance in using beamline ID29 and Ganesh Natrajan in using the beamline ID14eh1.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Thierry Prangé, Université Paris Descartes, Faculté de Pharmacie, 4 Avenue de l'Observatoire, 75006 Paris, France; e-mail: thierry.prange@univ-paris5.fr; fax: 33153739925.
Abbreviations: LTP, lipid transfer protein; PEG, polyethylene glycol; LPC18:0, lyso stearoyl phosphatidyl choline; LPG, L-α-palmitoyl-phosphatidyl-glycerol; MES, 2-morpholinoethane sulfonic acid; PPII, poly-proline type II helix; RMS, root mean squared; PDB, Protein Data Bank.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035972.108.
References
- Baud, F., Pebay-Peyroula, E., Cohen-Addad, C., Odani, S., Lehmann, M.S. Crystal structure of hydrophobic protein from soybean; a member of a new cysteine-rich family. J. Mol. Biol. 1993;231:877–887. doi: 10.1006/jmbi.1993.1334. [DOI] [PubMed] [Google Scholar]
- Blein, J.P., Coutos-Thevenot, P., Marion, D., Ponchet, M. From elicitins to lipid-transfer proteins: A new insight in cell signalling involved in plant defense mechanisms. Trends Plant Sci. 2002;7:293–296. doi: 10.1016/s1360-1385(02)02284-7. [DOI] [PubMed] [Google Scholar]
- Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M., Paciorek, W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Bruker. Bruker AXS Inc; Madison, Wisconsin: 1997. XPREP. [Google Scholar]
- Buhot, N., Douliez, J.P., Jacquemard, A., Marion, D., Tran, V., Maume, B.F., Milat, M.L., Ponchet, M., Mikes, V., Kader, J.C., et al. A lipid transfer protein binds to a receptor involved in the control of plant defence responses. FEBS Lett. 2001;509:27–30. doi: 10.1016/s0014-5793(01)03116-7. [DOI] [PubMed] [Google Scholar]
- Buhot, N., Gomes, E., Milat, M.L., Ponchet, M., Marion, D., Lequeu, J., Delrot, S., Coutos-Thevenot, P., Blein, J.P. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol. Biol. Cell. 2004;15:5047–5052. doi: 10.1091/mbc.E04-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, K., Zhang, K., Zhang, L., Morikis, D., Kim, S.T., Mollet, J.C., de la Rosa, N., Tan, K., Lord, E.M. Two SCA (stigma/style cysteine-rich adhesin) isoforms show structural differences that correlate with their levels of in vitro pollen tube adhesion activity. J. Biol. Chem. 2007;282:33845–33858. doi: 10.1074/jbc.M703997200. [DOI] [PubMed] [Google Scholar]
- Charvolin, D., Douliez, J.P., Marion, D., Cohen-Addad, C., Pebay-Peyroula, E. The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 Å resolution. Eur. J. Biochem. 1999;264:562–568. doi: 10.1046/j.1432-1327.1999.00667.x. [DOI] [PubMed] [Google Scholar]
- Cheng, C.S., Samuel, D., Liu, Y.J., Shyu, J.C., Lai, S.M., Lin, K.F., Lyu, P.C. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry. 2004;43:13628–13636. doi: 10.1021/bi048873j. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Da Silva, P., Landon, C., Industri, B., Marais, A., Marion, D., Ponchet, M., Vovelle, F. Solution structure of a tobacco lipid transfer protein exhibiting new biophysical and biological features. Proteins. 2005;59:356–367. doi: 10.1002/prot.20405. [DOI] [PubMed] [Google Scholar]
- de La Fortelle, E., Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Douliez, J.-P., Michon, T., Elmorjani, K., Marion, D. Mini review: Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 2000a;32:1–20. [Google Scholar]
- Douliez, J.-P., Michon, T., Marion, D. Steady-state tyrosine fluorescence to study the lipid-binding properties of a wheat nonspecific lipid-transfer protein (nsLTP1) Biochim. Biophys. Acta. 2000b;1467:65–72. [PubMed] [Google Scholar]
- Douliez, J.-P., Pato, C., Rabesona, H., Molle, D., Marion, D. Disulfide bond assignment, lipid transfer activity and secondary structure of a 7-kDa plant lipid transfer protein, LTP2. Eur. J. Biochem. 2001;268:1400–1403. doi: 10.1046/j.1432-1327.2001.02007.x. [DOI] [PubMed] [Google Scholar]
- Dubreil, L., Compoint, J.-P., Marion, D. Interaction of puroindolines with wheat flour polar lipids determines their foaming properties. J. Agric. Food Chem. 1997;45:108–116. [Google Scholar]
- Edqvist, J., Farbos, I. Characterization of germination-specific lipid transfer proteins from Euphorbia lagascae . Planta. 2002;215:41–50. doi: 10.1007/s00425-001-0717-x. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gincel, E., Simorre, J.P., Caille, A., Marion, D., Ptak, M., Vovelle, F. Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers. Eur. J. Biochem. 1994;226:413–422. doi: 10.1111/j.1432-1033.1994.tb20066.x. [DOI] [PubMed] [Google Scholar]
- Gomar, J., Petit, M.C., Sodano, P., Sy, D., Marion, D., Kader, J.C., Vovelle, F., Ptak, M. Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds. Protein Sci. 1996;5:565–577. doi: 10.1002/pro.5560050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet, P., Robert, X., Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G.W., Lee, J.Y., Song, H.K., Chang, C., Min, K., Moon, J., Shin, D.H., Kopka, M.L., Sawaya, M.R., Yuan, H.S., et al. Structural basis of nonspecific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 2001;308:263–278. doi: 10.1006/jmbi.2001.4559. [DOI] [PubMed] [Google Scholar]
- Heinemann, B., Andersen, K.V., Nielsen, P.R., Bech, L.M., Poulsen, F.M. Structure in solution of a four-helix lipid binding protein. Protein Sci. 1996;5:13–23. doi: 10.1002/pro.5560050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh, F., Pons, J.L., Gautier, M.F., de Lamotte, F., Dumas, C. Structure of a liganded type 2 nonspecific lipid-transfer protein from wheat and the molecular basis of lipid binding. Acta Crystallogr. D Biol. Crystallogr. 2005;61:397–406. doi: 10.1107/S0907444905000417. [DOI] [PubMed] [Google Scholar]
- Hollenbach, B., Schreiber, L., Hartung, W., Dietz, K.J. Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta. 1997;203:9–19. doi: 10.1007/s00050159. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Jones, T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Lascombe, M.B., Ponchet, M., Venard, P., Milat, M.L., Blein, J.P., Prange, T. The 1.45 Å resolution structure of the cryptogein-cholesterol complex: A close-up view of a sterol carrier protein (SCP) active site. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1442–1447. doi: 10.1107/S0907444902011745. [DOI] [PubMed] [Google Scholar]
- Lascombe, M.B., Buhot, N., Bakan, B., Marion, D., Blein, J.P., Lamb, C.J., Prange, T. Crystallization of DIR1, a LTP2-like resistance signalling protein from Arabidopsis thaliana . Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:702–704. doi: 10.1107/S1744309106023748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Min, K., Cha, H., Shin, D.H., Hwang, K.Y., Suh, S.W. Rice nonspecific lipid transfer protein: The 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. J. Mol. Biol. 1998;276:437–448. doi: 10.1006/jmbi.1997.1550. [DOI] [PubMed] [Google Scholar]
- Lerche, M.H., Kragelund, B.B., Bech, L.M., Poulsen, F.M. Barley lipid-transfer protein complexed with palmitoyl CoA: The structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure. 1997;5:291–306. doi: 10.1016/s0969-2126(97)00186-x. [DOI] [PubMed] [Google Scholar]
- Leslie, A.G. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- Li, S.S. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K.F., Liu, Y.N., Hsu, S.T., Samuel, D., Cheng, C.S., Bonvin, A.M., Lyu, P.C. Characterization and structural analyses of nonspecific lipid transfer protein 1 from mung bean. Biochemistry. 2005;44:5703–5712. doi: 10.1021/bi047608v. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis . Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Marion, D., Douliez, J.-P., Gautier, M.F., Elmorjani, K. Plant lipid transfer proteins: Relationships between allerginicity and structural, biological and technological properties. In: Mills E.N.C., Shewry P.R., editors. Plant food allergens. Blackwell Science; Oxford, UK: 2004. pp. 57–69. [Google Scholar]
- Molina, A., Segura, A., Garcia-Olmedo, F. Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 1993;316:119–122. doi: 10.1016/0014-5793(93)81198-9. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nieuwland, J., Feron, R., Huisman, B.A., Fasolino, A., Hilbers, C.W., Derksen, J., Mariani, C. Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell. 2005;17:2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, H., Vauthrin, S., Mikes, V., Milat, M.L., Panabieres, F., Marais, A., Brunie, S., Maume, B., Ponchet, M., Blein, J.P. Mediation of elicitin activity on tobacco is assumed by elicitin-sterol complexes. Mol. Biol. Cell. 2001;12:2825–2834. doi: 10.1091/mbc.12.9.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquato, N., Berni, R., Folli, C., Folloni, S., Cianci, M., Pantano, S., Helliwell, J.R., Zanotti, G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant nonspecific lipid transfer protein pan-allergens. J. Mol. Biol. 2006;356:684–694. doi: 10.1016/j.jmb.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Pearce, R.S., Houlston, C.E., Atherton, K.M., Rixon, J.E., Harrison, P., Hughes, M.A., Alison Dunn, M. Localization of expression of three cold-induced genes, blt101, blt4. 9, and blt14, in different tissues of the crown and developing leaves of cold-acclimated cultivated barley. Plant Physiol. 1998;117:787–795. doi: 10.1104/pp.117.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Ponchet, M., Panabieres, F., Milat, M.L., Mikes, V., Montillet, J.L., Suty, L., Triantaphylides, C., Tirilly, Y., Blein, J.P. Are elicitins cryptograms in plant-Oomycete communications? Cell. Mol. Life Sci. 1999;56:1020–1047. doi: 10.1007/s000180050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons, J.L., de Lamotte, F., Gautier, M.F., Delsuc, M.A. Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein. J. Biol. Chem. 2003;278:14249–14256. doi: 10.1074/jbc.M211683200. [DOI] [PubMed] [Google Scholar]
- Rodrigues, M.L., Archer, M., Martel, P., Miranda, S., Thomaz, M., Enguita, F.J., Baptista, R.P., Pinho e Melo, E., Sousa, N., Cravador, A., et al. Crystal structures of the free and sterol-bound forms of β-cinnamomin. Biochim. Biophys. Acta. 2006;1764:110–121. doi: 10.1016/j.bbapap.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Samuel, D., Liu, Y.J., Cheng, C.S., Lyu, P.C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa) J. Biol. Chem. 2002;277:35267–35273. doi: 10.1074/jbc.M203113200. [DOI] [PubMed] [Google Scholar]
- Schneider, T.R., Sheldrick, G.M. Substructure solution with SHELXD . Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Shin, D.H., Lee, J.Y., Hwang, K.Y., Kim, K.K., Suh, S.W. High-resolution crystal structure of the nonspecific lipid-transfer protein from maize seedlings. Structure. 1995;3:189–199. doi: 10.1016/s0969-2126(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Stapley, B.J., Creamer, T.P. A survey of left-handed polyproline II helices. Protein Sci. 1999;8:587–595. doi: 10.1110/ps.8.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, P., Booij, H., Schellekens, G.A., Van Kammen, A., De Vries, S.C. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 1991;3:907–921. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., Metraux, J.P. Systemic acquired resistance. Annu. Rev. Phytopathol. 1997;35:235–270. doi: 10.1146/annurev.phyto.35.1.235. [DOI] [PubMed] [Google Scholar]
- Tassin, S., Broekaert, W.F., Marion, D., Acland, D.P., Ptak, M., Vovelle, F., Sodano, P. Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry. 1998;37:3623–3637. doi: 10.1021/bi9723515. [DOI] [PubMed] [Google Scholar]
- Tassin-Moindrot, S., Caille, A., Douliez, J.P., Marion, D., Vovelle, F. The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 2000;267:1117–1124. doi: 10.1046/j.1432-1327.2000.01109.x. [DOI] [PubMed] [Google Scholar]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M.D., Isupov, M.N., Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Yubero-Serrano, E.M., Moyano, E., Medina-Escobar, N., Munoz-Blanco, J., Caballero, J.L. Identification of a strawberry gene encoding a nonspecific lipid transfer protein that responds to ABA, wounding and cold stress. J. Exp. Bot. 2003;54:1865–1877. doi: 10.1093/jxb/erg211. [DOI] [PubMed] [Google Scholar]