Abstract
Survivin is a member of the inhibitor of apoptosis protein (IAP) family that blocks cell death by inhibiting the caspase activation pathways. Overexpressed in all common human neoplasms but undetectable in most normal adult tissues, survivin confers tumor resistance to apoptosis and represents an ideal molecular target for therapeutic intervention. How survivin blocks apoptosis, however, has been a subject of intense debate, as evidenced by conflicting reports regarding whether or not survivin can directly bind and inactivate effector caspases. We chemically synthesized large amounts of highly pure human survivin of 142 amino acid residues using native chemical ligation and functionally compared synthetic survivin and a recombinant XIAP—the most intensively studied member of the IAP family. Inhibition assays showed that, while caspase-3 could be effectively inhibited by XIAP, survivin had no detectable inhibitory activity against the enzyme, even at concentrations several thousand-fold higher than XIAP. Our finding supports the premise that survivin does not directly inhibit effector caspases.
Keywords: enzymes, enzyme inhibitors, methods of protein and peptide synthesis, synthesis of peptides and proteins
Apoptosis is a genetically regulated cell death mechanism essential for the development and homeostasis of multicellular organisms. In mammalian cells, apoptosis can be triggered either by death ligand-induced cross-linking of the tumor necrosis factor (TNF) family of cell-surface receptors or by the release into the cytoplasm of mitochondrial proteins such as cytochrome c (Strasser et al. 2000). In spite of the different (intrinsic and extrinsic) pathways to cell death, all apoptosis signals converge on a common machinery of cell destruction by activated caspases—a family of cysteine proteases with aspartate substrate specificity (Salvesen and Dixit 1997; Thornberry and Lazebnik 1998). Upon stimulation by a death signal, the upstream “initiator” caspases (caspase-8 or -9) are activated, which, in turn, activate the downstream “effector” caspases (caspase-3 and -7), leading to the cleavage of vital intracellular proteins and, ultimately, to cell death.
Apoptosis is regulated by two families of proteins: the BCL2 family comprising both pro- and anti-apoptotic members (Cory and Adams 2002) and the inhibitor of apoptosis protein (IAP) family consisting only of anti-apoptotic molecules (Salvesen and Duckett 2002). To date, eight members of the IAP family have been identified in humans, among which survivin with 142 amino acid residues is the smallest. Expressed in most tumor cells but largely undetectable in normal differentiated tissues (Ambrosini et al. 1997), survivin confers malignant progression, poor prognosis, and tumor resistance to apoptosis. Ample evidence demonstrates that survivin suppression triggers caspase-dependent apoptosis both in vitro and in vivo, whereas expression of survivin is consistently associated with inhibition of induced cell death in cell culture systems and in transgenic animals as well (Altieri 2003). As a critical regulator of cell survival in tumors, survivin has emerged as a promising molecular target for anticancer therapy (Altieri 2003).
The most intensively studied member of the IAP family, XIAP, directly binds and inhibits caspase-3, -7, and -9 (Deveraux et al. 1997; Shi 2002). However, the mechanisms of inhibition of apoptosis by other members of the IAP family and by survivin, in particular, remain a matter of intense debate, as evidenced by several conflicting reports questioning whether or not survivin can directly bind and inactivate caspase-3 and -7 (Tamm et al. 1998; Banks et al. 2000; Conway et al. 2000; Shin et al. 2001). It has been suggested that the conflicting results are at least partially attributable to different forms of and purification methods for recombinant IAPs (Eckelman et al. 2006). To help further clarify the mode of action of survivin, obtaining large amounts of the protein from a different source would be of great advantage. We hereby describe total chemical synthesis of the full-length human survivin by the means of native chemical ligation and findings with synthetic survivin with respect to its inhibitory activity against human caspase-3.
Results and Discussion
Survivin can be chemically synthesized via native chemical ligation
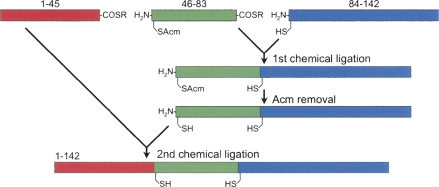
Native chemical ligation—a revolutionary synthetic methodology pioneered by Kent and co-workers (Dawson et al. 1994; Dawson and Kent 2000)—allows for facile synthetic access to large quantities of small- to medium-sized proteins in a highly efficient fashion. In a one-step NCL, the first peptide bearing a C-terminal thioester moiety chemoselectively reacts in aqueous solution with the second peptide containing an N-terminal Cys residue, resulting in a longer polypeptide chain linked by a new peptide bond. When multiple peptide segments are ligated either sequentially or convergently, a significantly longer polypeptide results. A two-step, sequential ligation strategy for the synthesis of survivin is illustrated in Figure 1.
Figure 1.
Strategy for the synthesis of human survivin by sequential native chemical ligation of three peptide fragments at ligation sites His45–Cys46 and Gly83–Cys84.
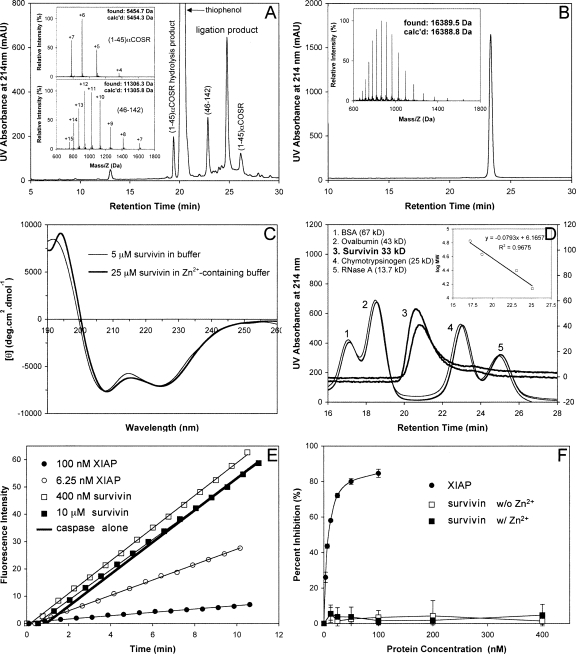
All three peptide fragments, (1–45)αCOSR (R = CH2CO-Leu-OH), (46–83)αCOSR, and (84–142), were individually synthesized on appropriate resin using Boc chemistry, purified by preparative reversed-phase HPLC to homogeneity, and verified by electrospray ionization mass spectrometry. To prevent an intramolecular ligation (head-to-tail backbone cyclization) within (46–83)αCOSR, the N-terminal residue Cys46 was orthogonally protected by acetamidomethyl (Acm). The first ligation reaction for (46–83)αCOSR and (84–142) was almost complete in 4 h, yielding (46–142) (11,376.6 Da found, 11,376.8 Da expected). Methods for Cys(Acm) deprotection in peptides are well established in the literature (Bang and Kent 2004). Nearly quantitative Cys46(Acm) deprotection was achieved by AgOAc treatment of the first ligation product (46–142) followed by DTT quenching. The second ligation between (1–45)αCOSR and Acm-free (46–142) proceeded to completion overnight (Fig. 2A). The determined molecular mass of 16,389.5 Da is within experimental error of the expected value of 16,388.8 Da calculated on the basis of the average isotopic compositions of the full-length protein (Fig. 2B). On a 0.25-mmol scale of synthesis, more than 100 mg of highly purified survivin was obtained.
Figure 2.
(A) Ligation of (1–45)αCOSR and (46–142) at 1.5 h. The reaction was monitored by analytical HPLC on a Waters XBridge C18 column (4.6 × 150 mm, 3.5 μM) running a 30-min gradient of 25%–45% acetonitrile containing 0.1% TFA at a flow rate of 1 mL/min. (Insets) Mass spectra of (1–45)αCOSR and (46–142) determined by ESI-MS. (B) Ligated full-length survivin characterized by C18 RP-HPLC and ESI-MS. HPLC conditions: Waters symmetry 300 C18 column (4.6 × 150 mm, 5 μM) running a 30-min gradient of 5%–65% acetonitrile containing 0.1% TFA at a flow rate of 1 mL/min. (C) CD spectra of synthetic survivin at 5 μM in 5 mM phosphate buffer containing 0.1 mM TCEP, pH 7.5 (thin line), and at 25 μM in 5 mM phosphate buffer containing 0.1 mM TCEP and 50 μM Zn2+, pH 7.5 (thick line). (D) Representative size-exclusion chromatograms of synthetic survivin (3) and molecular mass standards (1, 2, 4, 5). Linear regression analysis of the correlation between logarithmic M r and retention time is illustrated in the inset. (E) Representative raw data from the hydrolysis of Ac-DEVD-AMC by caspase-3 in the absence and presence of different concentrations of XIAP and synthetic survivin. (F) Dose-dependent percent inhibition of caspase-3 by XIAP (filled circles), synthetic survivin without Zn2+ (empty squares), and synthetic survivin in the presence of Zn2+ (filled squares). Each curve is the mean of three independent experiments.
Synthetic survivin adopts α-helical conformation and dimerizes in solution
Survivin is known to dimerize in solution. Crystallographic studies of human survivin confirmed that the protein consists of an N-terminal zinc-binding, baculovirus IAP repeat (BIR) domain and an elongated C-terminal amphipathic α-helix, arranged in a bow tie-shaped dimeric structure (Chantalat et al. 2000; Verdecia et al. 2000). The BIR domain is composed of a three-stranded antiparallel β-sheet packed against several short helices and stabilized globally by a Zn2+ ion tetrahedrally coordinated by Cys57, Cys60, Cys84, and His77. Dimerization of survivin is mediated primarily by hydrophobic interactions between two BIR domains and is thought to be functionally important.
Spontaneous folding of synthetic survivin was achieved by reconstituting the polypeptide into aqueous buffer. Shown in Figure 2C are near-UV CD spectra of synthetic survivin prepared at 5 μM in 5 mM phosphate buffer containing 0.1 mM TCEP, pH 7.5, and at 25 μM in the same buffer containing 50 μM Zn2+. The two CD spectra were similar, exhibiting double minima at 208 and 222 nm and a strong positive peak at ∼195 nm, characteristic of α-helical secondary structure and consistent with known structural features of the protein. In the absence of Zn2+, survivin, at 25 μM, showed signs of minor precipitation, and thus, its CD data were collected at a lower protein concentration (5 μM). This observation suggests that Zn2+ stabilizes the protein in solution. Interestingly, the mean residue ellipticity of synthetic survivin leveled off at ∼7000, indicative of an α-helical content significantly lower than implied by the crystal structure of human survivin (Verdecia et al. 2000). Our data suggest that the elongated amphipathic C-terminal α-helix observed crystallographically and involved directly in crystal packing contacts may not actually exist in solution. In fact, the crystal structure of a highly homologous mouse survivin and the NMR solution structure of a truncated human survivin (1–120) both showed that the C-terminal helix did not fully extend all the way to the end of the sequence (Muchmore et al. 2000; Sun et al. 2005). It is worth noting that a shower of small crystals of synthetic survivin was obtained from the conditions described by Verdecia et al. (2000). Large crystals were subsequently grown using the hanging-drop diffusion method at 4°C from 100 mM HEPES-Na buffer containing 200 mM trisodium citrate and 20% isopropyl alcohol. However, they diffracted poorly. Optimization of crystallization conditions is currently underway.
Additional evidence for the correct folding of synthetic survivin came from size-exclusion chromatography. Shown in Figure 2D is synthetic survivin along with four molecular mass standards on Superdex 75. Survivin eluted from the column between ovalbumin (43 kDa) and chymotrypsinogen (25 kDa). A calibration curve was obtained (Fig. 2D, inset), from which an apparent molecular mass of 33 kDa was derived for survivin, consistent with the predicted value of 32,778 Da for a homodimer of survivin. Notably, addition of EDTA in the buffer did not affect the retention time of survivin (data not shown), suggesting that dimerization of human survivin is not mediated by Zn2+ binding, a finding consistent with the structural features of human survivin (Verdecia et al. 2000).
Synthetic survivin does not directly inhibit caspase-3
We examined the proteolytic activity of caspase-3 to hydrolyze a fluorogenic substrate Ac-DEVD-AMC, in the absence and presence of synthetic survivin, in 50 mM HEPES containing 100 mM NaCl, 10 mM DTT, 10% glycerol, and 0.1% CHAPS, pH 7.4. Briefly, different concentrations of survivin were preincubated with a fixed concentration of caspase-3 (10 units, specific activity ≥2500 units/μg protein) at 25°C for 20 min, followed by addition to a total volume of 300 μL of assay solution of the substrate at a final concentration of 30 μM. The release of 7-amino-4-methylcoumarin (AMC) was monitored for at least 10 min on a fluorimeter with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. As shown in Figure 2, E and F, up to 10 μM survivin had little inhibitory activity against the enzyme. In sharp contrast, XIAP showed dose-dependent inhibition of caspase-3 at much lower protein concentrations (25% inhibition at 3.125 nM, 85% inhibition at 100 nM). Since trace amounts of EDTA (∼1 μM, originated from the caspase preparation) were present in the assay buffer, to eliminate the effect of Zn2+ or lack thereof on the inhibitory activity of synthetic survivin against caspase-3, we added Zn2+ in the assay buffer to a final concentration of ∼2 μM and repeated the dose-dependent inhibition assay under otherwise identical conditions. Our results clearly showed that synthetic survivin was inactive against caspase-3 independently of Zn2+ (Fig. 2F).
Reed and colleagues reported that human survivin bound specifically to caspase-3 and -7 in vitro and inhibited caspase activity and cell death in cells exposed to various apoptotic stimuli (Tamm et al. 1998). Shin et al. observed similar results with respect to caspase binding and inhibition using a recombinant human survivin (Shin et al. 2001). Two alternatively spliced forms of mouse survivin were also shown to inhibit caspase-3 activity by Conway et al. (2000). However, Altieri and colleagues sharply questioned the validity of the conclusions by Conway et al. (2000) by showing no measurable inhibitory activity of a recombinant mouse survivin and a native human survivin against caspase-3 (Banks et al. 2000). Growing evidence suggests that survivin suppresses apoptosis independently of direct caspase binding and inhibition (Suzuki et al. 2000; Marusawa et al. 2003; Song et al. 2003). In fact, structural studies indicate that survivin, unlike XIAP, lacks critical structural elements required for effective caspase inhibition (Eckelman et al. 2006). Our findings with synthetic survivin reinforce a growing consensus that XIAP is probably the only mammalian IAP that directly inhibits caspase activity (Eckelman et al. 2006). While the mechanisms by which survivin protects tumor cells from apoptosis remain unclear and await further examination, our facile synthetic access to large amounts of highly pure human survivin should provide a useful tool for elucidating the molecular basis of how this protein works in the anti-apoptotic pathways.
Materials and Methods
Materials
Chemicals used in peptide synthesis and purification have been described elsewhere (Li et al. 2005). EnzChek Caspase-3 assay kit #1 was purchased from Invitrogen; recombinant caspase-3 was obtained from Calbiochem and recombinant human XIAP, from R&D Systems.
Solid-phase synthesis of (1–45)αCOSR (R = CH2COLeu-OH), (46–83)αCOSR, and (84–142)
The amino acid sequence of the 142-residue survivin is shown in Figure 3. Two ligation sites, His45-Cys46 and Gly83-Cys84, are conveniently located in the amino acid sequence of survivin. The three peptide segments of survivin, (1–45)αCOSR, (46–83)αCOSR, and (84–142), were individually synthesized on HSCH2CO-Leu-OCH2-PAM and Boc-Asp(OBzl)-OCH2-PAM resin on an automated peptide synthesizer ABI 433A using an optimized HBTU activation/DIEA in situ neutralization protocol developed by Kent and co-workers for Boc chemistry (Schnolzer et al. 1992). Crude peptides, after HF cleavage and deprotection, were precipitated with cold ether and purified by preparative C18 reversed-phase (RP) HPLC, and their molecular masses ascertained by electrospray ionization mass spectrometry (ESI-MS).
Figure 3.
Amino acid sequence of the 142-residue survivin.
Native chemical ligation and spontaneous folding of survivin
Native chemical ligation was carried out in 0.1 M phosphate buffer containing 6 M guanidine hydrochloride and 2% thiophenol, pH 7.4. Removal of Acm was achieved by dissolving at 1 mg/mL the first ligation product, (46–142), in 50% acetonitrile containing 0.1% TFA, to which 300-fold molar excess of silver acetate (5 mg/mL stock) was added. The reaction proceeded for 1 h before being quenched by DTT, and the resultant product was purified by RP-HPLC to homogeneity for the second ligation. Folding of synthetic survivin was achieved by dissolving the polypeptide at 0.1 mg/mL in 25 mM Tris/HCl buffer containing 100 mM NaCl, 5 mM DTT, and 25 μM Zn2+, pH 7.5. The protein solution was quantified by UV measurements at 280 nm using a molar extinction coefficient of 16,500, calculated according to a published algorithm (Pace et al. 1995).
CD spectroscopy and size-exclusion chromatography
Far-UV CD spectra were obtained on a Jasco J-810 spectropolarimeter at 25°C using a 0.1-cm path length. Two survivin solutions were prepared: 5 μM in 5 mM phosphate buffer, 0.1 mM TCEP, pH 7.5, and 25 μM in 5 mM phosphate buffer, 0.1 mM TCEP and 50 μM Zn2+, pH 7.5.
The oligomerization state of synthetic survivin was determined by measuring the relative molecular mass (M r) by size-exclusion chromatography on an Amersham Biosciences Superdex 75 column (10 × 300 mm) running 25 mM Tris/HCl buffer containing 100 mM NaCl and 1 mM DTT, 25 μM Zn2+, pH 7.5, at a flow rate of 0.5 mL/min. Four proteins, bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen (25 kDa), and ribonuclease A (13.7 kDa) were chosen as standards. A calibration curve was obtained, i.e., log(M r) = −0.0793t (retention time) + 6.1657, from which the M r value of synthetic survivin was derived.
In vitro caspase inhibition assay
A spectrofluorometric assay of caspase-3 activity was carried out using the EnzChek Caspase-3 assay kit (Invitrogen) at room temperature in 50 mM HEPES, 100 mM NaCl, 10 mM DTT, 10% glycerol, and 0.1% CHAPS, pH 7.4. Caspase-3 and survivin were preincubated at 25°C for 20 min, and the mixture was transferred to a 96-well plate, followed by addition of the substrate Ac-DEVD-AMC. Activities were measured by the release of 7-amino-4-methylcoumarin (AMC) from the substrate using Varian (Cary) Eclipse fluorescence spectrophotometer accessorized with a Cary Eclipse microplate reader in kinetic mode with excitation and emission wavelengths of 360 and 460 nm, respectively. XIAP was used as a positive control.
Acknowledgments
We thank Dr. Xiangqun Li and Juahdi Monbo for technical help. This work was partially supported by a Research Scholar Grant CDD112858 from the American Cancer Society (to W.L.).
Footnotes
Reprint requests to: Wuyuan Lu, Institute of Human Virology, University of Maryland School of Medicine, 725 West Lombard Street, Baltimore, MD 21201, USA; e-mail: wlu@ihv.umaryland.edu; fax: (410) 706-7583.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036145.108.
References
- Altieri, D.C. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Ambrosini, G., Adida, C., Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Bang, D., Kent, S.B. A one-pot total synthesis of crambin. Angew. Chem. Int. Ed. Engl. 2004;43:2534–2538. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]
- Banks, D.P., Plescia, J., Altieri, D.C., Chen, J., Rosenberg, S.H., Zhang, H., Ng, S.C. Survivin does not inhibit caspase-3 activity. Blood. 2000;96:4002–4003. [PubMed] [Google Scholar]
- Chantalat, L., Skoufias, D.A., Kleman, J.P., Jung, B., Dideberg, O., Margolis, R.L. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual α-helical extensions. Mol. Cell. 2000;6:183–189. [PubMed] [Google Scholar]
- Conway, E.M., Pollefeyt, S., Cornelissen, J., DeBaere, I., Steiner-Mosonyi, M., Ong, K., Baens, M., Collen, D., Schuh, A.C. Three differentially expressed survivin cDNA variants encode proteins with distinct antiapoptotic functions. Blood. 2000;95:1435–1442. [PubMed] [Google Scholar]
- Cory, S., Adams, J.M. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Dawson, P.E., Kent, S.B. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- Dawson, P.E., Muir, T.W., Clark-Lewis, I., Kent, S.B. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Deveraux, Q.L., Takahashi, R., Salvesen, G.S., Reed, J.C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Eckelman, B.P., Salvesen, G.S., Scott, F.L. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., de Leeuw, E., Lu, W. Total chemical synthesis of human psoriasin by native chemical ligation. Biochemistry. 2005;44:14688–14694. doi: 10.1021/bi051519g. [DOI] [PubMed] [Google Scholar]
- Marusawa, H., Matsuzawa, S., Welsh, K., Zou, H., Armstrong, R., Tamm, I., Reed, J.C. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore, S.W., Chen, J., Jakob, C., Zakula, D., Matayoshi, E.D., Wu, W., Zhang, H., Li, F., Ng, S.C., Altieri, D.C. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell. 2000;6:173–182. [PubMed] [Google Scholar]
- Pace, C.N., Vajdos, F., Fee, L., Grimsley, G., Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen, G.S., Dixit, V.M. Caspases: Intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- Salvesen, G.S., Duckett, C.S. IAP proteins: Blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Schnolzer, M., Alewood, P., Jones, A., Alewood, D., Kent, S.B. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int. J. Pept. Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- Shi, Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shin, S., Sung, B.J., Cho, Y.S., Kim, H.J., Ha, N.C., Hwang, J.I., Chung, C.W., Jung, Y.K., Oh, B.H. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- Song, Z., Yao, X., Wu, M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during Taxol-induced apoptosis. J. Biol. Chem. 2003;278:23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- Strasser, A., O'Connor, L., Dixit, V.M. Apoptosis signaling. Annu. Rev. Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- Sun, C., Nettesheim, D., Liu, Z., Olejniczak, E.T. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry. 2005;44:11–17. doi: 10.1021/bi0485171. [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Ito, T., Kawano, H., Hayashida, M., Hayasaki, Y., Tsutomi, Y., Akahane, K., Nakano, T., Miura, M., Shiraki, K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- Tamm, I., Wang, Y., Sausville, E., Scudiero, D.A., Vigna, N., Oltersdorf, T., Reed, J.C. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- Thornberry, N.A., Lazebnik, Y. Caspases: Enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- Verdecia, M.A., Huang, H., Dutil, E., Kaiser, D.A., Hunter, T., Noel, J.P. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]