Abstract
With the aid of monoclonal antibody (mAb) 2625, raised against the lipopolysaccharide (LPS) of Legionella pneumophila serogroup 1, subgroup OLDA, we isolated mutant 811 from the virulent wild-type strain RC1. This mutant was not reactive with mAb 2625 and exhibited an unstable phenotype, since we observed an in vitro and in vivo switch of mutant 811 to the mAb 2625–positive phenotype, thus restoring the wild-type LPS. Bactericidal assays revealed that mutant 811 was lysed by serum complement components, whereas the parental strain RC1 was almost serum resistant. Moreover, mutant 811 was not able to replicate intracellularly in macrophage-like cell line HL-60. In the guinea pig animal model, mutant 811 exhibited significantly reduced ability to replicate. Among recovered bacteria, mAb 2625–positive revertants were increased by fourfold. The relevance of LPS phase switch for pathogenesis of Legionella infection was further corroborated by the observation that 5% of the bacteria recovered from the lungs of guinea pigs infected with the wild-type strain RC1 were negative for mAb 2625 binding. These findings strongly indicate that under in vivo conditions switching between two LPS phenotypes occurs and may promote adaptation and replication of L. pneumophila. This is the first description of phase-variable expression of Legionella LPS.
Keywords: Legionella pneumophila, LPS, phase-variation, serum resistance, virulence
Legionella pneumophila is the causative agent of Legionnaires' disease, a severe pneumonia with frequently fatal progression (1). The habitat of Legionella species are natural or man-made water reservoirs where the bacteria survive and multiply intracellularly in amebae (2–4) in tight association with biofilms (5–7). Infection of man occurs by inhalation of Legionella-containing aerosols, but person to person transmission has never been observed (1, 8). In the human lung attachment of L. pneumophila and internalization into alveolar-macrophages is mediated by the major outer membrane protein, MOMP,1 the complement factors C3b and iC3b and the corresponding receptors (9, 10). In phagocytes fusion of Legionella-containing phagosomes with lysosomes is prevented and L. pneumophila survives and multiplies within macrophages (11, 12).
Several virulence factors of L. pneumophila have been identified and characterized. The macrophage infectivity potentiator protein (Mip) plays an important role in infection of macrophages, although its precise function is unclear (13–20). The products of the icm and dot loci are required for intracellular multiplication. Again, their role in the pathogenesis of disease is unresolved (21–25). Likewise, LPS of L. pneumophila is considered a factor mediating pathogenicity (8). It is the major immunodominant antigen and represents the basis for the classification of serogroups (26–29). In contrast to enterobacterial LPS activation it has been shown that Legionella LPS is able to activate both the classical and the alternative complement pathway (30). Due to the exceptional chemical structure of the L. pneumophila LPS, it is likely that this molecule participates in a number of essential legionellae capabilities, such as adaptation to various environmental challenges (31). The L. pneumophila serogroup (SG) 1 (strain Philadelphia) LPS differs from that of other Gram-negative bacteria in that its lipid A section consists of long chain fatty acids which may account for the weak endotoxicity of the molecule (31). The O-specific chain is composed of an α-(2→ 4) interlinked 5-acetamidino-7-acetamido-8-O-acetyl-3,5,7,9-tetradeoxy-l-glycero-d-galacto-nonulosonic acid (legionaminic acid) homopolymer. This unusual sugar molecule completely lacks free hydroxyl groups and is thus very hydrophobic (31, 32). In addition, an isomer of legionaminic acid hypothesized to be a terminal sugar of the LPS O-chain has been recently detected (33). The outer core oligosaccharide also exhibits hydrophobic properties (34). Based on these findings, it can be assumed that L. pneumophila possesses a hydrophobic cell surface that may support concentration of the bacterium in aerosols as well as adherence to host cells (31, 35).
To further elucidate the role of the LPS molecule and the surface properties of L. pneumophila in adaptation to various exogenous conditions, we raised mAb against the LPS of L. pneumophila SG 1 (subgroup OLDA). In this study, we describe mAb 2625 which binds to this LPS. Moreover, we show that the O-chain as well as the core are required for binding of mAb 2625. With the aid of mAb 2625, we isolated an LPS mutant from the virulent patient isolate RC1 (subgroup OLDA). Here we report for the first time that the LPS structure appears to be a virulence determinant of L. pneumophila and that expression of L. pneumophila LPS occurs in a phase-variable manner.
Materials and Methods
Bacterial Strains and Cultivation.
L. pneumophila SG 1 strain RC1 (OLDA), a clinical isolate, was a generous gift from B. Wright (Rigshospitalet, Copenhagen, Denmark). All other Legionella strains were obtained from the American Type Culture Collection (Rockville, MD) and the National Collection of Type Cultures (London, UK), respectively. Strains and sources are listed in Table 1. Legionella strains were cultivated on charcoal yeast extract (CYE) agar supplemented with buffered charcoal yeast extract (BCYE) growth supplement and MWY selective supplement (Unipath-Oxoid, Wesel, Germany). Plates were incubated at 37°C under 5% CO2 for 48–72 h unless otherwise stated. Propagation in liquid media (1% wt/vol yeast extract supplemented with BCYE growth supplement) was carried out at 37°C under constant agitation.
Table 1.
Legionella Strains Used in this Study and Indication of the Source
| Strain | Source | Strain | Source | |||
|---|---|---|---|---|---|---|
| L. pneumophila SG 1 (OLDA) | ATCC 43109 | L. pneumophila SG 1 (Oxford) | ATCC 43110 | |||
| L. pneumophila SG 1 | ATCC 33152 | L. pneumophila SG 1 | ATCC 33153 | |||
| L. pneumophila SG 1 | ATCC 43108 | L. pneumophila SG 1 | ATCC 43112 | |||
| L. pneumophila SG 1 | ATCC 43106 | L. pneumophila SG 1 | ATCC 43107 | |||
| L. pneumophila SG 1 | NCTC 11191 | L. pneumophila SG 1 | NCTC 11193 | |||
| L. pneumophila SG 1 | NCTC 11201 | L. pneumophila SG 1 | NCTC 11231 | |||
| L. pneumophila SG 1 | NCTC 11378 | L. pneumophila SG 1 | NCTC 11404 | |||
| L. pneumophila SG 2 | ATCC 33154 | L. pneumophila SG 3 | ATCC 33155 | |||
| L. pneumophila SG 4 | ATCC 33156 | L. pneumophila SG 5 | ATCC 33216 | |||
| L. pneumophila SG 6 | ATCC 33215 | L. pneumophila SG 7 | ATCC 33823 | |||
| L. pneumophila SG 8 | ATCC 35096 | L. pneumophila SG 9 | ATCC 35289 | |||
| L. pneumophila SG 10 | ATCC 43283 | L. pneumophila SG 11 | ATCC 43130 | |||
| L. pneumophila SG 12 | ATCC 43290 | L. pneumophila SG 13 | ATCC 43736 | |||
| L. pneumophila SG 14 | ATCC 43703 | L. anisa | ATCC 35291 | |||
| L. cherrii | ATCC 35252 | L. erythra | ATCC 35303 | |||
| L. birminghamensis | ATCC 43702 | L. bozemanii | ATCC 33217 | |||
| L. dumoffii | ATCC 33279 | L. gormanii | ATCC 33297 | |||
| L. micdadei | ATCC 33204 |
Pseudomonas aeruginosa (ATCC 49266) was obtained from the American Type Culture Collection. The following strains were isolates from the Institut für Medizinische Mikrobiologie (Medizinische Hochschule Hannover, Germany): P. fluorescens, Bordetella pertussis, Acinetobacter lwoffii, and Escherichia coli.
Production of Monoclonal Antibodies.
6-wk-old female BALB/c mice (Zentralinstitut für Versuchstierkunde, Hannover, Germany) were immunized intraperitoneally for four times once a week with 2 × 108 L. pneumophila SG 1 strain RC1 viable cells as previously described (36). Before injection, bacteria were passaged once in a guinea pig as described below. At the end of the immunization regimen, mice were splenectomized and the spleen cells were fused with X63-Ag8.653 myeloma cells as described elsewhere (37). The culture supernatant fluids of growing clones were screened by ELISA with whole L. pneumophila SG 1 (strain RC1) cells as antigens. The resulting hybridomas were cloned by limiting dilution.
Immunoelectron Microscopy.
Bacteria were fixed with 0.5% formaldehyde and 0.2% glutaraldehyde (final concentrations) in 0.1 M PBS for 1 h on ice. After three washes with 0.1 M PBS containing 10 mM glycine to block free aldehyde groups, the cells were embedded by progressively lowering the temperature with Lowicryl K4M resin (38). The following modifications of the method were made: (a) after dehydration in 10% ethanol, the samples were treated with 0.5% uranyl acetate in 10% ethanol for 1 h on ice; (b) the infiltration step with 1 part ethanol and 1 part K4M resin was performed overnight; (c) the infiltration step with one part ethanol and two parts K4M resin lasted for 12 h; and (d) infiltration with pure K4M resin lasted for 2 d. After polymerization of the samples for 2 d at −35°C, samples were trimmed and polymerized for another day at room temperature. Ultrathin sections were incubated overnight with 200 μg of IgG per ml of mAb 2625 or mAb LPS-1 (39) at 4°C. mAb LPS-1 was purchased from Progen (Heidelberg, Germany). After washing with 0.1 M PBS, sections were incubated with protein A–gold complexes (10-nm diam; concentration giving an A520 of 0.02). The sections were subsequently rinsed with 0.1 M PBS containing 0.01% Tween 20 and then with distilled water. After air drying, the sections were counterstained with 4% aqueous uranyl acetate (pH 4.5) for 5 min. Samples were examined with a Zeiss EM 910 electron microscope at an acceleration voltage of 80 kV at calibrated magnifications.
Western Blot.
1 ml of bacterial cell suspensions (OD550nm 1.2) were centrifuged and the resulting pellet was resuspended in 100-μl sample solution (20% glycerol, 3% sodium dodecyl sulfate, 3% 2-mercaptoethanol, 1% bromphenol blue). The suspensions were heated to 100°C for 5 min before 5-μl aliquots were applied to 12.5% polyacrylamide gels. To ensure equal protein concentrations in each lane, control gels were stained with Coomassie Blue dye (Sigma Chemical Co., Deisenhofen, Germany). For analysis of LPS samples, 2 μg purified LPS was applied to gels after boiling in sample solution. Western blotting onto nitrocellulose filters was carried out as described by Towbin et al. (40). Filter membranes were blocked with 3% dried milk powder suspended in PBS. Immunostaining was performed with mAb 2625 and mAb LPS-1 (39), respectively, and subsequent incubation with alkaline phosphatase-labeled goat anti–mouse immunoglobulin (Dianova, Hamburg, Germany).
Colony Blot.
Nitrocellulose filters were soaked in sterile PBS before colonies grown on BCYE agar were blotted. Filter membranes were then placed for 5 minutes on paper filter sheets (Schleicher and Schuell, Dassel, Germany) soaked with 70% ethanol. Nitrocellulose filters were air-dried at room temperature and subsequently blocked with 3% dried milk powder suspended in PBS. Immunostaining of the filters was performed as described above.
Extraction of LPS and Isolation of O-Chain Polymers.
LPS of L. pneumophila SG 1 strain RC1 (subtype OLDA) and mutant 811 was isolated from dry cells by a modified phenol-chloroform- petroleum ether procedure as described (32, 41). Starting from washed and enzymatically degraded dried cells excellent yields were obtained ranging between 8.1% (wt/wt) for the wild-type and 9.9% (wt/wt) for the mutant, respectively. LPS (155 mg of the wild-type, 144 mg of the mutant) was degraded with 0.1 M NaOAc-HOAc buffer (pH 4.4, 30 ml) at 100°C for 4 h and the resulting precipitate was removed by centrifugation. The supernatant was freeze-dried and fractionated by gel permeation chromatography on a Sephadex G-50 (S) column (2.5 × 50 cm; Pharmacia Biotechnology Inc., Freiburg, Germany) using a pyridinium acetate buffer (pH 4.5) and monitoring with a Knauer differential refractometer. Appropriate fractions were pooled and lyophilized.
SDS-PAGE and LPS Silver Staining.
SDS-PAGE was carried out in 14% polyacrylamide gels using Mini-Protean II system (Bio-Rad Laboratories, München, Germany). LPS bands were visualized by the silver-staining technique as described elsewhere (42).
Chemical LPS Analysis by Gas–Liquid Chromatography and Nuclear Magnetic Resonance.
Gas–liquid chromatography (GLC) was performed with a Varian Model 3700 chromatograph equipped with a capillary column of SPB-5 using a temperature gradient 150→ 320°C at 5°C/min. GLC-mass spectrometry in both chemical ionization (CI, with ammonia) and electron impact (EI) modes was carried out with a Hewlett-Packard Model 5989 instrument equipped with a capillary column of HP-1 under the same chromatographic conditions as in GLC. Monosaccharides were analyzed by GLC after methanolysis with 2 M HCl/MeOH (120°C, 16 h) and acetylation with Ac2O in pyridine (70°C, 0.5 h). 1H- and 13C-nuclear magnetic resonance (NMR) spectra were obtained with a Bruker AM-360 spectrometer for solutions in D2O at 60°C with acetone (δH 2.225, δC 31.45) as internal standard. Standard Bruker software was used in all 1H- and 13C-NMR experiments.
Competition ELISA.
1 μg purified LPS from L. pneumophila strain RC1 in 20 μl 0.2 M sodium carbonate buffer, pH 9.6, per well was adsorbed to microtiter plates (Microlon, Greiner, Nürtingen, Germany) overnight at 4°C. Plates were subsequently blocked with 2% (wt/vol) dried milk powder in PBS for 1 h at room temperature. mAbs 2625 and LPS-1 (39), respectively, were preincubated with carbohydrate fractions from wild-type strain RC1 and mutant strain 811, respectively. These sugar fractions containing O-chain polysaccharide with attached core oligosaccharide were obtained from gel permeation chromatography as described above. The polysaccharides were redissolved in distilled water to a concentration of 50 μg/μl. 1 μl of serial dilutions of carbohydrate fractions was added to 19 μl dilutions of mAb 2625 and mAb LPS-1, respectively. Before addition of the sugars the antibodies were diluted to a suitable concentration in PBS. In control reactions 1 μl distilled water was added to the antibody dilutions instead of carbohydrate solutions. Coincubation of mAbs and carbohydrate fractions was performed at 4°C overnight. After blocking and three washing steps with PBS, microtiter plates were incubated with the antibody–carbohydrate mixture for 2 h at room temperature and subsequently washed with PBS for three times. Detection was carried out by incubation of the plates with peroxidase-labeled goat anti–mouse antibody (Dianova) for 1 h at room temperature. After three washing steps as before the substrate H2O2 and azino-di-ethylbenzthiazolinsulfonate (ABTS) was added and after a 30-min incubation at room temperature absorbance was determined in a microplate reader at 405 nm. Inhibition of antibody binding was calculated as percentage of the control reactions (no carbohydrates added). All reactions were carried out in duplicates.
Bactericidal Assay.
For investigation of serum resistance of L. pneumophila wild-type strain RC1 and mutant strain 811 normal human serum (NHS) was obtained from 10 healthy volunteers. Blood was allowed to clot for 30 min at room temperature. After centrifugation for 5 min at 2,000 g, the sera were pooled, quick-frozen in liquid nitrogen and stored at −80°C. Pool serum was negative for anti-L. pneumophila antibodies as determined by standard diagnostic serology methods (immune fluorescence test). Bacteria were plated on BCYE agar from frozen stocks and suspended in 0.9% saline after 40 h of incubation. Optical density was determined at 600 nm and bacterial suspensions were appropriately diluted in 0.9% saline. 40% NHS was incubated with 106 bacteria in a final reaction volume of 1 ml. The reaction mixture was incubated at 37°C in a water bath and stopped on ice at 0, 15, 30, and 60 min, respectively. Appropriate dilutions were plated on BCYE agar.
Infection of HL-60 Cells with L. pneumophila.
Infection of HL-60 cells was performed essentially as described (25, 43). Human macrophage-like cell line HL-60 was propagated in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (GIBCO BRL, Eggenstein, Germany). Differentiation of the cells was induced with PMA (Sigma Chemical Co.) at a final concentration of 10−8 M. After 48 h of incubation with PMA cells were washed and 0.5 × 106 adherent HL-60 cells per well were infected with 104 bacteria of the appropriate L. pneumophila strain. Bacteria were plated on BCYE agar from frozen stocks and harvested after 40 h incubation. The optical density of bacterial suspensions in 0.9% saline was determined at 600 nm and appropriate dilutions were prepared in RPMI medium. For determination of intracellular multiplication, at 0, 24, 48, and 72 h after infection culture supernatants were removed, HL-60 cells were lysed by suspending in ice-cold water. Suspended cells and supernatant were pooled and aliquots of serial dilutions were plated on BCYE agar.
In a second assay, extracellular remaining bacteria were killed by adding gentamicin (GIBCO BRL) to a final concentration of 40 μg/ml after 2 h of coincubation of HL-60 cells and bacteria. After a 2-h incubation time with gentamicin, cells were washed again and incubated in antibiotic-free medium. Under these conditions, HL-60 were infected with 106 bacteria per well. At time intervals of 0, 24, 48, and 72 h after gentamicin removal, appropriate dilutions were plated for determination of intracellular multiplication.
Intratracheal Infection of Guinea Pigs.
Male and female guinea pigs strain 2BS (400–600 g) obtained from the Zentrales Tierlaboratorium (Medizinische Hochschule Hannover, Germany) were infected with L. pneumophila by intratracheal application of the bacteria as described (15, 44). Animals were anesthetized by intramuscular injection of a mixture of 10 μl ketamine (10% solution) and 10 μl xylazinhydrochloride (2% solution) per 100 g body weight. Subsequently, a small skin incision was made in the ventral neck and the bacterial suspension was injected into the exposed trachea. Before inoculation, the appropriate L. pneumophila strains were plated on BCYE agar from frozen stocks and were incubated for 40 h. Bacteria were harvested and suspended in 0.9% saline to a concentration of 108/ml. 0.3 ml (corresponding to 3 × 107 CFU) of this suspension was injected into the tracheal lumen with a 24-gauge needle and the incision was closed with sutures. In previous experiments, 3 × 107 CFU had been determined as the 50% lethal dose (LD50) of strain RC1 to guinea pig strain 2BS within 3–5 d after infection. After the infection, animals were observed several times daily for signs of illness and respiratory disease. 48 h after infection animals were killed and the lungs were removed. Lung tissue was homogenized in 0.9% saline and aliquots of serial dilutions of the lung suspensions were plated on BCYE agar. For determination of recovered Legionella, CFU were counted after 72 h incubation. Aliquots of the lung suspensions were also plated on blood agar to control for contaminating bacteria. Binding of injected and recovered bacteria to mAb 2625 was in all experiments monitored by colony blot assay. Animal experiments were carried out with the permission and according to the guidelines of local authorities.
Results
Characterization of mAb 2625.
To generate mAbs, mice were immunized with whole cells of L. pneumophila SG 1 strain RC1 (OLDA). Before the immunization, strain RC1 was subjected to a single guinea pig passage. Screening of hybridoma supernatants in an ELISA with the same strain used for immunization led to the isolation of IgG3 mAb 2625. The reactivity of mAb 2625 was not eliminated after treatment with proteinase K, suggesting that a carbohydrate epitope was recognized. Binding of mAb 2625 occurred exclusively in strains belonging to the OLDA and Oxford subgroups, of L. pneumophila SG 1, respectively, as determined by ELISA and Western blot analysis. There was no binding reaction observed in any other of the 14 tested SG 1 strains. This was also the case for type strains from serogroup 2 to 14. Likewise, eight non-pneumophila strains as well as several bacteria from other genera were tested and did not bind to mAb 2625. All of the investigated strains are listed in Materials and Methods.
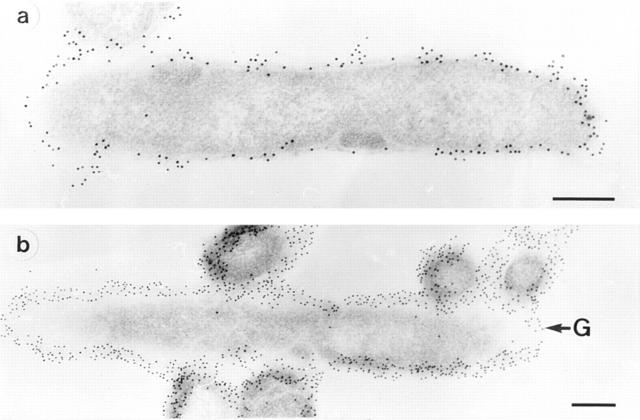
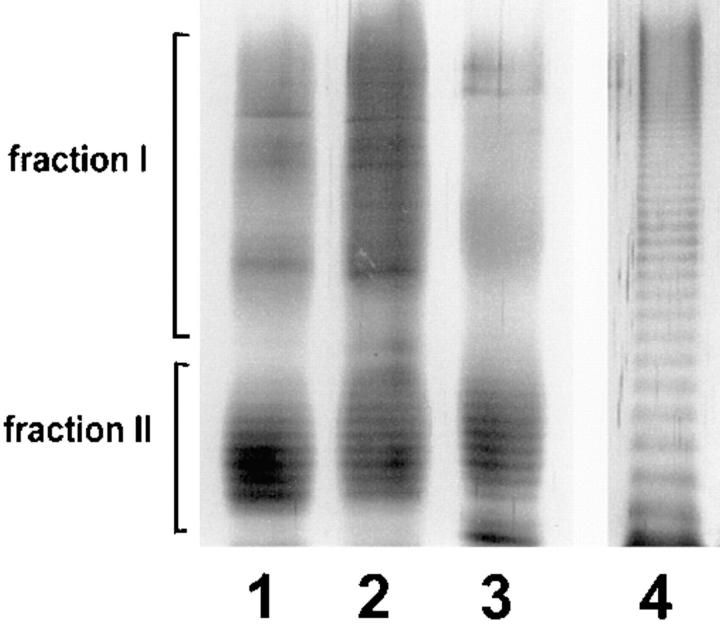
In electron microscopy experiments, the epitope on the antigen bound by mAb 2625 could be localized to the cell surface of L. pneumophila (Fig. 1). In Western blot analysis of whole cell lysates and purified LPS, mAb 2625 exhibited a ladder-like binding pattern characteristic of LPS (Fig. 2 A, lanes 1 and 3). With the aid of molecular mass standards for SDS-PAGE, the ladder-like bands were estimated to be in the range of 30–65 kD. This molecular mass range does not correspond to the molecular mass of L. pneumophila LPS, but permitted a relative comparison of LPS banding patterns in Western blots. In control experiments, we incubated Western blots with mAb LPS-1 (39), an mAb specific to serogroup 1 of L. pneumophila. As shown in Fig. 2 B (lanes 1 and 3), LPS-1 exhibited a ladder-like banding pattern in the range of 20–30 kD and in addition, faint bands in the range of 35–45 kD. This binding pattern corresponds well to the image of L. pneumophila LPS bands which become visible after silver staining (Fig. 3). From the binding characteristics of mAb 2625, that is the banding pattern in a higher molecular mass range than observed with mAb LPS-1, we conclude that mAb 2625 binds a conformational epitope, which is not generated before a distinct O-chain length is achieved.
Figure 1.
Immunoelectron microscopy of L. pneumophila SG 1 strain RC1 (OLDA). Staining of sections was performed with mAb 2625 (a). In control experiments sections were immunostained with mAb LPS-1 (39), which binds specifically to LPS of L. pneumophila SG 1 (b). G, gold-particle. Bar, 0.25 μm.
Figure 2.
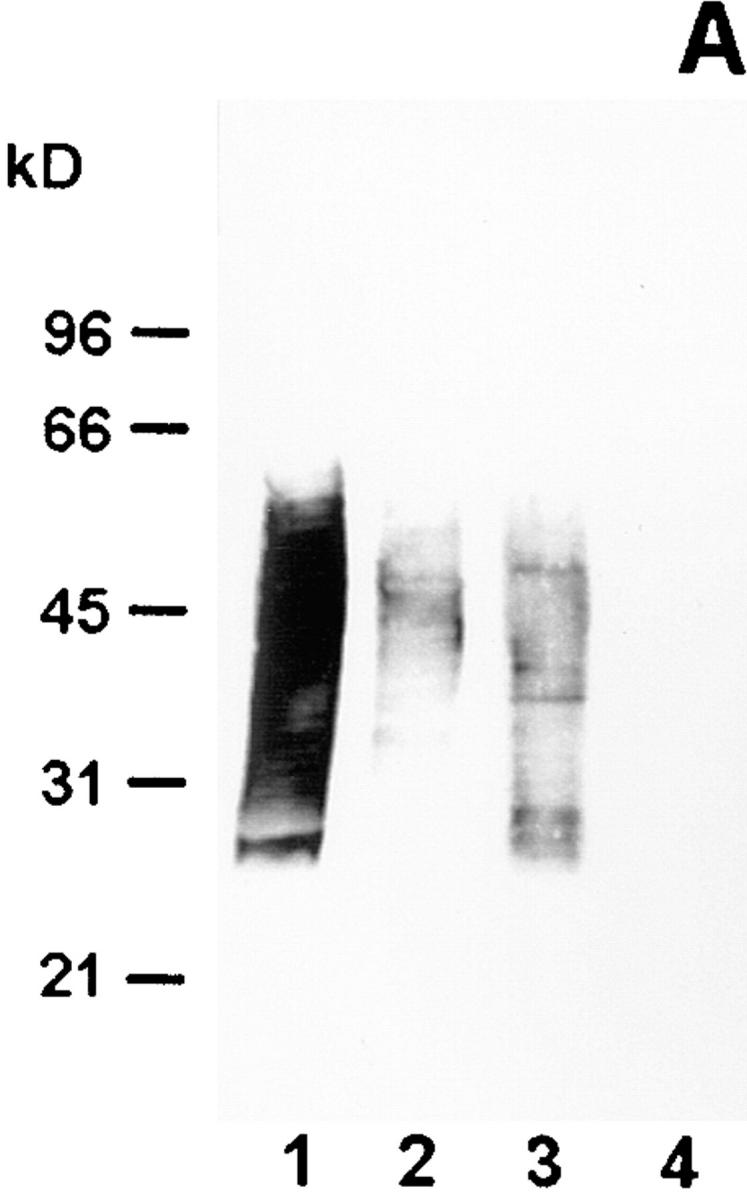
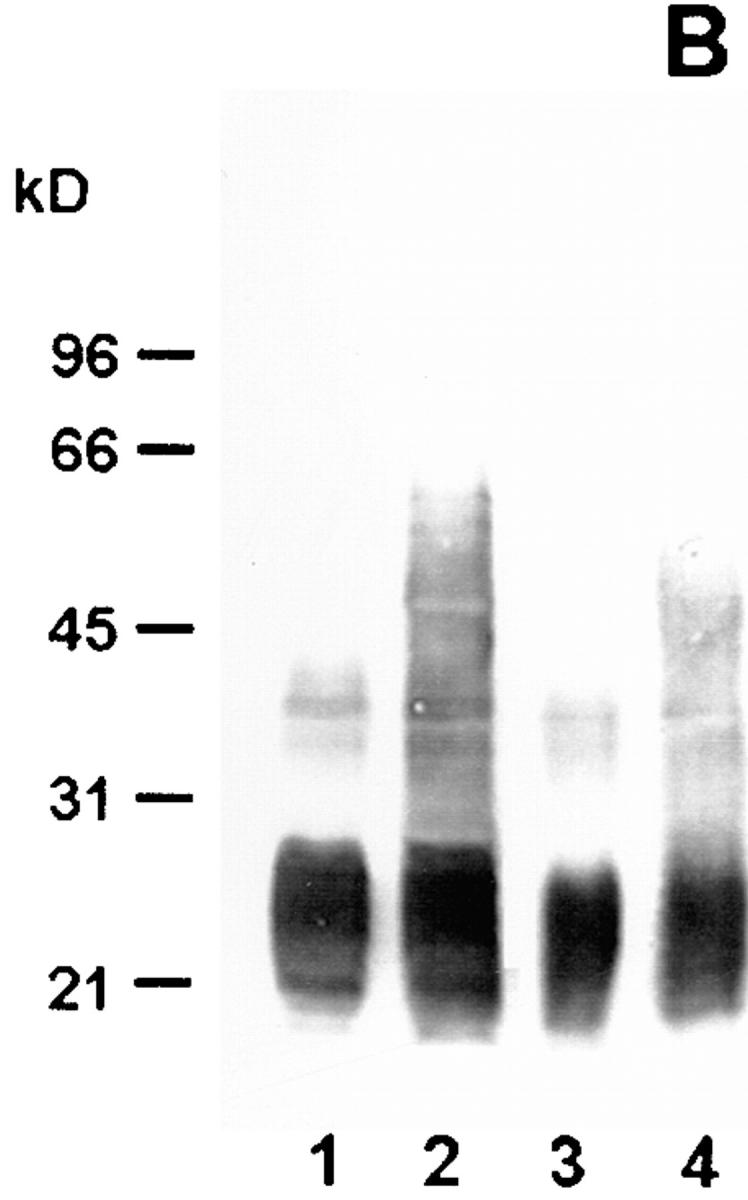
Western blot analysis of wild-type strain RC1 and mutant 811 with mAb 2625 (A) and mAb LPS-1 (B), respectively. Lane 1, wild-type RC1 whole cell lysate; lane 2, mutant 811 whole cell lysate; lane 3, wild-type RC1 2 μg purified LPS; lane 4, mutant 811 2 μg purified LPS. Numbers on the left side indicate molecular masses of a standard protein marker. The molecular mass of L. pneumophila LPS does not correspond to that of the marker proteins, but determination of a relative range of LPS bands was achieved by this method.
Figure 3.
SDS-PAGE and silver staining of LPS from L. pneumophila SG 1. 2 μg purified LPS were applied to each lane. Lane 1, mutant 811; lane 2, wild-type RC1; lane 3, strain Philadelphia. All strains exhibit a characteristic bimodular distribution of LPS O-chain representing 10–35 and 45–100 carbohydrate units, respectively. On the left side, LPS bands corresponding to fractions I and II of the gel filtration on Sephadex columns are indicated. These fractions were employed for epitope mapping by competition ELISA. As a control, 2 μg purified LPS from P. aeruginosa subgroup Fisher 2 was applied to lane 4.
Isolation of LPS-Mutant Strain 811.
Next, we wondered whether mAb 2625–reactive epitope is present on all bacteria within a population. Therefore, we performed colony blots on the virulent L. pneumophila strain RC1, a clinical isolate of the OLDA subtype of serogroup 1, with mAb 2625. Using this strategy, we could show that mAb 2625– negative colonies could be detected in a frequency of 10−4. We isolated one of the mAb 2625–negative colonies for further analysis. Interestingly, the phenotype of this LPS variant, termed 811, was unstable and exhibited a remarkable switching back to the LPS phenotype of the parent strain. This could be shown using mAb 2625. When a single colony of the mutant 811 with mAb 2625 nonreactivity confirmed by colony blot analysis was restreaked on BCYE agar, 80–100% of the grown colonies were again positive for binding of mAb 2625. Colony morphology of mutant strain 811 did not exhibit any differences compared with that of the wild-type strain, but the colony material appeared extremely viscous and sticky. By repeating the procedure of colony blots and restreaking single mAb 2625– negative colonies (three passages), we could reduce the proportion of mAb 2625–positive (wild-type) colonies of the mutant 811 to ∼10%. For all experiments described in the following paragraphs, the mutant 811 was subjected to this treatment. The ∼10% portion of wild-type cells in the mutant 811 population was controlled in all experiments and confirmed by colony blot analysis with mAb 2625. Mutant 811 replicated with the same growth rate in liquid media as the wild-type RC1. As could be expected from colony blot analysis, mutant 811 did not bind mAb 2625 in Western blot analysis (Fig. 2 A, lanes 2 and 4). The faint bands visible in lane 2 of Fig. 2 A are due to ∼10% wild-type cells present in the bacterial suspension. In contrast, mAb LPS-1 revealed enhanced binding to mutant 811 in comparison to the parental wild-type strain RC1, resulting in a more sensitive staining of high molecular mass LPS populations. It is conceivable that LPS of mutant 811 is altered in a way that the epitope bound by mAb LPS-1 becomes more accessible and antibody binding is thereby promoted.
Chemical Analysis of the LPS O-Chain of OLDA Wild-type RC1 and Mutant 811.
To determine the chemical alteration in LPS composition of mutant 811, which was indicated by the antibody binding characteristics, we analyzed the LPS O-chain as well as the core oligosaccharide of both strains. For this purpose, the LPS from strains RC1 and mutant 811 was isolated. In Western blot analysis, purified LPS from both strains exhibited the same binding characteristics with mAb 2625 and mAb LPS-1 as has been determined for cell lysates of the strains (Fig. 2, A and B, lanes 3 and 4). As recently published, the core oligosaccharide of L. pneumophila SG 1 strain Philadelphia is composed of Rha/ QuiNAc/GlcNAC/Man/Kdo in a relative ratio of ∼2:1:2: 2:2 (34, 45). Analysis of the core sugar components of strains RC1 and mutant 811 by GLC-MS revealed that the same sugars were present in both strains. However, complete analysis of the core structure has not yet been accomplished. In particular, modification of sugars, conformational structures, and ketosidic linkages within the core remain to be established. Chemical analysis of the carbohydrate composition of the O-chain was performed by 1H- and 13C-NMR spectra analysis. It was found that the O-specific chain was composed of a homopolymer of α-(2→ 4) interlinked 5-acetamidino-7-acetamido-3,5,7,9-tetradeoxy- l-glycero-d-galacto-nonulosonic acid (8-de-O-acyl derivative of legionaminic acid). No structural difference was observed between the 8-O-deacetylated legionaminic acid in the two strains investigated. In addition, length distribution of the LPS O-chain was identical in wild-type RC1 and mutant 811, as was determined by SDS-PAGE and gel permeation chromatography. SDS-PAGE of purified LPS from RC1 and 811 revealed a ladder-like banding pattern with a bimodular distribution of O-chain length which is typical for Legionella LPS (Fig. 3). No difference in banding pattern was observed between wild-type and mutant. Moreover, both strains exhibited a banding pattern very similar to that of L. pneumophila SG 1 strain Philadelphia. In conclusion, no difference in O-chain structure and length was observed between wild-type RC1 and mutant 811, further confirming chemical data indicating the O-chain not to be changed in the mutant.
Epitope Mapping of mAb 2625 and mAb LPS-1.
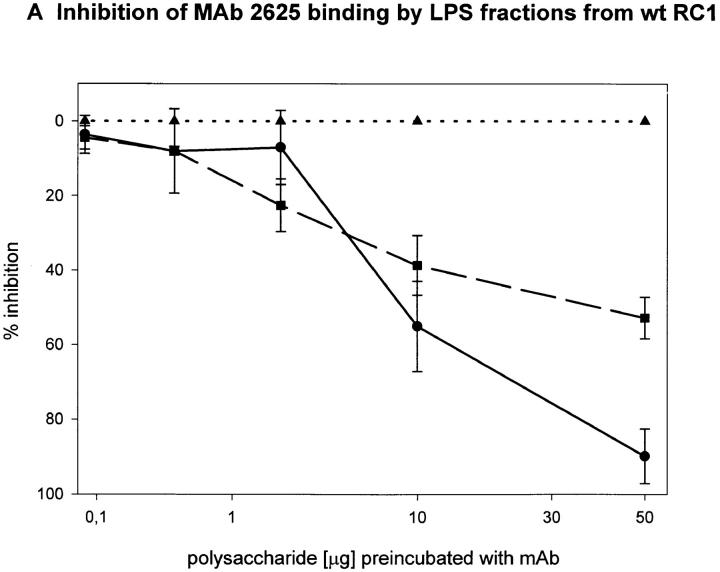
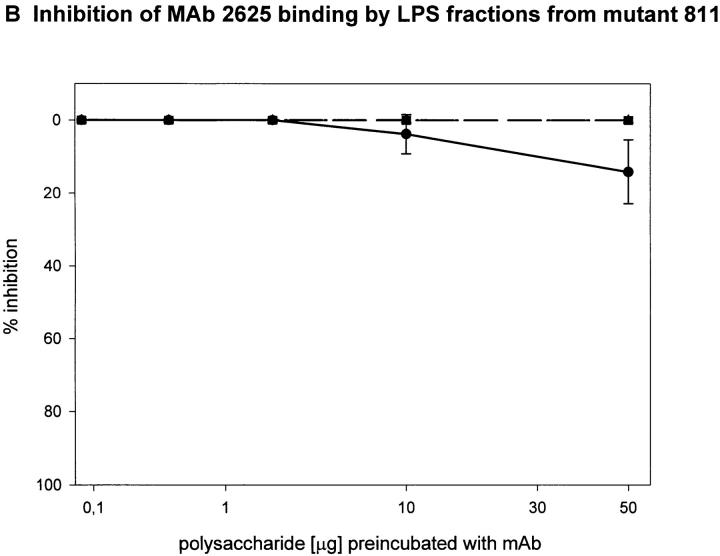
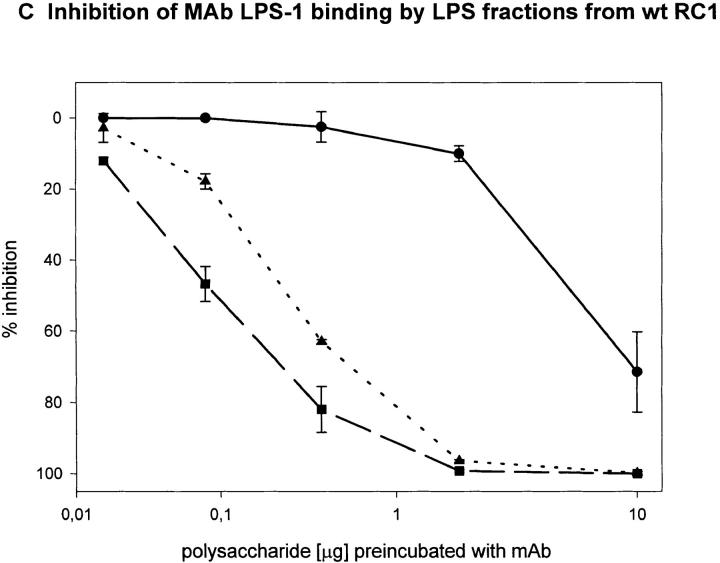
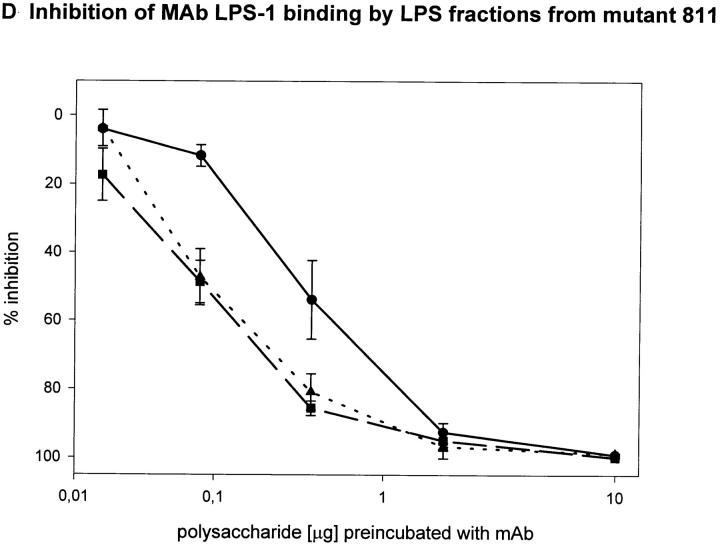
To determine the region of the LPS molecule where a structural difference between wild-type RC1 and mutant 811 could be located, we attempted to map the epitopes bound by mAb 2625 and mAb LPS-1, respectively. For this purpose we investigated the binding capacity of sugar fractions obtained from the gel filtration assay. Before separation on Sephadex columns, the lipid A moiety had been removed from LPS by mild acid hydrolysis. After gel filtration three fractions of O-chain with attached core oligosaccharide were eluted from the column: fraction I represented the long O-chain (45-100-mer); fraction II the short O-chain (10-35-mer); and fraction III the isolated core oligosaccharide. The LPS bands separated by SDS-PAGE, which correspond to fractions I and II, are indicated in Fig. 3. All fractions eluted from the column contained the carbohydrate moiety of LPS (O-specific chain attached to the core) alone and therefore could not be analyzed by Western blot or conventional ELISA techniques. For this reason we performed a competition ELISA which is based on the principle of inhibition of antibody binding to LPS by LPS carbohydrate fractions. Purified LPS from L. pneumophila SG 1 wild-type RC1 was adsorbed to microtiter plates. mAb 2625 and LPS-1, respectively, were preincubated with serial dilutions of sugar fractions obtained from Sephadex gel filtration. In this way we investigated fractions I and II from wild-type RC1 and mutant 811 as well as the core portion of both strains. The results of the competition ELISA are illustrated in Fig. 4. Binding of mAb 2625 is inhibited in a concentration dependent manner by carbohydrates from fractions I and II of wild-type RC1, but not by the core oligosaccharide, indicating that the epitope bound by mAb 2625 involves the O-specific chain. It is noteworthy, that carbohydrates from fraction II exhibited a decreased binding to mAb 2625 in comparison to those from fraction I. O-chain molecules of the required length are presumably only present as a minor part of fraction I. For mutant 811, weak binding of carbohydrates from fraction I to mAb 2625 is due to wild-type bacteria (∼10%, see above) that are inherent to the unstable character of 811. Inhibition of binding of mAb LPS-1 occurred in a likewise dose-dependent way. Carbohydrates from fractions I and II, as well as the core fractions from wild-type RC1 and mutant 811, were bound by mAb LPS-1. As already shown in Western blot analysis (see Fig. 2), binding of mAb LPS-1 to 811 is increased in comparison to wild-type RC1 (Fig. 4, C and D). In conclusion, mAb 2625 binds the LPS O-chain, but not the isolated core oligosaccharide. However, we cannot exclude that the core moiety is also required for mAb 2625 binding, since carbohydrates from fractions I and II contain the core portion as well as the O-chain. Since no differences in the O-antigen composition by chemical analysis could be observed, we conclude that the epitope bound by mAb 2625 is a conformational epitope which involves both the O-chain and the core. In contrast, the epitope bound by mAb LPS-1 is clearly located in the core oligosaccharide, and the O-chain is not required for binding. Together with the results obtained from chemical analysis, these data strongly support the idea, that alterations in the core oligosaccharide of mutant 811 are responsible for loss of mAb 2625 binding and increased binding to mAb LPS-1.
Figure 4.
Epitope mapping of mAb 2625 (A and B) and mAb LPS-1 (C and D), respectively. Inhibition of binding of mAb 2625 and mAb LPS-1 to LPS from wild-type RC1 by LPS fractions from wild-type RC1 (A and C) and mutant 811 (B and D) was analyzed by competition ELISA. Percentage of inhibition is given as means of duplicate values. Fraction I is represented by circles, fraction II by squares and the core portion by triangles.
Intracellular Replication of RC1 and 811 in HL-60 Cells.
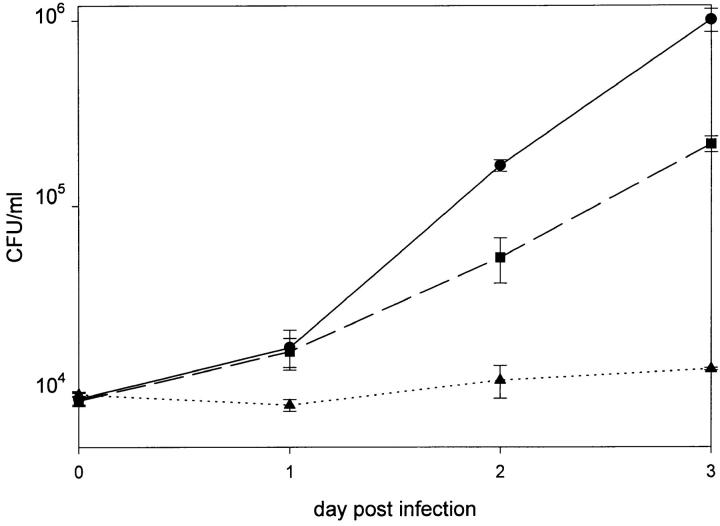
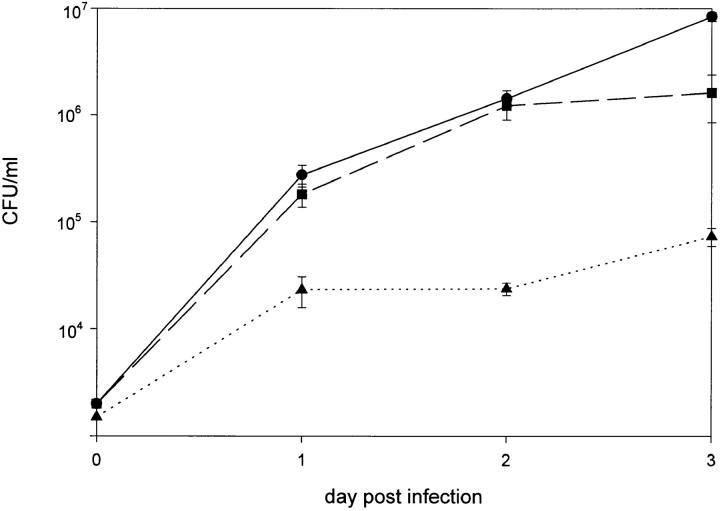
Virulence of mutant 811 in comparison to the parent strain RC1 was determined by infection of human macrophage-like cell line HL-60 and determination of CFU on days 1 to 3 after infection. 2 × 106 HL-60 cells were infected with 104 bacteria from frozen stocks. Wild-type strain RC1 proved to be virulent and replicated in HL-60 cells by 2 orders of magnitude within 72 h (Fig. 5). A mAb 2625– positive revertant of mutant 811 (811-rev.) served as a control in these experiments. It is well known that virulence of pathogenic bacteria is attenuated or even abolished after laboratory passage on artificial media, strain 811-rev. was therefore employed to identify those effects due to agar passage. In comparison with the animal passaged wild-type strain RC1, 811-rev. exhibited a slightly reduced growth rate, but proved to be virulent (Fig. 5). In contrast, mutant 811 failed to replicate intracellularly in HL-60 cells. The number of recovered bacteria at all time intervals remained in the range of the inoculum (Fig. 5). In conclusion, wild-type RC1 proved to be able to replicate in HL-60 cells, whereas mutant 811 showed no replication and consequently proved to be avirulent. Moreover, by investigating 811-rev., it became evident that switching of 811 to the wild-type LPS-phenotype (mAb 2625 binding) also restored virulence. These findings indicate that alteration of LPS carbohydrate moiety was the only mutation that had occurred in 811. When gentamicin was added to kill extracellular bacteria after 2 h of coincubation of host cells and bacteria, mutant 811 was found intracellularly to approximately the same extent as wild-type RC1 and 811-rev. (Fig. 6). These data show that mutant 811 entered HL-60 cells as efficiently as the wild-type strain, but was unable to replicate in the host cell. Colony blot analysis of mutant 811 revealed that the percentage of mAb 2625-positive revertants among bacteria recovered from the HL-60 infection assays was identical to that of the inoculum.
Figure 5.
Infection of HL-60 cells with L. pneumophila strains RC1 (circles), 811 (triangles), and 811-rev. (squares). CFU were determined at 24, 48, and 72 h post infection and are shown as means of duplicates.
Figure 6.
Infection of HL-60 cells with L. pneumophila. After 2 h coincubation of host cells and bacteria, extracellularly remaining bacteria were killed by gentamicin. The CFU determined at day 0 therefore represent intracellular bacteria. (Circles) Wild-type RC1; (triangles) mutant 811; (squares) 811-rev. CFU are shown as mean of duplicates.
Investigation of Serum Resistance.
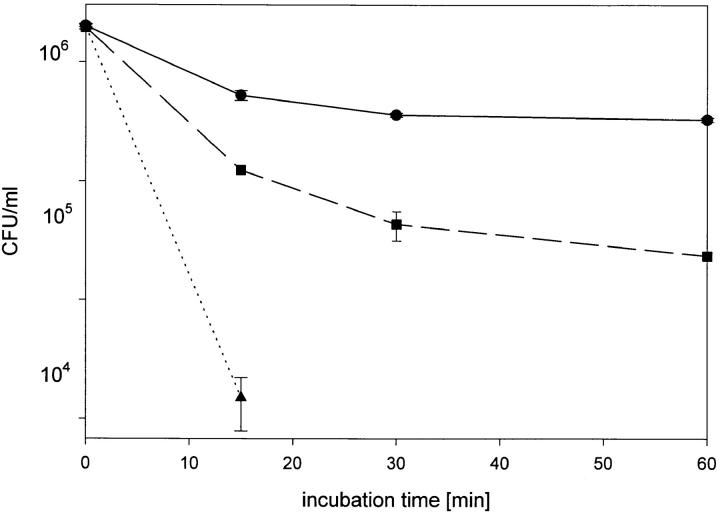
To determine the bactericidal activity of complement present in NHS on wild-type RC1, mutant 811 and 811-rev., we performed bactericidal assays as described in Materials and Methods. After a 1-h incubation in 40% serum, number of bacteria from wild-type strain RC1 declined by <1 log. Thus, wild-type RC1 was not completely, but almost serum resistant. In contrast, the number of viable bacteria from mutant 811 declined by 3 logs (from 6.1 log10 to 3.2 log10) within 15 min incubation in 40% NHS (Fig. 7). After 30 min no viable bacteria were recovered (detection limit 102 CFU/ml). Therefore, mutant 811 was serum-sensitive and the sensitivity is related to an altered LPS conformation. Incubation of 811-rev. in 40% NHS revealed that the number of viable bacteria was reduced by 2 logs within 1 h (Fig. 7). These findings indicate that agar passage does indeed have an influence on serum resistance, even though its effect is minor. From the results of the bactericidal assays, we conclude that the alteration in LPS carbohydrate moiety of mutant 811, which is presumably located in the core oligosaccharide, results in deprivation of resistance against serum complement activity. In addition, our data show that serum resistance of L. pneumophila is mediated by the LPS carbohydrate moiety.
Figure 7.
Bactericidal assay for determination of the lytic effects of serum complement on wild-type RC1 (circles), mutant 811 (triangles) and 811-rev. (squares). Bacteria were incubated with 40% normal human serum at 37°C. Aliquots of the reactions were plated at different time intervals for determination of viable bacteria. CFU are shown as mean of duplicates.
In Vivo Virulence of Wild-type RC1 and Mutant 811.
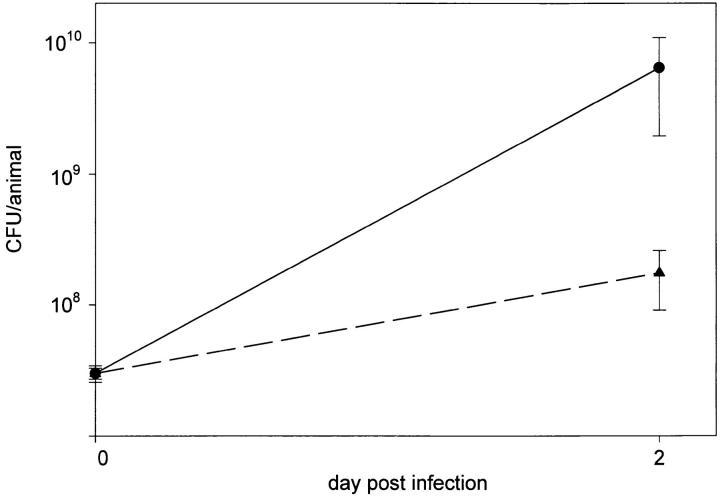
Next, we were interested to study virulence of mutant 811 under in vivo conditions in the guinea pig animal model. Animals (n = 4 for each bacterial strain) were infected by intratracheal injection of bacteria. On day 2 after infection, animals infected with the wild-type strain RC1 exhibited signs of severe illness as fever, ruffled fur, almost no motion and reaction and respiratory distress. Animals were killed on day 2 after infection and lungs were removed. The lungs appeared greatly enlarged and were completely hemorrhagic. After homogenization of the lungs, aliquots of the homogenates were plated for determination of the number of bacteria. 6.5 × 109 (mean value) bacteria were recovered from the animals (Fig. 8). The control blood agar plates were found to be sterile. In contrast, the animals infected with the mutant strain 811 showed moderate signs of illness, such as elevated body temperature and limited motion, but did not show signs of respiratory distress. These animals were also killed on day 2 after infection. Lungs were only slightly enlarged and hemorrhagic patches were visible, but were not distributed over the entire lung tissue. 1.8 × 108 viable bacteria (mean value) were recovered from the animals, a significantly lower number than was determined for the wild-type strain (Fig. 8). Interestingly, colony blot analysis of strain 811 isolated from animal lungs revealed that 35% of the recovered bacteria were positive for mAb 2625 binding, whereas only 8% of the inoculated bacteria were mAb 2625-positive. These results indicate that under in vivo selective pressure the 2625–positive phenotype (wild-type phenotype) is evidently advantageous over the mAb 2625–negative phenotype of mutant 811. It remains unclear if a preferential replication of the mAb 2625–positive portion of mutant 811 occurred or if switching back to the wild-type phenotype is promoted under in vivo conditions.
Figure 8.
Investigation of virulence of L. pneumophila wild-type strain RC1 (circle) and mutant 811 (triangle) in the guinea pig animal model. Animals were intratracheally infected with 3 × 107 bacteria. Number of recovered bacteria was determined 48 h after infection by plating aliquots of lung suspensions.
On the other hand, for the wild-type strain RC1 we observed a significantly increased switching frequency under in vivo conditions to the mAb 2625–negative phenotype. A frequency of 5 × 10−2 among recovered bacteria from guinea pig lungs was determined by colony blot assay, whereas mAb 2625–negative clones could be detected among agar plated bacteria in a frequency of only 10−4 (see above).
In conclusion, mutant 811 was not able to cause severe pneumonia as the wild-type RC1 did in the animal host and showed a significantly reduced replication in comparison to the wild-type strain. Even though mutant 811 was completely serum sensitive, it was not cleared from the animal lung. Moreover, our data strongly indicate that two LPS phases of L. pneumophila are expressed in vivo.
Discussion
We here describe the isolation and investigation of an LPS-mutant of Legionella pneumophila SG 1, subgroup OLDA. In comparison to other Gram-negative bacteria, L. pneumophila exhibits an unusual LPS structure (31, 32, 34). The chemical structure has recently been analyzed for L. pneumophila SG 1, subgroup Philadelphia. It was found that the lipid A moiety consists of long-chain fatty acids which may account for its low endotoxic activity (31). The core oligosaccharide lacks heptose and phosphate groups and exhibits hydrophobic properties due to the presence of four O-acetyl groups and three deoxy-sugars (Rha 2, QuiNAc) (31, 34), which has so far not been found in any other bacterial strain. The O-chain is composed of an unbranched homopolymer with α-(2→ 4) interlinked 5-acetamidino- 7-acetamido-8-O-acetyl-3,5,7,9-tetradeoxy-l- glycero-d-galacto-nonulosonic acid, termed legionaminic acid. Due to the lack of free hydroxyl groups and characteristic substituents, the O-chain is highly hydrophobic (31, 32). In this study we determined the chemical structure of the LPS O-antigen of L. pneumophila SG 1, subgroup OLDA. The O-chain was found to be of the same structure as the one from Philadelphia (31, 32), except that it lacks the 8-O-acetyl group and therefore is termed 8-O-deacetyl-legionaminic acid. The finding that OLDA strains all lack the 8-O-acetyl group was expected after the serological data gained by investigating antibody reactivities: the 8-O-acetyl group is known to be involved in binding of mAb 2 (27) and mAb 3/1 (46). In contrast, both antibodies do not bind to strains of the subgroup OLDA. The length of the O-chain of subgroup OLDA shows a bimodal distribution with maxima at 10–35 and 45–100 carbohydrate units, respectively. This banding pattern is very similar to that obtained for subgroup Philadelphia (32). LPS-mutant 811 did not show any differences in O-chain structure and length when compared with its parent wild-type strain RC1. Therefore, we conclude that LPS of mutant 811 is altered in the core oligosaccharide.
Mild acid hydrolysis in acetate buffer was used to cleave the lipid A moiety from the carbohydrate moiety of the LPS molecule. Under these conditions the ketosidic linkages of Kdo and iso-legionaminic acid are as well cleaved (32–34, 45), whereas O-acetyl groups remain intact (34, 45). It can be excluded that artifacts created by the acid hydrolysis treatment prevented the identification of modifications of the mutant 811 LPS compared with the wild-type LPS. Epitope mapping experiments showed that carbohydrate fractions isolated after mild acid hydrolysis and gel permeation chromatography still were able to compete for epitope binding by mAbs 2625 and LPS-1. If artifacts were generated by the degradation procedure, they did not interfere with those substituents important for formation of the epitopes bound by mAbs 2625 and LPS-1.
Our idea that the core sugar composition of mutant 811 differs from that of the parent wild-type RC1 is supported by the results of epitope mapping of mAb 2625 and mAb LPS-1. With the aid of a competition ELISA method we could show that mAb LPS-1 binds to the core oligosaccharide of L. pneumophila SG 1. mAb LPS-1 shows a stronger binding to mutant 811 than to wild-type RC1, indicating that alterations in the core structure of 811 enhance the accessibility of the LPS-1 epitope. In contrast, mAb 2625 binds to the LPS O-chain and the core oligosaccharide is presumably also involved in formation of the epitope. Moreover, short-chain polysaccharides showed a reduced binding to mAb 2625 in comparison to long-chain fractions. This finding supports our hypothesis that a distinct O-chain length is required for mAb 2625 reactivity.
In vitro and in vivo experiments revealed that virulence of mutant 811 was significantly reduced in comparison to the wild-type strain RC1. In in vitro assays, LPS-mutant 811 was unable to replicate intracellularly in macrophage-like cells even though the mutant bacteria were able to enter the host cells. In addition, mutant 811 was rapidly killed by serum complement factors, whereas the corresponding wild-type strain was almost resistant to complement lysis. By comparative analysis of LPS-mutant 811 and the parent wild-type RC1, these data show for the first time that resistance to serum complement of L. pneumophila is mediated by LPS carbohydrate moiety. However, we cannot exclude that the LPS variation of mutant 811 leads to additional modifications in the formation of the outer membrane. Such alterations could for example affect surface molecules which might as well be required for serum resistance and virulence of L. pneumophila. For many bacterial pathogens, surface polysaccharide structures such as capsules and LPS were found to be involved in complement inhibition (reviewed in references 47, 48). Currently we do not know whether killing of mutant 811 by serum complement factors is due to an increase in insertion of membrane attack complex (MAC) into the membrane of mutant 811 or if insertion of MAC occurs to the same extent in both, wild-type and mutant, but does not lyse wild-type cells. Therefore, future experiments should address the ability of wild-type RC1 and mutant 811 to prevent or promote insertion of MAC. In particular, activation of complement and deposition of complement factors C3b and C3bi on the cell surface is of special interest, since these molecules are known to mediate uptake into the host cells via complement receptors CR1 and CR3 (9, 10).
In the guinea pig animal model, mutant 811 was not able to cause severe pneumonia in guinea pigs infected with a dose that corresponds to the LD50 of wild-type strain RC1. Moreover, number of wild-type bacteria recovered from the lungs of infected animals exceeded the number of mutant bacteria recovered from infected animals by 1 to 2 orders of magnitude. Most interestingly, LPS-mutant 811 exhibited an unstable phenotype. The majority of cells from the originally isolated clone 811, which was negative for mAb 2625 binding, switched back to the wild-type phenotype and restored mAb 2625 binding. This phase variation between two LPS phenotypes was found to be immensely promoted in vivo in the animal host. Inoculated bacteria of mutant 811 included 8% cells positive for mAb 2625 binding of the wild-type phenotype. In contrast, among recovered bacteria from the infected animals the portion of wild-type bacteria was increased to 35%, indicating a selective advantage of the wild-type phenotype over the mutant phenotype. Bacteria recovered from animals infected with the wild-type RC1 contained 5% cells negative for mAb 2625 binding and therefore exhibiting the phenotype of mutant 811. We observed the same phase variation of wild-type RC1 and mutant 811 when the bacteria were incubated with heat inactivated human serum for 1 h (data not shown). Our results strongly indicate that phase variation of L. pneumophila LPS is induced and promoted by the animal host and by human serum. This is the first description of a phase-variable expression of surface polysaccharides of L. pneumophila. Diversity of surface carbohydrates achieved by means of high-frequency, reversible switching of sugar epitopes has been described and intensively studied in Haemophilus influenzae (49–58), Neisseria gonorrhoeae (59, 60) and N. meningitidis (61–64). In H. influenzae, expression of enzymes involved in LPS biosynthesis is controlled by multiple repeats of tetrameric nucleotides within the lic, lex2, and lgtC loci (50, 56, 58). A change in the number of tetrameric repeats, arising through slipped-strand mispairing, results in a frame shift mutation, thus preventing expression of the encoded enzyme. Generation of phenotypic LPS variation by these intragenic alterations is considered as a virulence mechanism, enabling the bacteria to adapt to different environmental conditions (55). Essentially the same slipped-strand mispairing mechanism is found in Neisseria. Genes of the lgt locus, encoding glycosyl-transferases responsible for LPS biosynthesis, are expressed or not depending on the number of guanosine residues within a poly-G stretch in the coding sequence (59–61). In N. meningitidis, expression of the terminal lacto-N-neotetraose on the LPS, which requires the glycosyl-transferase encoded by lgtA, is correlated with serum resistance, non-invasiveness and predominance in the blood of infected mice. In contrast, strains with the lacto-N-neotetraose negative LPS phenotype are serum-sensitive, invasive and predominantly found in the nasopharynx (61, 62, 65, 66). For L. pneumophila, it remains to be established by which molecular mechanism LPS phase variation is determined. A prerequisite to further investigate this question is the characterization of genes involved in LPS biosynthesis. Except for an O-acetyl-transferase gene (Mintz, C.S., unpublished data, accession number U32118) such genes have not yet been identified in Legionella.
Species of the genus Legionella do not express a capsule or an exopolysaccharide. Therefore the LPS carbohydrate moiety is the predominant molecule on the cell surface of these bacteria which contributes to the cell surface properties in an exceptionally important way. Nothing is known about adherence of legionellae to the lung epithelium. The ability of L. pneumophila to replicate in alveolar epithelial cells has been reported (67, 68). It is conceivable that adhesion and tight attachment to epithelial cells is a crucial step in infection before the target host cell can be invaded. Attachment and adhesion could be mediated by surface carbohydrates such as LPS, which has been suggested to act as an adhesin of numerous pathogenic bacteria (reviewed in reference 69). Moreover, LPS of L. pneumophila may also be involved in attachment to its host cell. In a very recent study, a 170-kD lectin of Hartmanella vermiformis has been identified as a potential receptor used by L. pneumophila to invade the protozoan cell (70). However, the ligand on the bacterial surface remains to be identified. Future studies should therefore focus on the role of LPS in attachment and adhesion to different host cells and environments that are exploited by L. pneumophila.
Acknowledgments
We are grateful to Bettina Brand for help with the HL-60 infection assay. Larry Phillips is acknowledged for critical suggestions on the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to E. Lüneberg.
Abbreviations used in this paper
- BYCE
buffered CYE
- CI
chemical ionization
- CYE
charcoal yeast extract
- EI
electron impact
- GLC
gas–liquid chromatography
- MAC
membrane attack complex
- MOMP
major outer membrane protein
- NHS
normal human serum
- NMR
nuclear magnetic resonance
- SG
serogroup
Footnotes
This work is dedicated to Dieter Bitter-Suermann on the occasion of his 60th birthday.
References
- 1.Winn WC., Jr Legionnaires disease: historical perspective. Clin Microbiol Rev. 1988;1:60–81. doi: 10.1128/cmr.1.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields BS. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 3.Fliermans CB. Ecology of Legionella: From data to knowledge with a little wisdom. Microb Ecol. 1996;32:203–228. doi: 10.1007/BF00185888. [DOI] [PubMed] [Google Scholar]
- 4.Rowbotham TJ. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 5.Marrao G, Verissimo A, Bowker RG, da Costa MS. Biofilms as major sources of Legionellaspp. in hydrothermal areas and their dispersion into stream water. FEMS Microbiol Ecol. 1993;12:25–33. [Google Scholar]
- 6.Rogers J, Keevil CW. Immunogold and fluorescein immunolabelling of Legionella pneumophilawithin an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl Environ Microbiol. 1992;58:2326–2330. doi: 10.1128/aem.58.7.2326-2330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker JT, Sonesson A, Keevil CW, White DC. Detection of Legionella pneumophilain biofilms containing a complex microbial consortium by gas chromatography-mass spectrometry analysis of genus-specific hydroxy fatty acids. FEMS Microbiol Lett. 1993;113:139–144. doi: 10.1111/j.1574-6968.1993.tb06504.x. [DOI] [PubMed] [Google Scholar]
- 8.Rechnitzer C. Pathogenetic aspects of Legionnaires' disease: interaction of Legionella pneumophilawith cellular host defences. APMIS Suppl. 1994;43:1–43. [PubMed] [Google Scholar]
- 9.Payne NR, Horwitz MA. Phagocytosis of Legionella pneumophilais mediated by human monocyte complement receptors. J Exp Med. 1987;166:1377–1389. doi: 10.1084/jem.166.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellinger Kawahara, C., and M.A. Horwitz. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophilaand mediates phagocytosis of liposome-MOMP complexes by human monocytes. J Exp Med. 1990;172:1201–1210. doi: 10.1084/jem.172.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowling JN, Saha AK, Glew RH. Virulence factors of the family Legionellaceae. . Microbiol Rev. 1992;56:32–60. doi: 10.1128/mr.56.1.32-60.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuman HA, Horwitz MA. Legionella pneumophilainvasion of mononuclear phagocytes. Curr Topics Microbiol. 1996;209:99–115. doi: 10.1007/978-3-642-85216-9_6. [DOI] [PubMed] [Google Scholar]
- 13.Cianciotto NP, Eisenstein BI, Mody CH, Toews GB, Engleberg NC. A Legionella pneumophilagene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engleberg NC, Carter C, Weber DR, Cianciotto NP, Eisenstein BI. DNA sequence of mip, a Legionella pneumophilagene associated with macrophage infectivity. Infect Immun. 1989;57:1263–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cianciotto NP, Eisenstein BI, Mody CH, Engleberg NC. A mutation in the mip gene results in an attenuation of Legionella pneumophilavirulence. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 16.Cianciotto NP, Fields BS. Legionella pneumophila mipgene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. Mip protein of Legionella pneumophilaexhibits peptidyl-prolyl-cis/trans isomerase (PPlase) activity. Mol Microbiol. 1992;6:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 18.Hacker J, Ott M, Wintermeyer E, Ludwig B, Fischer G. Analysis of virulence factors of Legionella pneumophila. . Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:348–358. doi: 10.1016/s0934-8840(11)80851-0. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig B, Rahfeld J, Schmidt B, Mann K, Wintermeyer E, Fischer G, Hacker J. Characterization of Mip proteins of Legionella pneumophila. . FEMS Microbiol Lett. 1994;118:23–30. doi: 10.1111/j.1574-6968.1994.tb06798.x. [DOI] [PubMed] [Google Scholar]
- 20.Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, Hacker J. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophilain eukaryotic cells. Infect Immun. 1995;63:4576–4583. doi: 10.1128/iai.63.12.4576-4583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophilalocus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. . Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 23.Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophilagenes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger KH, Merriam JJ, Isberg RR. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotAgene. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 25.Brand BC, Sadosky AB, Shuman HA. The Legionella pneumophila icmlocus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 26.Ciesielski CA, Blaser MJ, Wang WL. Serogroup specificity of Legionella pneumophilais related to lipopolysaccharide characteristics. Infect Immun. 1986;51:397–404. doi: 10.1128/iai.51.2.397-404.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joly JR, McKinney RM, Tobin JO, Bibb WF, Watkins ID, Ramsay D. Development of a standardized subgrouping scheme for Legionella pneumophilaserogroup 1 using monoclonal antibodies. J Clin Microbiol. 1986;23:768–771. doi: 10.1128/jcm.23.4.768-771.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolte FS, Conlin CA, Motley MA. Electrophoretic and serological characterization of the lipopolysaccharides of Legionella pneumophila. . Infect Immun. 1986;52:676–681. doi: 10.1128/iai.52.3.676-681.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otten S, Iyer S, Johnson W, Montgomery R. Serospecific antigens of Legionella pneumophila. . J Bacteriol. 1986;167:893–904. doi: 10.1128/jb.167.3.893-904.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mintz CS, Schultz DR, Arnold PI, Johnson W. Legionella pneumophilalipopolysaccharide activates the classical complement pathway. Infect Immun. 1992;60:2769–2776. doi: 10.1128/iai.60.7.2769-2776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zähringer U, Knirel YA, Lindner B, Helbig JH, Sonesson A, Marre R, Rietschel ET. The lipopolysaccharide of Legionella pneumophilaserogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog Clin Biol Res. 1995;392:113–139. [PubMed] [Google Scholar]
- 32.Knirel YA, Rietschel ET, Marre R, Zähringer U. The structure of the O-specific chain of Legionella pneumophilaserogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- 33.Knirel YA, Moll H, Helbig JH, Zähringer U. Chemical characterization of a new 5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid released by mild acid hydrolysis of the Legionella pneumophilaserogroup 1 lipopolysaccharide. Carbohydr Res. 1997;304:77–79. doi: 10.1016/s0008-6215(97)00211-5. [DOI] [PubMed] [Google Scholar]
- 34.Knirel YA, Moll H, Zähringer U. Structural study of a highly O-acetylated core of Legionella pneumophilaserogroup 1 lipopolysaccharide. Carbohydr Res. 1996;293:223–234. doi: 10.1016/0008-6215(96)00194-2. [DOI] [PubMed] [Google Scholar]
- 35.Colbourne JS, Dennis PJ, Lee JV, Bailey MR. Legionnaires' disease: reduction in risks associated with foaming in evaporative cooling towers. Lancet. 1987;1:684. doi: 10.1016/s0140-6736(87)90445-4. [DOI] [PubMed] [Google Scholar]
- 36.Steinmetz I, Rheinheimer C, Hübner I, Bitter D, Suermann Genus-specific epitope on the 60-kilodalton Legionellaheat shock protein recognized by a monoclonal antibody. J Clin Microbiol. 1991;29:346–354. doi: 10.1128/jcm.29.2.346-354.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters H, Jurs M, Jann B, Jann K, Timmis KN, Bitter-Suermann D. Monoclonal antibodies to enterobacterial common antigen and to Escherichia coli lipopolysaccharide outer core: demonstration of an antigenic determinant shared by enterobacterial common antigen and E. coliK5 capsular polysaccharide. Infect Immun. 1985;50:459–466. doi: 10.1128/iai.50.2.459-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth J, Bendayan M, Carlemalm E, Villiger W, Garavito M. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J Histochem Cytochem. 1981;29:663–671. doi: 10.1177/29.5.6166664. [DOI] [PubMed] [Google Scholar]
- 39.Sethi KK, Brandis H. Establishment of hybridoma cell lines secreting anti-Legionella pneumophilaserogroup 1 monoclonal antibodies with immunodiagnostic potential. Zentralbl Bakteriol Mikrobiol Hyg A. 1983;255:294–298. [PubMed] [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology. 1992;24:145–149. [PubMed] [Google Scholar]
- 41.Moll H, Sonesson A, Jantzen E, Marre R, Zähringer U. Identification of 27-oxo-octacosanoic acid and heptacosane-1,27-dioic acid in Legionella pneumophila. . FEMS Microbiol Lett. 1992;76:1–6. doi: 10.1016/0378-1097(92)90354-q. [DOI] [PubMed] [Google Scholar]
- 42.Fomsgaard A, Freudenberg MA, Galanos C. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol. 1990;28:2627–2631. doi: 10.1128/jcm.28.12.2627-2631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marra A, Horwitz MA, Shuman HA. The HL-60 model for the interaction of human macrophages with the Legionnaires' disease bacterium. J Immunol. 1990;144:2738–2744. [PubMed] [Google Scholar]
- 44.Winn WC, Jr, Davis GS, Gump DW, Craighead JE, Beaty HN. Legionnaires' pneumonia after intratracheal inoculation of guinea pigs and rats. Lab Invest. 1982;47:568–578. [PubMed] [Google Scholar]
- 45.Moll H, Knirel YA, Helbig JH, Zähringer U. Identification of an α-d-Manp-(1-8)-Kdo disaccharide in the inner core region and the structure of the complete core region of the Legionella pneumophilaserogroup 1 lipopolysaccharide. Carbohydr Res. 1997;304:91–95. doi: 10.1016/s0008-6215(97)00210-3. [DOI] [PubMed] [Google Scholar]
- 46.Helbig JH, Lück PC, Knirel YA, Witzleb W, Zähringer U. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophilaserogroup 1. Epidemiol Infect. 1995;115:71–78. doi: 10.1017/s0950268800058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joiner KA. Complement evasion by bacteria and parasites. Annu Rev Microbiol. 1988;42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 48.Horstmann RD. Target recognition failure by the nonspecific defense system: surface constituents of pathogens interfere with the alternative pathway of complement activation. Infect Immun. 1992;60:721–727. doi: 10.1128/iai.60.3.721-727.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiser JN, Lindberg AA, Manning EJ, Hansen EJ, Moxon ER. Identification of a chromosomal locus for expression of lipopolysaccharide epitopes in Haemophilus influenzae. . Infect Immun. 1989;57:3045–3052. doi: 10.1128/iai.57.10.3045-3052.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzaelipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 51.Weiser JN, Maskell DJ, Butler PD, Lindberg AA, Moxon ER. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzaelipopolysaccharide. J Bacteriol. 1990;172:3304–3309. doi: 10.1128/jb.172.6.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiser JN, Williams A, Moxon ER. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. . Infect Immun. 1990;58:3455–3457. doi: 10.1128/iai.58.10.3455-3457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maskell DJ, Szabo MJ, Butler PD, Williams AE, Moxon ER. Molecular analysis of a complex locus from Haemophilus influenzaeinvolved in phase-variable lipopolysaccharide biosynthesis. Mol Microbiol. 1991;5:1013–1022. doi: 10.1111/j.1365-2958.1991.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 54.Maskell, D.J., M.J. Szabo, P.D. Butler, A.E. Williams, and E.R. Moxon. 1992. Molecular biology of phase-variable lipopolysaccharide biosynthesis by Haemophilus influenzae. J. Infect. Dis. 165(Suppl. 1):90–92. [DOI] [PubMed]
- 55.Roche RJ, Moxon ER. Phenotypic variation of carbohydrate surface antigens and the pathogenesis of Haemophilus influenzaeinfections. Trends Microbiol. 1995;3:304–309. doi: 10.1016/s0966-842x(00)88959-3. [DOI] [PubMed] [Google Scholar]
- 56.Jarosik GP, Hansen EJ. Identification of a new locus involved in expression of Haemophilus influenzaetype b lipooligosaccharide. Infect Immun. 1994;62:4861–4867. doi: 10.1128/iai.62.11.4861-4867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roche RJ, Moxon ER. Phenotypic variation in Haemophilus influenzae: the interrelationship of colony opacity, capsule and lipopolysaccharide. Microb Pathog. 1995;18:129–140. doi: 10.1016/s0882-4010(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 58.Hood DW, Deadman ME, Allen T, Masoud H, Martin A, Brisson JR, Fleischmann R, Venter JC, Richards JC, Moxon ER. Use of the complete genome sequence information of Haemophilus influenzaestrain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- 59.Gotschlich EC. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeaelipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang QL, Gotschlich EC. Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgtgenes encoding glycosyl transferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jennings MP, Hood DW, Peak IR, Virji M, Moxon ER. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. . Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 62.Virji M, Makepeace K, Peak IR, Ferguson DJ, Jennings MP, Moxon ER. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol Microbiol. 1995;18:741–754. doi: 10.1111/j.1365-2958.1995.mmi_18040741.x. [DOI] [PubMed] [Google Scholar]
- 63.Hammerschmidt S, Müller A, Sillmann H, Mühlenhoff M, Borrow R, Fox A, van Putten J, Zollinger WD, Gerardy R, Schahn, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 64.Hammerschmidt S, Hilse R, van Putten JP, Gerardy R, Schahn, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidisvia a transposable genetic element. EMBO (Eur Mol Biol Organ) J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 65.Jones DM, Borrow R, Fox AJ, Gray S, Cartwright KA, Poolman JT. The lipooligosaccharide immunotype as a virulence determinant in Neisseria meningitidis. . Microb Pathog. 1992;13:219–224. doi: 10.1016/0882-4010(92)90022-g. [DOI] [PubMed] [Google Scholar]
- 66.Moran EE, Brandt BL, Zollinger WD. Expression of the L8 lipopolysaccharide determinant increases the sensitivity of Neisseria meningitidisto serum bactericidal activity. Infect Immun. 1994;62:5290–5295. doi: 10.1128/iai.62.12.5290-5295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mody CH, Paine R, III, Shahrabadi MS, Simon RH, Pearlman E, Eisenstein BI, Toews GB. Legionella pneumophilareplicates within rat alveolar epithelial cells. J Infect Dis. 1993;167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 68.Cianciotto NP, Stamos JK, Kamp DW. Infectivity of Legionella pneumophila mipmutant for alveolar epithelial cells. Curr Microbiol. 1995;30:247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- 69.Jacques M. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 1996;4:408–409. doi: 10.1016/0966-842X(96)10054-8. [DOI] [PubMed] [Google Scholar]
- 70.Venkataraman C, Haack BJ, Bondada S, Abu Y, Kwaik Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformisas a potential receptor for attachment and invasion by the Legionnaires' disease bacterium. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]