Abstract
General methods to engineer genetically encoded, reversible, light-mediated control over protein function would be useful in many areas of biomedical research and technology. We describe a system that yields such photo-control over actin assembly. We fused the Rho family GTPase Cdc42 in its GDP-bound form to the photosensory domain of phytochrome B (PhyB) and fused the Cdc42 effector, the Wiskott-Aldrich Syndrome Protein (WASP), to the light-dependent PhyB-binding domain of phytochrome interacting factor 3 (Pif3). Upon red light illumination, the fusion proteins bind each other, activating WASP, and consequently stimulating actin assembly by the WASP target, the Arp2/3 complex. Binding and WASP activation are reversed by far-red illumination. Our approach, in which the biochemical specificity of the nucleotide switch in Cdc42 is overridden by the light-dependent PhyB-Pif3 interaction, should be generally applicable to other GTPase-effector pairs.
Keywords: cytoskeleton, photoswitch, phytochrome, signal transduction, protein engineering
Biological processes occur over a wide range of scales in length and time. The development of organisms depends on the formation of macroscopic tissues that grow over a period of days. Micrometer-size cells divide and differentiate on a timescale of minutes to hours. Nanometer scale signaling assemblies reorganize in seconds after cell stimulation. To understand biology, tools are required that enable observation and perturbation of processes on these different length- and timescales. In recent years, tools for observation have grown rapidly, particularly with the advent of GFP-based approaches (1). However, tools for perturbation, particularly on the smallest scales, have lagged behind.
Classical genetic techniques target a desired gene of interest and examine the resulting perturbation over the lifetime of an organism. Thus, these classical methods are well suited to study events, such as development, that occur over longer timescales. Chemical genetics and pharmacology enable perturbations on a faster timescale of minutes and with greater temporal control through the introduction of cell-permeable small molecules (2). These methods are advantageous, because they also allow for the conditional regulation of activity through noncovalent and reversible interactions, which is convenient for studies at the cellular level. Other chemical perturbation methods, such as use of photolabile “caged” compounds, can increase resolution further by affecting subcellular targets in seconds (3, 4). Chemical photoswitches such as azobenzene can further offer reversible photo-control when attached to macromolecules (5). These photo-sensitive reagents afford precise spatial and temporal regulation of subcellular processes, because molecular activity is controlled directly by the time and location of a light stimulus. The utility of these highest resolution methods has been limited in biological investigations, however, because of the requirement to introduce exogenous, chemically modified materials into cells. The recently discovered light-regulated ion channel, channelrhodopsin, surmounts this shortcoming, because the protein is genetically encoded and its retinal chromophore is naturally occurring in most eukaryotic cells (6). Thus, channelrhodopsin promises to have major impact across biology and, in particular, in neuroscience. Despite these advances, no technology is currently available that can conditionally modulate protein function in a general fashion with the combined characteristics of specificity of genetic techniques, rapid reversibility of small molecule-based strategies, and high spatial and temporal precision afforded by light control.
Nature has developed photoregulatory systems in plants and bacteria in which light control of protein activity is genetically encoded (7–9). In these systems, absorption of light by photoreceptor proteins is coupled to changes in protein conformation and/or dynamics that alter activity toward downstream effector molecules. Thus, light is transduced to intracellular signaling. The phytochrome proteins (PhyA to PhyE in Arabidopsis thaliana) constitute a major class of photoreceptors (7–9). In plants, the phytochromes regulate growth and development in response to red and far-red light (7, 8). Plant phytochromes are ≈125 kDa proteins that use a covalently attached tetrapyrrole chromophore, phytochromobilin (PΦB). Phytochromes interconvert between two states: the red light absorbing form, Pr, which is inactive in most cases, and the far-red absorbing form, Pfr, which is typically active (8, 9). Pr is predominant in the dark. Red light absorption converts Pr to Pfr, which then transmits signals through binding to transcription factors such as phytochrome interacting factor 3 (Pif3), to regulate photomorphogenesis (7, 9–12). Thermal reversion of Pfr to Pr occurs on a timescale of hours (13, 14). However, irradiation with far-red light can rapidly switch phytochrome back into the inactive Pr form in seconds, causing its dissociation from transcription factors. Recent studies have demonstrated that holo-phytochrome B (holo-PhyB) can be heterologously produced in E. coli through coexpression with two enzymes, heme oxygenase and bilin reductase, that together generate PΦB from heme (15, 16). Similarly, holo-PhyB can be generated in yeast through growth of strains expressing the apo-protein on media containing the phycocyanobilin chromophore (PCB, a close analog of PΦB) isolated from blue-green algae (17, 18). Together, these properties of phytochrome make it an ideal candidate for engineering a reversible, photoswitchable signaling system that combines elements of speed, spatial precision, and reversibility to control the location and timing of protein function.
Here, we describe a genetically encoded signaling system where we can rapidly and reversibly modulate protein activity with light. We have engineered chimeras of PhyB and Pif3 fused to the Wiskott-Aldrich Syndrome protein (WASP) and to its activator, the Rho family GTPase Cdc42 (19, 20). In normal signaling, upstream cues load Cdc42 with the GTP nucleotide, enabling the protein to bind WASP with high affinity. The Cdc42(GTP)-WASP complex can then activate the actin nucleation factor, the Arp2/3 complex, causing assembly of new actin filaments in vitro and in vivo (21–24). Cdc42(GDP) can also activate WASP, but with much lower affinity (25). Here, we show that the Pfr form of PhyB [(Pfr)PhyB] binds to Pif3 with high affinity (KD = 575 nM) and that the interaction can be reversed upon irradiation with far-red light. In our chimeras, the PhyB-Pif3 interaction can override the nucleotide switch, enabling Cdc42(GDP) to activate WASP toward Arp2/3 complex with high potency after stimulation with red light. Far-red illumination rapidly reverses WASP activity. (Pfr)PhyB-Cdc42(GDP) produces a level of Pif3-WASP activity that is appreciably higher than that achievable by even saturating Cdc42(GDP). When implemented in vivo, this system should enable detailed exploration of the relationship between dynamics of the Cdc42-WASP pathway and dynamics of actin assembly and cell morphology. Our method of nucleotide switch override is likely to be generally applicable to other GTPase-effector systems and should provide a method to probe a variety of cellular functions in a way that is inaccessible by current approaches.
Results
Design Rationale for a Photoswitchable Cdc42/WASP System.
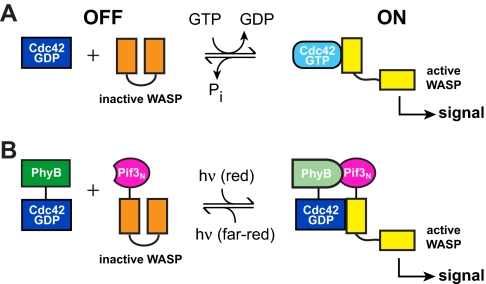
The Rho GTPase Cdc42 is a central regulator of actin cytoskeletal structure and dynamic. Cdc42 functions as a molecular switch, cycling between an inactive GDP-bound state and an active GTP-bound state. In the GTP-bound state, Cdc42 binds with high affinity to the GTPase binding domain (GBD) of its autoinhibited effector, WASP (19). Binding causes a conformational change in WASP, which enables the C-terminal VCA domain (for Verprolin homology, Central hydrophobic and Acidic domain) of the protein to activate the ubiquitous actin nucleation factor, the Arp2/3 complex (21, 23). The growth of new actin filaments generated by Arp2/3 complex provides the mechanical force required for cell growth, motility, and cytoskeletal rearrangements (26, 27). We previously demonstrated that although Cdc42(GTP) has higher affinity for WASP, Cdc42(GDP) is also able to drive substantial (≈50%) WASP activation at sufficiently high concentrations (i.e., approximating saturation) (25). Thus, high Cdc42 concentration is able to partially override the biochemical specificity of the nucleotide switch. On this basis, we reasoned that if Cdc42 and WASP were fused separately to two strongly interacting partner proteins, high local concentrations in the partner-mediated complex would drive WASP activation even by Cdc42(GDP) (Fig. 1). This design concept was borne out in preliminary experiments with a chemically mediated dimerization system [supporting information (SI) Text, Figs. S1 and S2 and Table S1], and we proceeded to determine whether it could be applied to light regulation as well.
Fig. 1.
Engineering a genetically encoded, reversible light-controlled signaling system. (A) The Cdc42-WASP interaction is normally controlled by nucleotide exchange and hydrolysis. (B) Cellular control can be overridden by fusing Cdc42 and WASP to PhyB and Pif3N, respectively, rendering WASP activity sensitive to light.
High Affinity Association of PhyB with Pif3 Can Be Reversibly Controlled by Light.
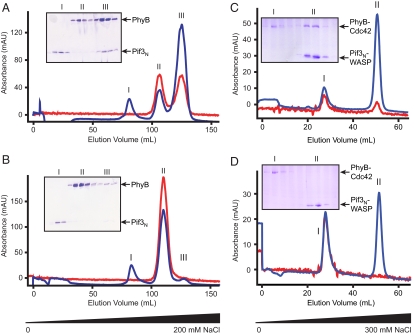
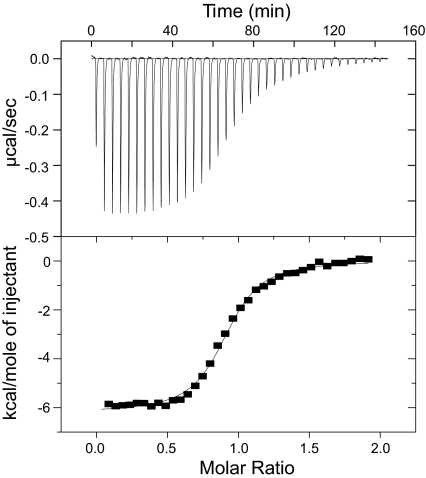
Based on previous domain mapping of the PhyB-Pif3 interaction (10, 11), we generated an N-terminal fragment of holo-PhyB (residues 1–651 covalently attached to PΦB; referred to hereafter simply as PhyB) in bacteria by coexpression with Synechocystis sp. PCC6803 heme oxygenase (Ho1) and a truncated form of the Arabidopsis thaliana bilin reductase, PΦB synthase (HY2) (Figs. S1 and Table S1) (15, 16). Attachment of the chromophore to PhyB was verified by zinc blot analysis and by UV absorbance at both 280 nm and 665 nm throughout several steps of chromatographic purification (Figs. S3 and S4). To evaluate the functionality of the purified PhyB, we examined its interactions with a previously identified binding domain of Pif3 (residues 1–100; referred to hereafter as Pif3N). After dark adaptation or far-red irradiation, PhyB and Pif3N eluted from anion exchange columns at two distinct salt concentrations upon coinjection (Fig. S5). Upon irradiation with red light, the two proteins formed a tight complex that eluted at higher salt concentrations than either free component (Fig. 2A and Fig. S5D). The complex could be largely dissociated by subsequent irradiation with far-red light, which severely decreased the chromatographic peak corresponding to the PhyB/Pif3N complex and concomitantly increased those corresponding to free Pif3N and PhyB (Fig. 2B). Although red light-induced complex formation is substantial, we were never able to achieve complete conversion to the dimer even upon prolonged illumination. This probably reflects the photostationary state of the system under the irradiation conditions used here and some dissociation of the complex during chromatography. However, we could achieve virtually complete dissociation of the complex with far-red illumination, again reflecting our photo-excitation system and the absorbance spectra of Pr and Pfr (16). We used isothermal titration calorimetry (ITC) to further characterize and quantify the interaction between PhyB and Pif3N. As shown in Fig. 3, after red illumination, PhyB and Pif3N formed a 1:1 complex with a KD of 575 ± 96 nM. Under the same conditions, the KD of PhyB for Pif3N in the Pr state could not be quantified by ITC because of the absence of measurable heat of binding (data not shown). Together with the chromatographic data, this result suggests a change of affinity of ≈20-fold or greater upon absorption of far-red light.
Fig. 2.
Light-controlled association of PhyB and Pif3N. (A) A mixture of PhyB and Pif3N was irradiated with red light and separated by anion exchange chromatography using a NaCl gradient. The three peaks correspond to Pif3N alone (I), PhyB alone (II), and PhyB- Pif3N complex (III). (B) All protein-containing fractions from (A) were pooled, diluted to low salt, irradiated with far-red light, and reseparated identically to (A). (C and D) Same experiments as in (A and B), but using the PhyB-Cdc42 and Pif3N-WASP fusions. The two peaks correspond to PhyB-Cdc42 alone (I), and either the Pif3N-WASP:PhyB-Cdc42 complex (C) or Pif3N-WASP alone (D) (II). Chromatograms show protein absorbance at 280 nm (blue) and chromophore absorbance at 665 nm (red). Insets show Coomassie-Blue stained SDS-PAGE of corresponding peak fractions in each chromatographic run.
Fig. 3.
(Pfr)PhyB binds Pif3N with high affinity. Representative ITC data for (Pfr)PhyB-WASP binding to Pif3N. Raw heats versus time (Upper) and the integrated heats of reaction versus molar ratio of ligand to receptor (Lower) are plotted. Fitted values: stoichiometry, 0.91 ± 0.01 : 1; KD, 575 ± 96 nM; ΔH, −6300 ± 120 cal·mol−1; ΔS, 7.77 ± 0.40 cal·mol−1·K−1.
PhyB-Cdc42(GDP) Activates Pif3N-WASP in a Light-Dependent Fashion.
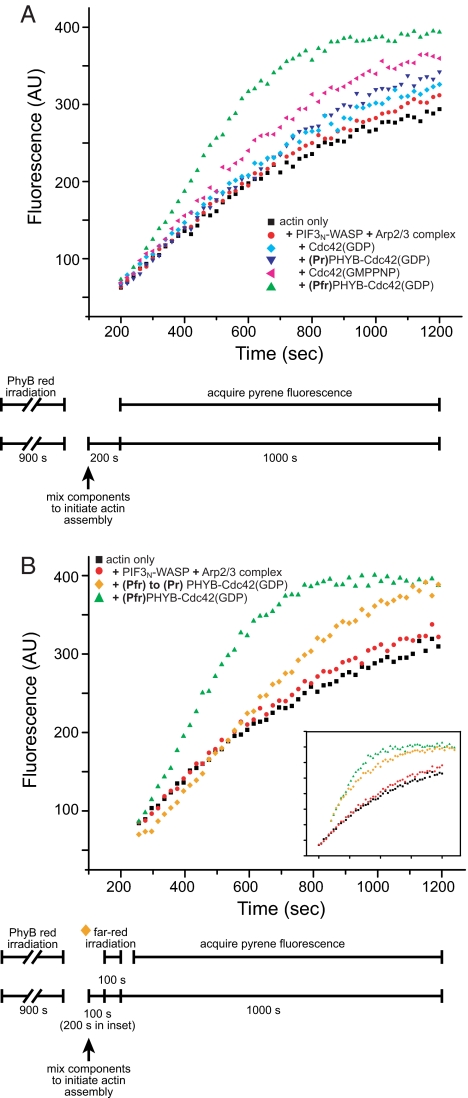
We next evaluated whether the WASP/Cdc42(GDP) interaction in the fusion proteins could be photo-regulated through the PhyB-Pif3N interaction. Because we have no knowledge of the structural organization of the (Pfr)PhyB-Pif3N complex, we produced four pairs of fusion constructs with different domain organizations (and Table S1 and Fig. S6). We found that PhyB can only be fused N-terminally to WASP or Cdc42; constructs with PhyB at the C terminus did not express in bacteria or expressed very poorly with reduced chromophore incorporation. We assayed the functionality of PhyB and Pif3N in the various constructs chromatographically to ensure that fusion to Cdc42 or WASP did not alter the intrinsic light-dependent binding properties of the domains (Fig. 2 C and D). We then measured WASP activity toward Arp2/3 complex in the PhyB or Pif3N fusions using a pyrene-actin assembly assay performed under green safe light conditions. In this assay, an increase in pyrene fluorescence reports on actin polymerization into filaments. Activation of WASP (and consequent activation of Arp2/3 complex) was indicated by a decreased lag time and increased rate of filament formation. As detailed in Methods, for assays involving (Pfr)PhyB, the PhyB fusion was excited before addition to the reaction, and no red light illumination was applied during the time course of the assay. We initially examined PhyB-WASP and found that it had low basal activity toward Arp2/3 complex in both the Pr and Pfr states, similar to WASP alone. Activity was not increased by addition of 1 μM Pif3N-Cdc42(GDP) or 1 μM Cdc42(GDP)-Pif3N, even in the Pfr state, where the proteins should form appreciable complex based on the (Pfr)PhyB-Pif3N affinity (Fig. S6A and B). Thus, either the domains in these constructs were improperly oriented to enhance WASP activation by Cdc42(GDP) or the PhyB-Pif3N interaction was significantly weakened in these particular fusions. In the opposite construction, both Pif3N-WASP and WASP-Pif3N also had basal activity identical to control (Fig. 4A and Fig. S6C). However, in contrast to the previous case, both constructs were significantly activated by 500 nM (Pfr)PhyB-Cdc42(GDP). This activation was much greater than that induced by the same concentration of (Pr)PhyB-Cdc42(GDP) (compare green triangles and inverted blue triangles, Fig. 4A and Figs. S6 and S7). As shown in Fig. S7, for actin preparations where the baseline assembly rate is slower (probably because of more efficient removal of small filaments during purification), activation by Pfr was largely unchanged from Fig. 4A, but activation by Pr was decreased substantially. Thus, in these experiments the difference between Pfr- and Pr-mediated activation was even larger than seen in Fig. 4A. Pif3N-WASP appears to be the preferred construct, because its level of activation by (Pfr)PhyB-Cdc42(GDP) was significantly higher than that of WASP-Pif3N (compare Fig. S6 and Fig. S4A). The degree of light-dependent WASP activity in this system was very substantial. That is, 500 nM (Pfr)PhyB-Cdc42(GDP) activated Pif3N-WASP more strongly than 5–10 μM Cdc42(GDP) (Figs. 4A and Fig. S7 and S8) and even 5–20 μM Cdc42(GMPPNP) (Fig. 4 and Fig. S8), the latter mimicking Cdc42(GTP). These results establish that the PhyB-Pif3N interaction can be used to drive potent, light-dependent activation of WASP.
Fig. 4.
Reversible, light-controlled activation of Pif3N-WASP by PhyB-Cdc42(GDP). (A) Pyrene-actin assembly assays contained 4 μM actin (19% pyrene-labeled), 10 nM Arp2/3 complex, 50 nM Pif3N-WASP (red circles), except actin-only control (black squares). Assays additionally contained 5 μM Cdc42(GDP) (cyan diamond), 500 nM (Pr)PhyB-Cdc42(GDP) (blue inverted triangle), 5 μM Cdc42(GMPPNP) (magenta triangle), or 500 nM (Pfr)PhyB-Cdc42(GDP) (green triangle). (B) Assays performed as in A, except orange diamonds. There, assay was initiated for 100 s (200 s in Inset) with (Pfr)PhyB-Cdc42(GDP), irradiated with far-red light for 100 s, and then allowed to complete normally. Reversal of the PhyB-Cdc42(GDP) photoswitch in the first 200 s of assembly eliminated Pif3N-WASP activation. In both panels, the timing of biochemical and optical aspects of the assays are shown in the schematic below the graph.
An ideal light-controlled system will enable not only activation of protein function but also inactivation as well. In the pyrene-actin assembly assay, the vast majority of Arp2/3-mediated nucleation occurs early in the time course, when total filament content (and, thus, deviation of fluorescence intensity from baseline) is low. Once initiated, filaments grow independent of continued WASP/Arp2/3 activity, producing strong signal in the assay tens of seconds later. For this reason, it would be extremely difficult to observe switching of WASP activity and, thus, nucleation by Arp2/3 complex in real time. Thus, we examined photoswitching indirectly. We initiated polymerization reactions with (Pfr)PhyB-Cdc42(GDP) and Pif3N-WASP, allowed them to proceed for a period τ, exposed the reaction to far-red light for 100 s, and then acquired pyrene fluorescence data normally. As shown in Fig. 4B, when τ = 100 s, the far-red illumination decreased Pif3N-WASP activity substantially (compare orange diamonds to green triangles). Control reactions of WASP activation by Cdc42(GMPPNP) were not affected by far-red illumination, demonstrating that the decrease results from effects on the photosensory system (data not shown). Similarly, when τ = 200 s, the assembly time course was altered only slightly, because the majority of nucleation occurred before Pif3N-WASP activity was reversed (Fig. 4B Inset). Thus, the Cdc42-WASP system can be reversibly photoactivated with red and far-red light.
Discussion
We have shown here that the nucleotide switch in Cdc42 can be overridden through artificial dimerization of the GTPase with WASP. This has led to a system in which the activity of WASP can be controlled with light in reversible fashion using genetically encoded components. Such an approach should be generally applicable, provided that other GDP-bound GTPases can also activate their effectors when at high concentration. Several pieces of data suggest that this will likely be the case. Most directly, the GTP- and GDP-bound forms of GSα have been shown to activate adenylyl cyclase to similar levels, although the latter requires higher concentrations to do so (28). This has been explained by a model in which GSα exists in equilibrium between an inactive conformation (G) and an active conformation (G*), which are differentially favored by GDP- and GTP-binding, respectively. G* has higher affinity for adenylyl cyclase and can activate it. Because both nucleotide states of GSα can access the G* conformation, both are capable of activating adenylyl cyclase. However, because the GDP state favors G, its affinity for adenylyl cyclase is lower than that of the GTP state. Reciprocally, adenylyl cyclase binding shifts the allosteric equilibrium in GSα toward G*. A large body of spectroscopic data indicates that the Ras GTPase also exists in an analogous equilibrium between two conformations in both the GDP- and GTP-bound states (29–31). In addition, binding of the effector Raf strongly shifts the equilibrium to one state (29, 31). Although it has not been tested experimentally, these data suggest that like Cdc42 and GSα, Ras(GDP) can sample a GTP-like conformation, which could act on Raf similarly to Ras(GTP). Finally, two-state behavior has also been observed for Ran(GMPPNP) spectroscopically (29), and Ran(GDP) has been found capable of binding its effector RanBP1 with micromolar affinity (32). These observations on several distantly related family members argue that many GDP-bound GTPases should be capable of activating their effectors at sufficiently high concentrations. Thus, our approach to generating light-controlled GTPase-effector systems by overriding the nucleotide switch should have broad applicability. Given that many biological signaling events are driven by changes in the affinity of protein–protein interactions, this approach to light control may have utility in numerous other systems as well.
A number of hurdles must be overcome to implement PhyB-Pif3-based photoswitches of the type that we describe here in vivo. Principal among these is the need to generate holo-PhyB in eukaryotic systems. This may be accomplished as we have here, through expression of Ho1 and HY2, albeit at the expense of significant cell engineering. Alternatively, it may be possible to simply add the PCB chromophore (isolated from algae) to cells expressing the PhyB fusion. Because yeast readily take up PCB in this manner and produce holo-PhyB (17, 18), this may ultimately prove to be the more straightforward approach in higher eukaryotes as well. It will also be important to control both the relative and absolute levels of the PhyB and Pif3 fusions to produce quantitatively defined light-dependent activity, based on the affinities of the two components of the switch. This difficulty may be lessened somewhat through designing reporters of activity/binding directly into the photoswitch, for example by attaching CFP and YFP fluorophores to the two components of the switch. In such a design, intermolecular FRET would independently report on the binding status of the switch (1). Finally, as with all photo-controlled systems, spatial precision will be decreased by diffusion of the components after illumination. However, in contrast to photo-activation systems, where the active form remains active while it diffuses, in PhyB-based photoswitchable systems, it may be possible to achieve highly precise activation through local red light illumination coupled with global far-red illumination. If these difficulties can be overcome, the PhyB-Pif3 system could have great utility in understanding biology through the rapidly reversible spatial and temporal control of protein activity.
Methods
Generation of holo-PhyB.
The expression of holo-PhyB required the construction of expression vectors for three genes: Heme oxygenase 1 (Ho1), Phytochromobilin (PΦB) synthase lacking the transit peptide (ΔHY2) (16), and Phytochrome B (PhyB). Ho1 was obtained from Synechocystis sp. PCC6803 (gift of C. Lagarias) and subcloned into a modified pACYC expression vector (Novagen). ΔHY2 was obtained from Arabidopsis thaliana cDNA (TAIR; Stanford, CA) and subcloned into pCDFDuet-1 (Novagen). The N-terminal 651 residues of Arabidopsis thaliana PHYB were subcloned into a modified pET28a vector (Novagen) containing a C-terminal His6 tag preceded by a TEV protease cleavage site. All three plasmids were simultaneously transformed into E. coli strain BL21(DE3) by electroporation and plated onto LB agar containing kanamycin (30 μg/ml), chloramphenicol (34 μg/ml), and spectinomycin (50 μg/ml). Bacteria were grown at 37°C in LB media containing all three antibiotics. Cells were induced with 0.5 mM IPTG at OD600 nm = 0.6 and cultured overnight at 18°C in the dark. Cell lysis, and purification and handling of holo-PhyB were performed under green safe light (Rosco, no. 89). PhyB-His6 was purified from cleared bacterial lysate by sequential Ni2+-affinity (Ni-Sepharose Fast Flow, GE Healthcare) and anion exchange (Source 15Q, GE Healthcare) chromatographies. The His6-tag was removed by Tev protease cleavage, followed by Ni2+-affinity chromatography. Flow through from the nickel affinity column was concentrated and purified by gel filtration chromatography (SD200, GE Healthcare). PhyB fusions with WASP (GBD-VCA) or Cdc42 (residues 1–179) through a 20 residue linker composed of Gly and Ser residues were obtained using a similar expression and purification protocol. The molar absorption coefficient for PhyB-WASP at 280 nm was 180,156 M−1·cm−1, as determined from amino acid analysis (Keck AAA & Protein Sequencing Lab, Yale University). We note that the accurate molar absorption coefficient derived from amino acid analysis is significantly different from the value estimated from the combined absorbances of Trp/Tyr residues and the free PΦB or PCB chromophore, but produces 1:1 stoichiometry of the PhyB-Pif3 complex determined by ITC, consistent with retention times on gel filtration chromatography columns.
Generation of Pif3N.
The N-terminal 100 residues of Pif3 was amplified from Arabidopsis thaliana cDNA (TAIR; Stanford, CA) and subcloned alone or fused to Cdc42 or WASP GBD-VCA, connected by a 20 residue linker composed of Gly and Ser residues, into a modified pMAL-C vector (New England Biolabs) containing N-terminal maltose binding protein (MBP) and C-terminal His6 tags with TEV protease restriction sites for the removal of both tags. Proteins were expressed and purified identically to the PhyB proteins above, except that the MBP and His6 tags were removed after TEV cleavage by successive amylose and Ni2+ affinity chromatographies.
Chromatographic Assays.
Anion exchange chromatography performed under green safe light was used to examine the binding of PhyB to Pif3N. A mixture of PhyB with Pif3N in 20 mM Tris pH 8, 50 mM NaCl, and 5 mM β-mercaptoethanol was exposed for 15 min to ≈1 mW red light (narrow bandpass interference filter 656 nm; Edmund Optics, Barrington, NJ) from a 200 W Thermo Oriel Hg lamp (Newport, Stratford, CT) before loading onto an 8-ml Source 15Q 10/100 column (GE Healthcare), which was eluted with a linear gradient to 300 mM NaCl in buffer A over 30 column volumes. Peak fractions containing Pif3N, PhyB, and the complex were pooled, exposed to ≤1 mW far-red light (narrow bandpass interference filter 766 nm; Edmund Optics, Barrington, NJ) for 15 min, and reseparated identically. Later analyses of actin assembly (Fig. 4B) and absorbance (data not shown) showed that photoswitching is complete in <100 s using these irradiation conditions.
Isothermal Titration Calorimetry (ITC).
Calorimetry was performed at 30°C using a VP-ITC microcalorimeter (Microcal Corp., Northampton, MA). Both PhyB-WASP and Pif3N were dialyzed against 1L of 25 mM phosphate pH 7, 150 mM NaCl, and 5 mM β-mercaptoethanol for 24 h before use. Heats of reaction were measured in the dark for up to 36 injections of 200 μM Pif3N into a 2.1-ml cell chamber containing 28 μM PhyB-WASP, which was previously exposed to red light for 15 min. The concentration of PhyB-WASP was calculated based upon the molar absorption coefficient determined from amino acid analysis. The total time to generate and record a binding isotherm was ≈2.5 h, which is less than the half life of the PhyB-Pif3N complex (>4 h) based upon a time course of complex dissociation performed using a pulldown assay (data not shown). Raw calorimetry data were processed, baseline corrected by subtracting heats from Pif3N injection into buffer, and fit to yield number of binding sites, KD, and binding enthalpy using ORIGIN software. Reported values are averages of two experiments.
Pyrene-Actin Polymerization Assays.
Pyrene-actin assembly assays were performed as described in ref. 33. Pyrene fluorescence was monitored at 407 nm (λex = 465 nm) on a Fluorolog 3–11 fluorimeter (Jobin Yvon Horiba, Edison, NJ). Assays were performed in buffer containing 10 mM imidazole pH 7, 50 mM KCl, 1 mM MgCl2, 10 mM EGTA, 4 μM actin (6% pyrene-labeled for assays with FKBP; 19% pyrene for assays with PhyB), and 10 nM Arp2/3 complex. In addition to the major absorbance maxima at 660 nm and 730 nm, Pr and Pfr also have smaller maxima at 375 nm and 390 nm, respectively (Fig. S4) (16). Thus, for assays including PhyB, reactions were carried out under green safe light, λex was adjusted to 460 nm, and data were acquired for 0.1 s every 20 s with either a 200, 250, or 350 s initial delay (Fig. 4). These changes decrease the sensitivity of fluorescence detection, necessitating the use of higher (19%) pyrene labeling of actin. In the absence of such precautions, pyrene excitation caused significant (>30%) conversion of (Pfr) to (Pr)PhyB during the course of the assay.
Supplementary Material
Acknowledgments.
We thank Drs. Kevin Gardner, Ulrich Genick, Xiaolan Yao, and Michael Roth for advice and discussion; Dr. J. Clark Lagarias (University of California, Davis, CA) for providing a clone of Ho1, Kevin Gardner and J. Clark Lagarias for critical reading of the manuscript, Ayman Ismail for sharing reagents for the actin assembly assays, and Inga Nelson for assisting with the initial cloning of phytochrome constructs. Work was supported by the Howard Hughes Medical Institute, Welch Foundation Grant I-1544 (to M.K.R.), and National Institutes of Health Grant GM52413 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801232105/DCSupplemental.
References
- 1.Muller-Taubenberger A, Anderson KI. Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol. 2007;77:1–12. doi: 10.1007/s00253-007-1131-5. [DOI] [PubMed] [Google Scholar]
- 2.Banaszynski LA, Wandless TJ. Conditional control of protein function. Chem Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Rothman DM, Shults MD, Imperiali B. Chemical approaches for investigating phosphorylation in signal transduction networks. Trends Cell Biol. 2005;15:502–510. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Adams SR, Tsien RY. Controlling cell chemistry with caged compounds. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 5.Renner C, Moroder L. Azobenzene as conformational switch in model peptides. Chembiochem. 2006;7:868–878. doi: 10.1002/cbic.200500531. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 8.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 9.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix–loop–helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 12.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 13.Oka Y, Matsushita T, Mochizuki N, Suzuki T, Tokutomi S, Nagatani A. Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell. 2004;16:2104–2116. doi: 10.1105/tpc.104.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98:10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukougawa K, Kanamoto H, Kobayashi T, Yokota A, Kohchi T. Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Lett. 2006;580:1333–1338. doi: 10.1016/j.febslet.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Lagarias JC. Phytochrome assembly in living cells of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:12535–12539. doi: 10.1073/pnas.91.26.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 19.Higgs HN, Pollard TD. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- 20.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi H, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung DW, Rosen MK. The nucleotide switch in Cdc42 modulates coupling between the GTPase-binding and allosteric equilibria of Wiskott-Aldrich syndrome protein. Proc Natl Acad Sci USA. 2005;102:5685–5690. doi: 10.1073/pnas.0406472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 27.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 28.Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R. Allosteric determinants in guanine nucleotide-binding proteins. Proc Natl Acad Sci USA. 2003;100:14445–14450. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, Kalbitzer HR. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35:10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- 30.Rohrer M, et al. Structure of the metal-water complex in Ras x GDP studied by high-field EPR spectroscopy and 31P NMR spectroscopy. Biochemistry. 2001;40:1884–1889. doi: 10.1021/bi002164y. [DOI] [PubMed] [Google Scholar]
- 31.Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA. 2001;98:4944–4949. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhlmann J, Macara I, Wittinghofer A. Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- 33.Leung DW, Morgan DM, Rosen MK. Biochemical properties and inhibitors of (N-)WASP. Methods Enzymol. 2006;406:281–296. doi: 10.1016/S0076-6879(06)06021-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.