Abstract
Imprinted genes are important in development and their allelic expression is mediated by imprinting control regions (ICRs). On their DNA-methylated allele, ICRs are marked by trimethylation at H3 Lys 9 (H3K9me3) and H4 Lys 20 (H4K20me3), similar to pericentric heterochromatin. Here, we investigate which histone methyltransferases control this methylation of histone at ICRs. We found that inactivation of SUV4-20H leads to the loss of H4K20me3 and increased levels of its substrate, H4K20me1. H4K20me1 is controlled by PR-SET7 and is detected on both parental alleles. The disruption of SUV4-20H or PR-SET7 does not affect methylation of DNA at ICRs but influences precipitation of H3K9me3, which is suggestive of a trans-histone change. Unlike at pericentric heterochromatin, however, H3K9me3 at ICRs does not depend on SUV39H. Our data show not only new similarities but also differences between ICRs and heterochromatin, both of which show constitutive maintenance of methylation of DNA in somatic cells.
Keywords: genomic imprinting, SUV4-20H, PR-SET7, SUV39H, HP1
Introduction
Genomic imprinting is an epigenetic phenomenon that leads to the mono-allelic expression of genes, depending on the parental origin of the allele. Almost one hundred imprinted genes have been discovered, many of which are important in development (Morison et al, 2005). Most imprinted genes are organized into chromosomal domains, and allelic repression is controlled by essential sequence elements called ICRs (imprinting control regions; Delaval & Feil, 2004). ICRs are marked by methylation of DNA on either their maternally or their paternally inherited allele. This provides parental allele-specific function to the ICRs and thereby mediates imprinted gene expression. A well-studied imprinted domain is the insulin-like growth factor 2 (Igf2)-H19 locus on mouse chromosome 7, which is controlled by a paternally methylated ICR, the H19 ICR (Thorvaldsen et al, 1998). Directly adjacent is the Kcnq1-imprinted domain, which is controlled by a maternally methylated ICR, the KvDMR1 (Kvlqt1 differentially methylated region 1; Fitzpatrick et al, 2002). Perturbation of differential methylation at these ICRs is involved in growth-related diseases and also occurs in cancer. This raises the important question of how the differential methylation at ICRs is maintained throughout development, and whether chromatin features are important in this process. Recently, it was found that the methylated alleles of ICRs are consistently marked by trimethylation at H3 Lys 9 (H3K9me3) and H4 Lys 20 (H4K20me3; Wu et al, 2006; Delaval et al, 2007; Regha et al, 2007; Wagschal et al, 2008), as has been reported previously for pericentric heterochromatin (Lehnertz et al, 2003; Schotta et al, 2004). To unravel the role of this histone methylation, it is important to determine which histone methyltransferases (HMTs) are involved. This was determined by exploring candidate SET domain proteins with small interfering RNA and gene targeting approaches. Our findings highlight a sequential pathway that implicates PR-SET7 in the bi-allelic induction of mono-methylation of H4K20 (H4K20me1) at ICRs, and recruitment of SUV4-20H specifically to the DNA-methylated allele, to promote trimethylation of Lys 20. Our study identifies not only new similarities but also differences between ICRs and pericentric heterochromatin.
Results And Discussion
SUV4-20H controls H4K20me3 at ICRs
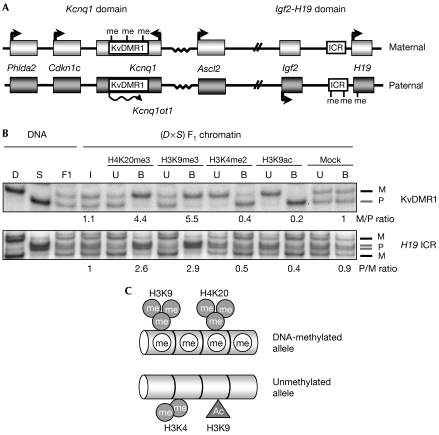
First, we determined whether ICRs show differential H4K20me3 in mouse embryonic fibroblasts (MEFs), the model system of this study. As an example of an ICR with methylation of maternal DNA, we studied the KvDMR1 and, as a model of a paternal ICR, the H19 ICR (Fig 1A). Chromatin immunoprecipitation (ChIP) on native chromatin was performed on primary MEFs derived from embryos that were (C57BL/6 × Mus spretus)F1 for distal chromosome 7 (Fig 1B). Using PCR amplification followed by allelic discrimination using single strand conformation polymorphisms, we determined the relative abundance of maternal and paternal alleles in the antibody-bound and unbound fractions. Chromatin on the DNA-methylated alleles of the ICRs was enriched in both H4K20me3 and H3K9me3. The unmethylated allele, by contrast, was associated with H3K4me2 and H3K9 acetylation (H3K9ac; Fig 1B,C). The same differential modifications were detected in immortalized MEFs (data not shown).
Figure 1.
Methylation of H4 Lys 20 at the H19 and KvDMR1 imprinting control regions. (A) Schematic presentation of the mouse Kcnq1 and Igf2-H19 domains. Boxes indicate genes and methylation of DNA (me) is shown. (B) ChIP on primary (C57BL/6 × SDP711)F1 MEFs followed by SSCP-mediated discrimination of the parental alleles. D, C57BL/6J control; S, SDP711 control with distal 7 of Mus spretus; F1, (C57BL/6 × SDP711)F1 control; I, input; U, unbound fraction; B, bound fraction; M, maternal allele; P, paternal allele. Allelic ratios are indicated below the relevant lanes. (C) Summary of histone modifications at ICRs. Ac, acetylation; ChIP, chromatin immunoprecipitation; ICR, imprinting control region; MEF, mouse embryonic fibroblast; SSCP, single strand conformation polymorphism.
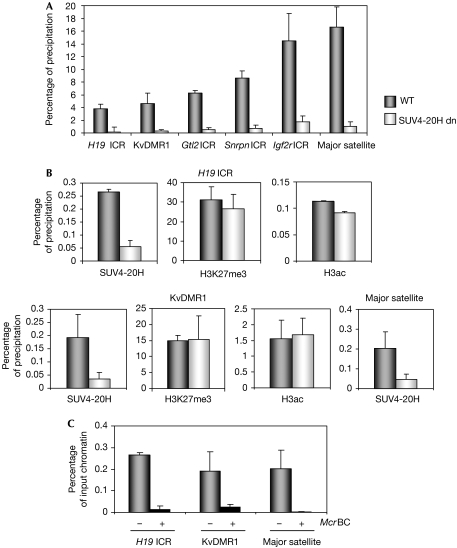
The HMTs that regulate H4K20me3 at pericentric heterochromatin, SUV4-20H1 and SUV4-20H2, are almost identical (Schotta et al, 2004). To determine whether they could control H4K20me3 at ICRs, we performed native ChIP on MEFs lacking both SUV4-20H1 and SUV4-20H2 (SUV4-20H dn cells). By using real-time PCR amplification, we measured the precipitation levels of H4K20me3 in SUV4-20H dn and wild-type MEFs at the H19 and KvDMR1 ICRs, as well as at the gene-trap locus 2 (Gtl2; Ig-DMR; Lin et al, 2003), Snrpn (Shemer et al, 1997) and Igf2 receptor (Igf2r; Regha et al, 2007) ICRs. At all five ICRs and at the major satellite DNA (at pericentric heterochromatin), we observed a strong reduction in the levels of H4K20me3 in SUV4-20H dn compared with wild-type MEFs. This result suggests that SUV4-20H maintains H4K20me3 at ICRs as in pericentric heterochromatin (Fig 2A).
Figure 2.
Allelic SUV4-20H recruitment controls H4K20me3 at imprinting control regions. (A) Loss of H4K20me3 at ICRs and at the major satellite DNA in immortalized SUV4-20H-deficient MEFs (SUV4-20H dn) and corresponding WT cells. Quantification of bound fractions was by qPCR amplification. Error bars represent standard deviation between three or more different ChIP experiments on independent chromatin preparations. (B) qPCR quantification of SUV4-20H, H3K27me3 and H3K9ac precipitation in SUV4-20H dn MEFs, and the corresponding WT MEFs at the H19 and KvDMR1 ICRs and at the major satellite DNA. (C) DNA was extracted from the WT chromatin precipitated with the antiserum against SUV4-20H, and digested with (+) or without (−) McrBC (which cuts methylated DNA only), followed by qPCR amplification. ChIP, chromatin immunoprecipitation; ICR, imprinting control region; MEF, mouse embryonic fibroblast; qPCR, quantitative PCR; WT, wild type.
Next, we explored whether SUV4-20H physically associates with chromatin at the H19 and KvDMR1 ICRs by using ChIP on crosslinked chromatin. We used an antiserum that binds to both SUV4-20H1 and SUV4-20H2, and, as a control for the quality of the chromatin preparations, we analysed H3K27me3 and H3K9ac (Delaval et al, 2007). We quantified the precipitated fractions by using real-time PCR amplification (Fig 2B) and corrected (in all ChIP experiments of this study) the obtained values against background precipitation with a non-immune IgG control antiserum. We found that SUV4-20H is recruited to the ICRs in wild-type cells. Furthermore, we demonstrated that this association occurred preferentially on the DNA-methylated allele by digesting the precipitated chromatin DNA with McrBC, a restriction endonuclease that is active only on methylated DNA. We then quantified the non-digested fraction of the DNA by real-time PCR amplification (Fig 2C). We also observed association of SUV4-20H with major satellite, which is DNA-methylated.
PR-SET7 mediates mono-methylation of H4K20 at ICRs
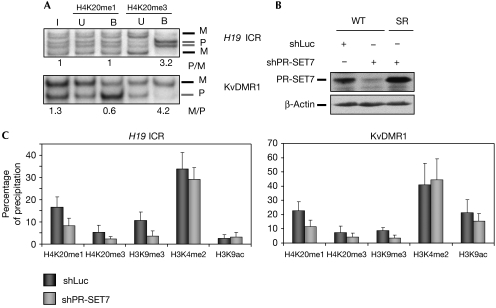
By analogy with the HMTs that regulate H3K9me3 (Peters et al, 2003), SUV4-20H could use H4K20me1 as a substrate to generate H4K20me3. Therefore, we explored whether H4K20me1 was also allele-specifically enriched at ICRs. At the H19 ICR and KvDMR1, we did not detect precipitation of H4K20me1 specifically on the DNA-methylated allele. Rather, precipitation was from both parental chromosomes and, at the KvDMR1, precipitation was even stronger on the unmethylated allele (Fig 3A). We also tested a third methylation state of H4K20, H4K20me2, which was reported to be abundant in human cells (Yang et al, 2008), but we did not observe any significant precipitation above the background at ICRs (data not shown).
Figure 3.
H4K20me1 at imprinting control regions is bi-allelic and controlled by PR-SET7. (A) Allelic analysis of H4K20me1 and H4K20me3 at the H19 and KvDMR1 ICRs in primary (C57BL/6 × SDP711)F1 MEFs. B, bound fraction; I, input; M, maternal; P, paternal; U, unbound fraction. (B) Immunoblot analysis of WT MEFs and MEFs synthesizing the PR-SET7 SR recombinant protein (SR), 5 days after luciferase (shLuc) or PR-SET7 (shPR-SET7) shRNA infection. β-Actin immunostaining was used as a loading control. (C) ChIP on WT and shPR-SET7 knockdown primary MEFs. qPCR amplification was performed on precipitated chromatin fractions to determine the percentage of precipitation. ChIP, chromatin immunoprecipitation; ICR, imprinting control region; MEF, mouse embryonic fibroblast; qPCR, quantitative PCR; WT, wild type.
Recently, we and others have reported that the HMT PR-SET7 (also known as SET8) controls the levels of H4K20me1 in human cells (Jørgensen et al, 2007; Tardat et al, 2007). To determine whether this HMT regulates H4K20me1 in the mouse as well, in particular at ICRs, we inhibited its expression in MEFs using an RNA-mediated interference strategy. A short hairpin RNA (shRNA) designed against PR-SET7 (shPR-SET7) was stably expressed in MEFs after retroviral infection and reduced the expression of this protein by about 50% (Fig 3B). This partial reduction in PR-SET7 did not affect cellular proliferation of the MEFs (data not shown). The expression of a luciferase-specific shRNA (shLuc), as a negative control, did not affect the expression of PR-SET7. In both cases, β-actin protein levels were not affected. The specificity of shPR-SET7 was verified by an shRNA-based protein replacement approach, in which shRNA-resistant wild-type PR-SET7 protein (SR) was expressed at physiological levels. This established that shPR-SET7 specifically downregulated the PR-SET7 protein (Fig 3B).
Next, we performed ChIP on the shPR-SET7 and shLuc cells. In agreement with the approximate 50% reduction in PR-SET7, a decrease of around 50% in H4K20me1 was observed at the H19 ICR and KvDMR1 in the knockdown cells, showing that PR-SET7 controls at least in part the mono-methylation of H4K20 (Fig 3C). The reduction in H4K20me1 correlated with a similar decrease in the levels of H4K20me3. This result suggests that H4K20me1 is converted into H4K20me3. Moreover, we obtained similar levels of precipitation of H3K4me2 and H3K9ac in shPR-SET7 and shLuc cells, a result that emphasizes the reproducibility of ChIP between cell lines (Fig 3C). Unexpectedly, however, in the shPR-SET7 cells, we observed a reduced precipitation of H3K9me3. To verify that the altered histone modifications were caused entirely by downregulation of PR-SET7, we performed ChIP on PR-SET7 knockdown cells in which we had restored the physiological levels of PR-SET7. In these cells (shPR-SET7+PR-SET7SR), the levels of H4K20me1 and H4K20me3 were again comparable to those in the shLuc control MEFs (supplementary Fig 1 online).
Methylation of DNA in the absence of H4K20me3
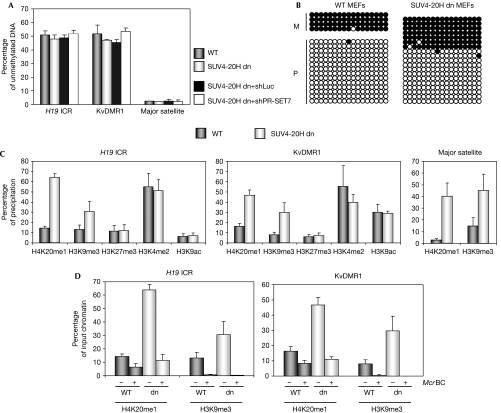
Although H4K20me3 is generally not enriched at the promoters of inactive genes (Mikkelsen et al, 2007), it is consistently associated with the methylated DNA of ICRs and pericentric heterochromatin. This raises the question of whether the maintenance of methylation of DNA could be linked to that of H4K20me3. We analysed methylation of DNA in SUV4-20H dn cells in comparison with primary (data not shown) and immortalized wild-type MEFs (Fig 4A), by purifying genomic DNA and digesting it with McrBC. Then, we quantified the non-digested fraction of the DNA by using real-time PCR amplification (Fig 4A), and observed that in both wild-type and SUV4-20H cells half of the chromosomes at the H19 and KvDMR1 ICRs were unmethylated, as expected. Furthermore, at the major satellite DNA, we detected complete digestion by McrBC, indicating that it had retained full methylation of DNA in the SUV4-20H dn cells. For KvDMR1, we performed bisulphite sequencing and found that on the methylated allele all CpG dinucleotides had remained methylated in the double knockout cells (Fig 4B). These results indicate that H4K20me3 is dispensable for the maintenance of methylation of DNA at the ICRs. Similarly, downregulation of PR-SET7, which causes a reduction in H4K20me3, did not alter methylation of DNA at the H19 and KvDMR1 ICRs (supplementary Fig 2A online). In addition, we downregulated PR-SET7 in the SUV4-20H dn MEFs to perturb further the methylation of H4K20 (supplementary Fig 3 online). Nevertheless, methylation of DNA remained unaltered at the ICRs (Fig 4A). Taken together, these data establish that H4K20me3 is dispensable for the maintenance of methylation of DNA in cultured MEFs. It is possible, however, that in combination with other histone modifications, it contributes to the faithful maintenance of methylation imprints in vivo.
Figure 4.
SUV4-20H deficiency alters methylation of H4-K20 and H3-K9, but not methylation of DNA. (A) Analysis of DNA methylation at H19 ICR and KvDMR1, in WT, SUV4-20H dn, SUV4-20H dn+shPR-SET7 and SUV4-20H dn+shLuc MEFs, respectively. Samples were digested with McrBC and analysed by qPCR. (B) Bisulphite sequencing analysis of KvDMR1 in (C57BL/6 × SDP711)F1 WT and SUV4-20H dn MEFs (in which the parental alleles cannot be distinguished by polymorphisms). Each row represents an individual chromosome. Methylated CpGs are shown as black circles and unmethylated CpGs as open circles. M, maternal; P, paternal. (C) qPCR quantification of precipitations after ChIP on SUv4-20H dn MEFs at the H19 and KvDMR1 ICRs and at the major satellites. (D) DNA obtained following precipitation of H4K20me1 and H3K9me3 was digested with (+) or without (−) McrBC. DNAs were then quantified as part of the original input chromatin, after ChIP, or after ChIP followed by digestion with McrBC. ChIP, chromatin immunoprecipitation; ICR, imprinting control region; MEF, mouse embryonic fibroblast; qPCR, quantitative PCR; WT, wild type.
Impact of methylation of H4K20 on chromatin
Next, we explored the impact of H4K20me3 depletion on other histone modifications. In SUV4-20H dn cells, we detected augmented precipitation of H4K20me1 at the H19 and KvDMR1 ICRs, and at the major satellite DNA (Fig 4C). The increase in H4K20me1 occurred entirely on the DNA-methylated allele, the allele to which SUV4-20H is recruited in wild-type cells (Fig 4D). Previously, we reported that in human cells, a global decrease in H4K20me1 is associated with a decrease in global levels of H4K20me3 on down-regulation of PR-SET7 (Tardat et al, 2007). Thus, H4K20me1 is a main substrate of SUV4-20H to promote H4K20me3, which explains the accumulation of H4K20me1 in the SUV4-20H dn MEFs (Fig 4C; supplementary Fig 3 online).
Interestingly, in SUV4-20H dn MEFs, we observed increased precipitation levels of H3K9me3 at ICRs, whereas H3K4me2 and H3K9ac remained unchanged. This higher precipitation of H3K9me3 in unfixed chromatin occurred entirely on the DNA-methylated alleles of the ICRs (Fig 4D). This was in agreement with our finding that in wild-type cells, H3K9me3 is deposited on the same parental allele as H4K20me3 (Fig 1B). At present, we are unable to explain clearly this increased precipitation of H3K9me3. One hypothesis would be that H4K20me1 is linked to H3K9me1 and that the latter is the substrate for conversion into H3K9me3. In agreement with this, it was reported that in MEFs, H4K20me1 and H3K9me1 are enriched at the same chromosomal regions and that these mono-methylations are present on the same nucleosomes (Sims et al, 2006). Conversely, the reduction in H4K20me3 could have led to a conformational change in chromatin (Benetti et al, 2007), providing a higher accessibility of the H3K9me3 antibody to its epitope. However, in the shPR-SET7 cells, a decrease in H4K20me3 correlated with reduced H3K9me3 precipitation (Fig 3), a finding that does not support this explanation. To explore this question further, we studied another H3 modification, H3K27me3, which is present on the same parental allele as H3K9me3. We did not obtain evidence for altered H3K27me3 precipitation in the SUV4-20H dn MEFs, and the same was observed for H3K4me2 and H3K9ac (Fig 4C). Nevertheless, we did not observe a clear change in the precipitation of H3K9me3 in formaldehyde crosslinked chromatin of SUV4-20H dn cells (data not shown), indicating that whatever the nature of the alteration on histone H3, this is shown best in native chromatin.
Whatever their precise inter-relationship, the observed alterations in the methylation of H4K20 and H3K9 did not affect the maintenance of differential DNA methylation at ICRs in cultured fibroblasts (Fig 4; supplementary Fig 2A online). As expected, we did not observe changes in allelic gene expression at the Igf2-H19- and Kcnq1-imprinted domains in the shPR-SET7 cells (supplementary Fig 2B online). The question remains as to whether imprinted expression is also faithfully maintained when PR-SET7 and SUV4-20H are disrupted in the in vivo context of the developing embryo.
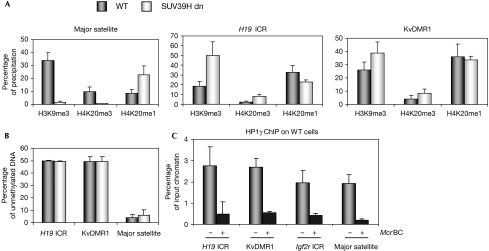
As PR-SET7 and SUV4-20H affected not only methylation of H4K20 but also the precipitation of H3K9me3, we explored which enzymes could regulate H3K9me3 at ICRs. As SUV39H1 and SUV39H2 control H3K9me3 at pericentric heterochromatin, we focused our attention on these HMTs (Peters et al, 2003). In SUV39H1 and SUV39H2 double-knockout MEFs (SUV39H dn; Lehnertz et al, 2003), we could not detect H3K9me3 and H4K20me3 at the major satellite DNA, whereas H4K20me1 was increased (Fig 5A), confirming our earlier work on heterochromatin (Schotta et al, 2004). At the H19 and KvDMR1 ICRs, by contrast, there was no reduction in H3K9me3 (or H4K20me1 or H4K20me3) in SUV39H dn MEFs (Fig 5A), as was recently reported for the Igf2r ICR (Regha et al, 2007). Methylation of DNA was also unaltered at ICRs in the SUV39H dn cells (Fig 5B). These data indicate that a yet unknown HMT controls H3K9me3 at ICRs.
Figure 5.
H3K9me3 at imprinting control regions is not controlled by SUV39H. (A) SUV39H dn cells were compared with WT MEFs by ChIP on native chromatin. At the pericentric major satellite, absence of SUV39H leads to loss of H3K9me3 and H4K20me3 and increased levels of H4K9me1. At the H19 and KvDMR1 ICRs, there is no loss of H3K9me3 (or H4K20me3) in SUV39H dn cells. (B) Unaltered methylation of DNA at the H19 and KvDMR1 ICRs. Genomic DNA of SUV39H dn and WT MEFs was digested with McrBC, followed by qPCR. (C) ChIP on crosslinked chromatin indicates precipitation of HP1γ at ICRs. Digestion with McrBC shows that precipitation occurred mostly on the DNA-methylated allele. ChIP, chromatin immunoprecipitation; HP1, heterochromatin protein 1; ICR, imprinting control region; MEF, mouse embryonic fibroblast; qPCR, quantitative PCR; WT, wild type.
What, then, dictates the allelic recruitment of SUV4-20H specifically to the DNA-methylated allele of ICRs? As this is not determined by H4K20me1, which is bi-allelic, recruitment could be achieved through the methylated DNA sequences themselves. Alternatively, it could occur through H3K9me3 and its recognition by heterochromatin protein 1 (HP1) proteins (Schotta et al, 2004; Benetti et al, 2007). In particular, we analysed Hp1γ, an HP1 protein detected by immunofluorescence at both heterochromatic and euchromatic regions (Dialynas et al, 2008). An antibody against Hp1γ gave significant precipitation at the H19 and KvDMR1 ICRs, and at the Igf2r ICR, as reported previously by Regha et al (2007). The precipitation of HP1γ at ICRs seemed preferential as it was detected mostly on the DNA-methylated allele (Fig 5C). Another non-exclusive hypothesis for the recruitment of SUV4-20H follows from the finding that deficiency in the pocket protein(s) retinoblastoma protein (pRb) disrupts H4K20me3 (Gonzalo & Blasco, 2005). pRb family members interact with many proteins that repress heterochromatin, and in particular with SUV4-20H and DNMT1 (DNA methyltransferase 1; Nielsen et al, 2001; Gonzalo & Blasco, 2005; Isaac et al, 2006). Two recently identified pRb-binding proteins (Rbbp1 and Rbbp1-like1) contribute to the maintenance of both H4K20me3 and H3K9me3 at the Snrpn ICR (Wu et al, 2006). Our study defines the first HMTs involved in methylation of histone at ICRs, and shows mechanistic similarities and differences between ICRs and heterochromatin (summarized in supplementary Fig 4 online). It is important to further unravel the regulation of methylation of histone at ICRs, a prerequisite for understanding their perturbation in human diseases and cancer. In future research, it would also be interesting to explore the extent to which these specialized sequence elements comprising (imperfect) tandem repeats contribute to the unusual organization of chromatin at ICRs.
Methods
Cell lines. SUV4-20H dn primary and immortalized MEFs were derived from Suv4-20h1/h2 double-knockout embryos (Schotta et al, 2008). SUV39H dn MEFs were derived from Suv39h1/h2 double-knockout embryos (Lehnertz et al, 2003). Both knockout MEF lines, and the corresponding control wild-type MEFs, were on a mixed genetic background of 129/SV and C57BL/6J origin.
ChIP on native and crosslinked chromatin. ChIP on native and crosslinked chromatin was performed as described previously (Umlauf et al, 2004). All ChIP experiments were performed three or more times on independent chromatin preparations. Details of the antibodies used are described in the supplementary information online.
shRNA preparation and infection. Wild-type MEFs were derived from fetuses that were (C57BL/6 × M. spretus)F1 for mouse distal chromosome 7 (SDP711 mice; Wagschal et al, 2008). A PR-SET7 shRNA was designed to target only PR-SET7 mRNA. A firefly luciferase nonspecific control shRNA was also used (supplementary Table 2 online). The shRNA sequences were introduced into the retroviral vector RNA-mediated interference Ready pSiren (BD BioSciences, Le Pont-de-Claix, France). Cells were infected with the corresponding retroviral particles, selection with puromycin (10 μg/ml) was started 48 h later, and the cells were collected 5 days after infection. PR-SET7 SR constructs were prepared by introducing silent mutations into PR-Set7 cDNA (Tardat et al, 2007). For stable expression of PR-SET7 SR, the cells were infected and selected with hygromycin B (100 μg/ml). Western blotting was as described previously (Tardat et al, 2007) and antisera are described in the supplementary information online.
Methylation of DNA and gene expression. Genomic DNA was isolated from cells, digested with McrBC and then analysed by quantitative PCR amplification together with undigested DNA, as described in Wagschal et al (2008). Bisulphite sequencing was performed as described previously (Wagschal et al, 2008). Oligonucleotides used are given in supplementary Table 1 online. For allelic gene expression analysis, we applied single-nucleotide and restriction fragment length polymorphisms (supplementary Table 2 online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge grant support from the Association pour la Recherche sur le Cancer (to R.F., C.S. and E.J.), the Institut National du Cancer (to R.F., C.S. and E.J.), the Agence National de la Recherche (to R.F.), the Association for International Cancer Research (to R.F. and C.S.) and the Cancéropôle Grand Sud-Ouest (to R.F.). T.J. and R.F. thank the 6th Framework NoE EPIGENOME for support and encouragement.
Footnotes
The authors declare that they have no conflict of interest.
References
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA (2007) Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K, Feil R (2004) Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 14: 188–195 [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R (2007) Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26: 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas GK et al. (2008) Plasticity of HP1 proteins in mammalian cells. J Cell Sci 120: 3415–3424 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins M (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Blasco MA (2005) Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 4: 752–755 [DOI] [PubMed] [Google Scholar]
- Isaac CE et al. (2006) The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol 26: 3659–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S et al. (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B et al. (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lin SP et al. (2003) Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1–Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 35: 97–102 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21: 457–465 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Peters AH et al. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Regha K et al. (2007) Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell 27: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G et al. (2008) A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs AD, Razin A (1997) Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94: 10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Houston SI, Magazinnik T, Rice JC (2006) A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem 281: 12760–12766 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR-Set7 dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12: 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D et al. (2004) Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Wagschal A et al. (2008) G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol 28: 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Tsai TF, Beaudet AL (2006) Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev 20: 2859–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H et al. (2008) Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem 283: 12085–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information