Abstract
We demonstrate that CD161 is a highly up-regulated gene in human interleukin (IL) 17 T helper cell (Th17) clones and that all IL-17–producing cells are contained in the CD161+ fraction of CD4+ T cells present in the circulation or in inflamed tissues, although they are not CD1-restricted natural killer T cells. More importantly, we show that all IL-17–producing cells originate from CD161+ naive CD4+ T cells of umbilical cord blood, as well as of the postnatal thymus, in response to the combined activity of IL-1β and IL-23. These findings implicate CD161 as a novel surface marker for human Th17 cells and demonstrate the exclusive origin of these cells from a CD161+CD4+ T cell progenitor.
Classically, naive CD4+ T cells have been thought to differentiate into two main lineages, Th1 and Th2 cells (1, 2). Th1 cells produce the signature cytokine IFN-γ, with the cytokine IL-12 produced by DCs being critical for their differentiation by inducing the activation of STAT-4 and up-regulation of T-bet transcription factor. In contrast, Th2 cells produce IL-4, IL-5, IL-9, and IL-13, and their development is usually started by IL-4 signaling with the participation of the transcription factors STAT-6 and GATA-3 (3). More recently, a third subset of CD4+ effector T cells that produce IL-17, named Th17, has been described in mice (4, 5). These cells express a transcription factor different from those of Th1 and Th2 cells, the retinoic acid–related orphan receptor γt (RORγt) (6). Human Th17 cells were found to be different from mouse Th17 cells not only with regard to some phenotypic and functional features, but mainly because of their different mechanisms of development (7, 8). Several papers agree that mouse Th17 cells originate from naive CD4+ T cells in the presence of IL-6 and TGF-β, and their development is then stabilized and/or amplified by IL-23 and IL-21 (9–11). In contrast, human Th17 cells seem to originate in response to the combined activity of IL-1β and IL-6 (12), or the activity of IL-1β or IL-23 alone (13), whereas the combined activity of IL-1β and IL-23 was not shown to have any additive or synergistic effect (13). Moreover, the addition of TGF-β to human naive or memory CD4+ T cells was found to inhibit the development of Th17 cells (12–14). In another study, it was reported that IL-1β and IL-6 up-regulated RORγt expression but did not induce Th17 differentiation in human adult naive CD4+ T cells, whereas IL-23 was a powerful up-regulator of its own receptor and was an important inducer of IL-17 and IL-22 (15). The discrepancy among results between humans and mice, and among different human studies, with regard to the development of Th17 cells has been attributed to the difficulty to ensure a truly naive cell population in humans (16). More recently, the combined activity of IL-1α or IL-1β and IL-23 was found to be required for the enhancement of IL-17–producing human memory T cells; however, in the same study, the differentiation of human naive T cells into Th17 cells could not be achieved (17). In a previous paper, we showed that in addition to human memory T cells producing IL-17 alone (Th17), there are several T cells in both blood and tissues that coproduce IL-17 and IFN-γ (Th17/Th1) (8). We also found that these two cell types express both RORγt and T-bet and that Th17 cells could be shifted to Th1 by the addition of IL-12, an effect that was partially antagonized by IL-23, suggesting a flexibility of human Th17 cells and their possible common developmental origin with Th1 cells (8). Furthermore, we and others identified IL-23R and CCR6 as surface molecules expressed by human Th17 cells (7, 8).
In this study, by using a microarray assay we found that CD161 was one of the most up-regulated genes in human Th17 clones compared with Th1 or Th2 clones. Accordingly, T blasts from all Th17 clones expressed CD161 on their surface, whereas all Th1 or Th2 clones examined were CD161−. All IL-17–producing cells were found to be included within the CD161+ fraction of adult circulating CD4+ T cells; they were not CD1d-restricted natural killer T (NKT) cells, but NKT-like cells. When CD161+ or CD161− cells were sorted from umbilical cord blood (UCB)–naive CD4+ T cells or from single-positive CD4+CD8− thymocytes and activated in the presence of IL-1β plus IL-23, Th17, Th17/Th1, or Th1, cells developed from the CD161+ fraction, whereas CD161− cells could never been induced to differentiate into IL-17–producing cells. In contrast, in the presence of IL-12, both CD161+ and CD161− cells only differentiated into Th1 cells. These findings indicate that CD161 is a novel surface marker for human IL-17–producing cells and demonstrate that human Th17 and Th17/Th1 cells exclusively originate from an NKT-like CD161+CD4+ T cell precursor present in the UCB, in the presence of IL-1β and IL-23 as polarizing cytokines.
RESULTS
Human Th17, but neither Th1 nor Th2, clones express CD161 on their surface
In a previous study, we generated a series of Th1, Th2, and Th17 clones from human circulating CD4+ T cells (8). We performed a microarray analysis to identify possible differences in gene expression by Th17 clones compared with Th1 or Th2 clones. Among genes that fulfilled the criteria described in Materials and methods as up- or down-regulated genes in Th17 versus Th1 or Th2 clones, in addition to those expected on the basis of previous knowledge, such as IL-17, IL-26, IL-23R, RORγt, and CCR6 (7, 8, 13), there was also CD161 (Fig. 1 A). All genes showing a log2 fold increase or decrease of ≥2.5 in Th17 versus Th1 and Th2 clones are shown in Tables S1–S4 (available at http://www.jem.org/cgi/content/full/jem.20080397/DC1). Therefore, CD161 expression was analyzed by either quantitative PCR or flow cytometry on a series of human Th1, Th2, or Th17 clones derived from healthy subjects and different from those used for the microarray assay. As shown in Fig. 1 B, Th17 clones expressed higher levels of CD161 mRNA than Th1 and Th2 clones, and virtually all of their T blasts expressed CD161, whereas T blasts from all Th1 and Th2 clones examined did not (Fig. 1 C). CD161 expression by resting Th17 clones was not affected by anti-CD3 plus anti-CD28 mAb stimulation in the presence of IL-2, and it was not induced on Th1 and Th2 clones (Fig. S1).
Figure 1.
Human Th17 clones express the NKT cell marker CD161. (A) Volcano plot representation (log2 fold change vs. t test p-value) between Th17 and Th1 (left) or Th2 (right) cell gene expression profiles. (B) CD161 mRNA levels were measured by real-time quantitative RT-PCR in Th1, Th2, and Th17 clones. Bars represent mean values ± SE obtained in 10 clones from each type different from those used for the microarray assay. (C) Representative flow cytometric analysis of CD161 surface expression for each type of clone. The gates were placed on the basis of an isotype-matched control mAb. Percentages of gated cells are shown.
All circulating Th17 cells from adult healthy subjects and Th17 cells from inflamed tissues are contained in the CD161+ fraction
We then asked whether the CD161 expression observed in Th17 clones was also shared by freshly derived circulating Th17 cells. To this end, CD4+ T cells purified from adult PBMCs of healthy subjects were sorted into CD161+ and CD161− cells, and the sorted populations were assessed by flow cytometry for their phenotypic features, as well as for their ability to produce IFN-γ or IL-17 in response to PMA plus ionomycin stimulation. Virtually all CD161+ cells were contained within the CD45RA−CD45RO+CD31− population (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20080397/DC1), suggesting their nature as memory T cells. Moreover, the cells of the CD161+ fraction produced IFN-γ, IL-17, or both, whereas IL-17–producing cells were virtually absent in the CD161− fraction (Fig. 2 A). Using the same technique, the assessment of the production of IL-21 and IL-22 showed that both CD161− and CD161+ cells were able to produce IL-21, whereas the ability to produce IL-22 was virtually restricted to CD161+ cells (Fig. S3). The CD161+ and CD161− fractions were also tested for RORγt, IL-23R, and CCR6 mRNA expression. CD161+ T cells expressed higher levels of mRNA for RORγt, IL-23R, and CCR6 than CD161− cells (Fig. 2 A). Because human Th17 cells have been shown to express CCR6 (7, 8), the association between CCR6 and CD161 was assessed. As shown in Fig. 2 B, the majority of CD161+ T cells also expressed CCR6, and when CD161+ cells were sorted into CCR6− and CCR6+ cells, the great majority of IL-17–producing cells appeared to be included in the CCR6+ fraction. In contrast, the CD161− fraction also contained CCR6+ cells, but they were not capable of producing IL-17, whereas they produced IFN-γ (Fig. 2 B). Accordingly, RORγt and IL-23R mRNA expression appeared to be restricted to the CD161+CCR6+ cell fraction (Fig. 2 B).
Figure 2.
Circulating IL-17–producing cells are contained in the CD161+CCR6+ fraction of circulating CD4+ T cells from adult subjects. (A) Detection of IL-17 and IFN-γ production and expression of RORγt, IL-23R, and CCR6 mRNA in CD161-depleted (black plot and bars) and CD161-enriched (red plot and bars) circulating CD4+ T cells from healthy adult subjects. Representative flow cytometric analysis (top) and mean values of cytokine-producing cell percentages ± SE obtained in seven different donors (bottom) are shown. (B) IL-17 and IFN-γ production in circulating CD161−CCR6−, CD161−CCR6+, CD161+CCR6−, and CD161+CCR6+ CD4+ T cells from one representative experiment out of three performed is shown. RORγt and IL-23R mRNA expression in the indicated cell populations is shown. Bars represent mean values ± SE obtained in three separate experiments. Percentages of gated cells are shown.
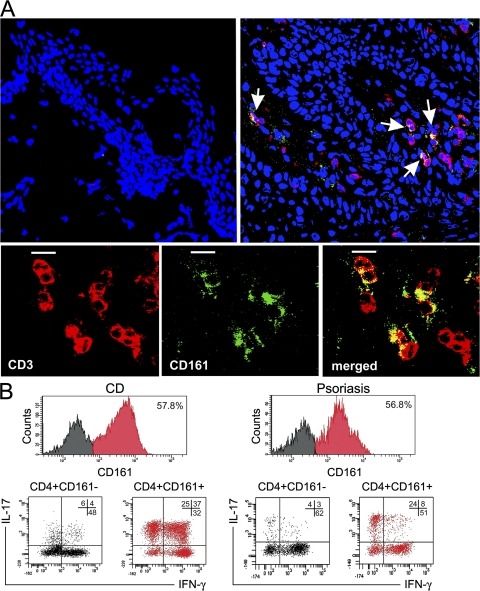
To provide evidence of the presence of CD161+ T cells, even in the inflamed tissues where Th17 cells are suspected to play a pathogenic role (18, 19), analysis by confocal microscopy on biopsy specimens of skin from psoriatic patients was also performed. As shown in Fig. 3 A, high percentages of T (CD3+) cells infiltrating the skin of psoriatic patients also coexpressed CD161+, whereas virtually no T cells were observed in control skin specimens of healthy subjects. To further support the results of the confocal microscopy analysis, infiltrating cells were recovered from both the inflamed gut of subjects with Crohn's disease (CD) and the skin of subjects suffering from psoriasis, and expanded in vitro for 7 d with anti-CD3 plus anti-CD28 mAb. Cultured cells were then stimulated with PMA plus ionomycin and examined for the expression of CD161 and for their ability to produce IFN-γ or IL-17. Virtually all Th17 and Th17/Th1 cells were present in the CD3+CD8−CD161+-gated T cells of both the gut from subjects with CD and the skin from subjects with psoriasis (Fig. 3 B).
Figure 3.
Detection of CD161+ T cells in inflamed tissues and demonstration of their ability to produce IL-17. (A) Evaluation of the expression of CD3 (red) and CD161 (green) in skin from a healthy donor (top left) or in lesional skin from a psoriatic patient (top right) by confocal microscopy. Arrows point to cells showing double labeling for CD3 and CD161 (yellow) in the skin of the psoriatic patient. TO-PRO-3 (Invitrogen) counterstained nuclei. (bottom) A close-up of CD3+CD161+ double-positive cells (merged; yellow) is shown. Images obtained in the skin of one out of three healthy or psoriatic donors are depicted. Bars, 10 μm. (B) Infiltrating T cells recovered from gut areas of subjects with CD or skin biopsies of subjects with psoriasis were expanded in vitro for 1 wk and assessed for CD161 expression, as well as for their ability to produce IFN-γ and IL-17 after stimulation with PMA plus ionomycin. Representative flow cytometric analysis obtained in one out of three subjects with CD and in one out of three subjects with psoriasis are shown. Percentages of gated cells are shown.
Circulating CD161+ Th17 cells are not NKT cells, but NKT-like cells
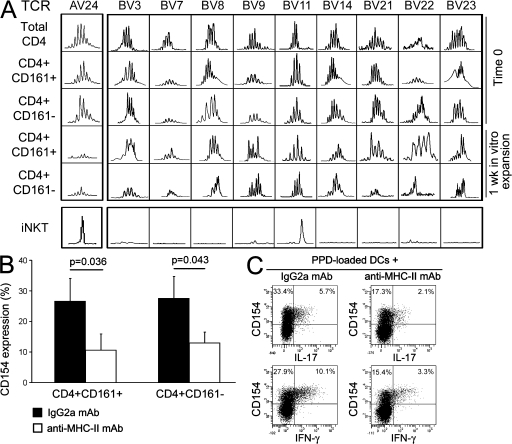
Because CD161 has been reported to be expressed by different cell types, including type I or II NKT cells or NKT-like cells (either CD4+ or CD8+ T cells; reference 20), we asked whether CD161+ Th17 cells belonged to the NKT or the NKT-like T cell population. Circulating CD161+CD4+ T cells containing IL-17–producing cells were not type I (or classical) invariant NKT (iNKT) cells because they had a polyclonal Vα24 TCR chain and exhibited a broad TCR Vβ repertoire, as shown by spectratyping analysis of both freshly isolated and 1-wk in vitro–expanded cells (Fig. 4 A). In addition, the presence of a broad TCR Vβ repertoire also excluded the possibility that these cells belong to the type II (nonclassical) NKT cells. Moreover, the response of purified CD161+CD4+ circulating T cells to allogeneic stimulation, as assessed by CD154 expression, was comparable to that of the CD4+CD161− cell fraction, and, more importantly it could be inhibited by the addition in culture of a blocking anti–MHC class II mAb (Fig. 4 B). Furthermore, the expression of IFN-γ and IL-17 in a purified protein derivative (PPD)–specific short-term CD4+ T cell line stimulated with PPD-loaded autologous DCs was equally inhibited by an anti–MHC class II mAb, as assessed by flow cytometry (Fig. 4 C).
Figure 4.
Circulating CD4+CD161+ Th17 cells are not NKT, but NKT-like, cells. (A) The TCR Vα24 and TCR Vβ repertoires were determined by CDR3 length analysis evaluated by spectratyping. The TCR Vα24 and TCR Vβ repertoires were assessed on freshly isolated (total CD4+, CD4+CD161+, and CD4+CD161− T cells) and on 1-wk in vitro–expanded CD4+CD161+ and CD4+CD161− T cells. Representative families of different Vβ chains assessed (BV3, BV7, BV8, BV9, BV11, BV14, BV21, BV22, and BV23) are depicted. One representative out of three different experiments is shown. CDR3 length analysis of Vα24 and Vβ11 in iNKT cells is also shown. (B) CD4+CD161+ and CD4+CD161− T cells were stimulated for 8 h with allogeneic DCs in the presence of an anti–MHC class II or an isotype control mAb, and were then assessed for CD154 expression. Bars represent mean values ± SE of percentages of CD154+ cells obtained in four different experiments. (C) A PPD-specific short-term T cell line highly enriched in IL-17–producing cells was stimulated for 8 h with PPD-loaded autologous DCs in the presence of an anti–MHC class II or an isotype control mAb. CD154 and cytokine expression were assessed by flow cytometry. Percentages of gated cells are shown.
IL-17–producing cells develop from UCB-naive T cells only in the presence of IL-1β plus IL-23
To identify conditions permissive for the differentiation of human naive T cells into IL-17–producing cells, we first stimulated purified CD45RA+RO− cells obtained from CD4+ T cells of adult subjects for 7 d with anti-CD3 and anti-CD28 mAbs, in the absence or presence of single cytokines (IL-1β, IL-6, IL-21, IL-23, and TGF-β) or of combinations of them. We used flow cytometry to analyze the production of IL-17 after stimulation with PMA plus ionomycin. Under any of the experimental conditions used, cells producing IL-17 could never be observed (unpublished data).
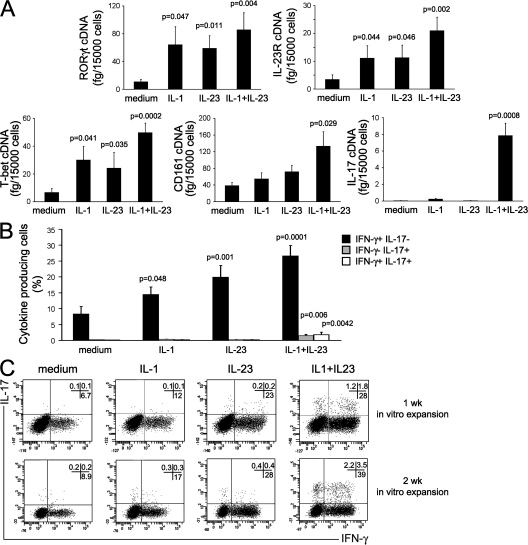
UCB CD4+ T cells were then used as source of naive T cells. These cells were cultured under the same conditions described in the previous paragraph, and their capacity to express mRNA for RORγt, T-bet, IL-23R, CD161, and IL-17 was assessed. Either IL-1β or IL-23 significantly up-regulated the expression of mRNA for RORγt, T-bet, and IL-23R, but not of CD161 or IL-17 (Fig. 5 A). When IL-1β and IL-23 were added together, there was a further up-regulation of T-bet, RORγt, and IL-23R, as well as of CD161, mRNA expression in comparison with cultures containing IL-1β or IL-23 alone; in addition, IL-17 mRNA expression became clearly detectable (Fig. 5 A). In contrast, none of the other cytokines added to the cultures, alone or in different combinations, was able to increase RORγt, IL-23R, IL-17, or CD161 mRNA expression (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20080397/DC1). Intracellular expression of IFN-γ and IL-17 was also evaluated on the same CD4+ T cell cultures by flow cytometry after stimulation with PMA plus ionomycin. Virtually no cells producing IL-17 and only a few IFN-γ–producing cells were observed in the absence of cytokines. The addition of either IL-1β or IL-23, or both, induced a significant increase in the proportion of CD4+ T cells able to produce IFN-γ (Fig. 5, B and C), but only the combination of both IL-1β and IL-23 enabled UCB CD4+ T cells to produce IL-17, partly in association with IFN-γ (Fig. 5, B and C).
Figure 5.
IL-17–producing cells can develop from UCB T cells in the presence of IL-1β plus IL-23. CD4+ T cells purified from UCB were stimulated for 7 d with anti-CD3 plus anti-CD28 mAb in the presence or absence of the indicated cytokines. (A) RORγt, T-bet, IL-23R, CD161, and IL-17 mRNA levels were measured by real-time quantitative RT-PCR. Bars represent mean values ± SE obtained from 13 donors. (B) Cells were also assessed by flow cytometry for their ability to produce IFN-γ alone (black bars), IL-17 alone (gray bars), or both (white bars) after stimulation with PMA plus ionomycin. Bars represent mean values ± SE obtained from 13 donors. (C) Representative flow cytometric analysis on UCB CD4+ T cells after stimulation with anti-CD3 plus anti-CD28 mAb, in the presence or absence of the indicated cytokines, performed after 1 wk (top) or 2 wk (bottom) of in vitro expansion. p-values refer to the comparison between cultures performed in the presence of the indicated cytokines versus those performed in the presence of medium alone. Percentages of gated cells are shown.
IL-17–producing cells exclusively originate from a CD161+ naive CD4+ T cell precursor
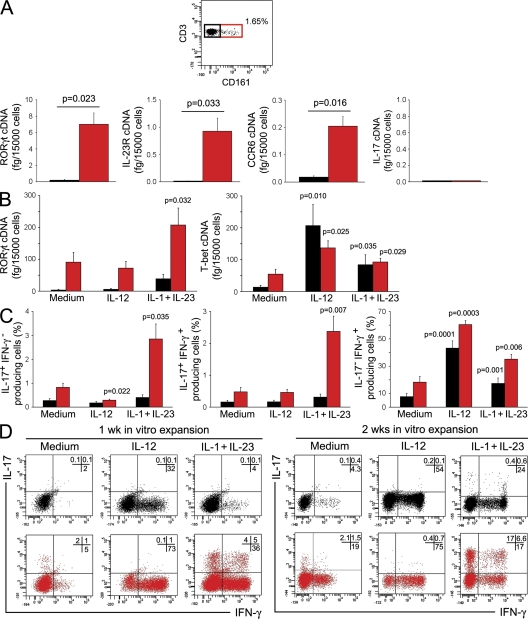
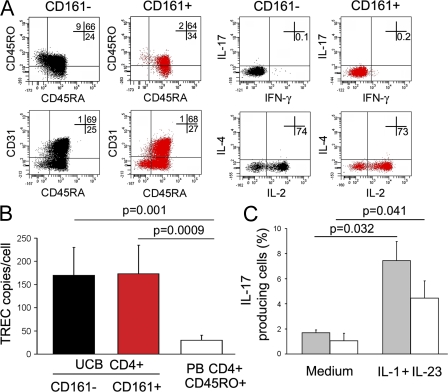
To investigate whether Th17 cells originated from a naive CD161+ precursor, UCB CD4+ T cells were first assessed for the expression of CD161. As shown in Fig. 6 A, flow cytometric analysis of UCB mononuclear cells (MNCs) showed the presence of a small but clearly detectable (0.76 ± 0.1% SE; n = 18) CD161+ cell population among CD4+ T cells. Therefore, UCB CD4+ T cells were sorted into CD161− and CD161+ cells. The purity of sorted populations was consistently >95%, as shown in Fig. S5 (available at http://www.jem.org/cgi/content/full/jem.20080397/DC1). The two sorted cell populations were then assessed for the expression of RORγt, IL-23R, CCR6, and IL-17 mRNA. As shown in Fig. 6 A, only CD161+ cells showed high levels of mRNA for RORγt, IL-23R, and CCR6, suggesting the constitutive expression of these molecules in the CD161+, but not in the CD161−, fraction. However, neither CD161+ nor CD161− cells expressed the IL-17 transcript (Fig. 6 A). Therefore, sorted CD161+ and CD161− UCB CD4+ T cells were stimulated for 7 d with anti-CD3 plus anti-CD28 mAb in presence of IL-1β plus IL-23, or of IL-12 alone, and the expression of RORγt and T-bet, as well as the type of cytokine produced, was assessed. The addition of IL-1β plus IL-23 resulted in the up-regulation of T-bet and RORγt, as well as the appearance of Th1 cells, in both fractions (Fig. 6, B and C), but the development of IL-17–producing cells was only observed in the CD161+ fraction (Fig. 6 C). In contrast, the addition of IL-12 induced in either CD161+ or CD161− CD4+ T cells the expression of high levels of T-bet, but not of RORγt (Fig. 6 B), and induced both cell fractions to differentiate into Th1 cells (Fig. 6 C). One representative flow cytometric picture is shown in Fig. 6 D (left). When CD161+ and CD161− naive CD4+ T cells were restimulated for an additional 7 d with anti-CD3 plus anti-CD28 mAb in the presence or absence of the polarizing cytokines, the proportion of both Th17 and Th17/Th1 cells was largely increased (Fig. 6 D, right). As already observed for total UCB CD4+ T cells (Fig. S4), neither IL-1β nor IL-23 alone, nor any other cytokine or cytokine combination, including IL-6 plus TGF-β, showed the ability to induce the appearance of IL-17–producing cells in either the CD161+ or the CD161− fraction of UCB CD4+ T cells (unpublished data).
Figure 6.
UCB CD161+CD4+ T cells constitutively express RORγt, IL-23R, and CCR6 mRNA and differentiate into Th17 cells in response to IL-1β plus IL-23. (A) Flow cytometric detection of CD161+ cells among UCB CD4+ T cells and measurement of RORγt, IL-23R, CCR6, and IL-17 mRNA levels in CD161-depleted (black bars) and CD161-enriched (red bars) unstimulated UCB CD4+ T cells. Bars represent mean values ± SE obtained in seven different donors. (B) CD4+CD161+ (red bars) and CD4+CD161− (black bars) T cells were stimulated for 7 d with anti-CD3 plus anti-CD28 mAb in the presence or absence of the indicated cytokines. RORγt and T-bet mRNA levels were measured by real-time quantitative RT-PCR. Bars represent mean values ± SE obtained from six donors. (C) The same cell fractions as in B were assessed by flow cytometry for their ability to produce IFN-γ and IL-17 after stimulation with PMA plus ionomycin. Bars represent mean values ± SE obtained from six donors. (D) Representative flow cytometric analysis of cytokine production by CD4+CD161+ (red plots) and CD4+CD161− (black plots) T cells after 1 wk (left) or 2 wk (right) of in vitro expansion. p-values refer to the comparison between cultures performed in the presence of the indicated cytokines versus those performed in the presence of medium alone. Percentages of gated cells are shown.
Finally, we asked whether the CD161+ UCB CD4+ T cells that differentiated into Th17 cells were really naive T cells or a small subset of contaminating memory T cells. To this end, freshly derived UCB CD4+ T cells were sorted into CD161+ and CD161− cells, and the expression of some phenotypic markers, as well as the ability to produce cytokines after stimulation with PMA plus ionomycin, were examined. Virtually all purified CD161+CD4+, as well as CD161−, UCB T cells expressed CD45RA (even if often in association with CD45RO) and CD31 (Fig. 7 A, left), and when stimulated with PMA plus ionomycin, they did not show the ability to produce cytokines (except IL-2; Fig. 7 A, right). Moreover, TCR excision circle (TREC) levels were examined in the two sorted populations and compared with those found in purified CD45RA−CD45RO+ adult circulating CD4+ T cells. As shown in Fig. 7 B, TREC levels were quite comparable in CD161+ and CD161− UCB CD4+ T cells and significantly higher than in purified CD45RA−CD45RO+ adult circulating CD4+ T cells. To provide additional evidence that UCB CD161+ T cells that gave origin to Th17 cells were not contaminating memory T cells, the CD161+ fraction of UCB CD4+ T cells was depleted of all CD45RO+ cells (including the double-positive CD45RA+CD45RO+ cells), and the remaining population, consisting only of CD45RA+CD45RO− cells, was stimulated with anti-CD3 plus anti-CD28 mAb in the presence of IL-1β plus IL-23. As shown in Fig. 7 C, despite the depletion of all cells expressing CD45RO, the remaining CD45RA+CD161+ UCB T cells maintained the ability to differentiate into IL-17–producing cells.
Figure 7.
UCB CD161+CD4+ T cells that differentiate into Th17 are naive T cells. (A) Representative flow cytometric analysis of CD45RA, CD45RO, and CD31 (left) and cytokine production after stimulation with PMA plus ionomycin (right) by freshly isolated CD161+ (red plots) and CD161− (black plots) UCB CD4+ T cells. Percentages of gated cells are shown. (B) TREC analysis was performed by quantitative PCR on freshly isolated CD4+CD161+ (red bar) or CD4+CD161− (black bar) UCB CD4+ T cells and on circulating CD45RA−CD45RO+ CD4+ T cells from adult donors (white bar). Bars represent mean values ± SE obtained in three different UCB or PB adult donors. (C) UCB CD4+ T cells were depleted of CD45RO+ cells (including double-positive CD45RA+CD45RO+ cells) by FACS using an anti-CD45RO–APC mAb. The fraction containing only CD45RA+CD45RO− (gray bars) and the fraction containing the remaining CD45RO+ cells (white bars) were activated with anti-CD3/CD28 mAb in the absence (medium) or presence of IL-1β plus IL-23. Bars represent mean values ± SE obtained in three different UCB donors.
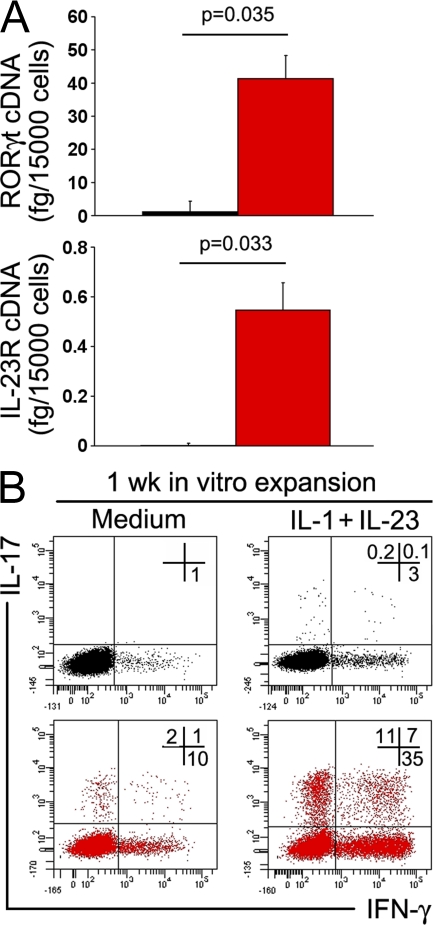
To definitively demonstrate the existence of a CD4+CD161+ precursor of human Th17 cells, experiments using T cells from postnatal human thymuses were also performed. A few (0.65 ± 0.15% SE) single-positive CD4+CD8− thymocytes were found to express CD161 and could be separated from CD161− CD4+CD8− thymocytes. As shown in Fig. 8, only purified CD4+CD161+ thymocytes constitutively expressed both RORγt and IL-23R mRNA (Fig. 8 A) and could be induced to differentiate into IL-17–producing cells after polyclonal activation in the presence of IL-1β and IL-23, whereas CD4+CD161− thymocytes could not (Fig. 8 B).
Figure 8.
CD161+ but not CD161− thymocytes constitutively express RORγt and IL-23R and can be induced to differentiate to Th17 cells. (A) RORγt and IL-23R mRNA levels detected in CD161− (black bars) and CD161+ (red bars) cells sorted from single-positive CD4+CD8− human thymocyte suspensions. Bars represent mean values + SE obtained in three different donors. (B) Representative flow cytometric analysis of IL-17 and IFN-γ production by CD4+CD161+ (red plots) and CD4+CD161− (black plots) thymocytes after 1 wk of in vitro expansion with anti-CD3/CD28 mAb in the absence (medium) or presence of IL-1β plus IL-23. Comparable results were obtained with CD161− and CD161+ thymocytes from the other two postnatal human thymuses. Percentages of gated cells are shown.
DISCUSSION
This study demonstrates that, at least in humans, IL-17–producing CD4+ T cells consistently express CD161 as an unexpected surface marker and originate from an NKT-like CD4+ T cell precursor. These observations originated from the results of a microarray analysis performed on human CD4+ T cell clones. This analysis showed that, in addition to up-regulated genes coding for molecules already known to be expressed by human Th17 cells, such as IL-17, IL-26, IL-23R, RORγt, and CCR6 (7, 8, 13), the CD161 gene was also highly up-regulated on Th17 compared with either Th1 or Th2 clones. More importantly, all IL-17–producing cells were contained within the CD161+ fraction of adult circulating CD4+ T cells. Of note, virtually all CD161+ cells were contained in the CD45RA−CD45RO+CD31− population, suggesting their nature as memory T cells. Moreover, among these cells there was a strict association between the expression of CD161 and CCR6, a chemokine receptor which has been found to be prevalently expressed by human Th17 cells (7, 8), such that virtually all IL-17–producing cells appeared to be contained in the CCR6+ fraction of the CD161+ population. Accordingly, IL-23R and RORγt mRNA expression was restricted to the CD161+CCR6+ cell fraction. The ability to produce IL-22 after stimulation was virtually restricted to the CD161+ circulating CD4+ T cells, whereas the production of IL-21 was shared by both CD161+ and CD161− circulating CD4+ T cells. The latter findings are in keeping with previous observations showing that IL-21 production is not restricted to Th17 cells (21), whereas IL-22 and IL-17 are coexpressed by Th17 cells (22).
CD161 (or NKR-P1A) is the human homologue of the mouse NK1.1 (23), which is expressed not only on almost all NK cells but also by a subset of T cells that have been named NKT cells (24, 25). The classical, type I NKT cells are α-galactosylceramide reactive and CD1d restricted, and use an invariant TCR α chain consisting of Vα14-Jα18 and a limited β chain repertoire dominated by Vβ 8.2, Vβ7, or Vβ2 in mice, and of Vα24-Jα18 with Vβ11 in humans (24, 25). The iNKT cells, at least in mice, may be CD4+ or double-negative and are able to produce large amounts of different cytokines after stimulation (24, 25). In addition to iNKT, NKT cells not bearing the Vα24-Jα18 (human) or the Vα14-Jα18 (mice) TCR, but still dependent on CD1d antigen presentation, have also been identified and named as nonclassical or type II NKT cells (20). These cells can express diverse but few (Vα3.2-Jα9 and Vα8) TCR α chains and can be identified by CD1d tetramers (26, 27). However, other cell types that express CD161 but are restricted by conventional MHC molecules instead of CD1d and, therefore, exhibit noninvariant TCRs, have been described in humans and named as NKT-like CD4+ or CD8+ T cells (20, 28). CD161+ cells were found to be a minority (about one out of five) among human circulating CD4+ T cells (29), with the great majority of them expressing CD45RO, which suggests their nature as memory T cells (24, 30). In contrast, the percentage of iNKT cells among human circulating CD4+ T cells is <1% (29), which is in contrast with the observations in mice, where the majority of circulating NK1.1+ cells are iNKT cells (24–32). The IL-17–producing CD161+CD4+ T cells described in this study were not CD1d-restricted NKT cells because they had a TCR Vα and Vβ repertoire heterogeneity incompatible with CD1d restriction, and their response to allogeneic stimulation or stimulation with a soluble antigen, such as PPD, was inhibited by blocking MHC class II. Despite the fact that human IL-17–producing cells are not iNKT but NKT-like cells, our findings may appear in disagreement with those reported in mice showing that a subset of iNKT cells produces IL-17, but curiously, whereas most mouse iNKT cells express NK1.1 (NKR-P1C), the IL-17–producing iNKT cells are NK1.1− (33, 34). However, by using the microarray assay on mouse NKT cells, production of IL-17 by both NK1.1+ and NK1.1− cells upon activation in vitro has also been reported (35).
The most intriguing observation emerging from this study was the demonstration that not only do human Th17 cells express CD161 during their life, but they can also exclusively originate from a CD161+ naive CD4+ T cell precursor. In agreement with the results previously reported by van Beelen et al. (17), we were unable to induce the development of Th17 cells from the purified CD45RA+CD45RO− (naive) circulating CD4+ T cells of adult subjects. Accordingly, virtually no CD161+ cells were found by us within the CD45RA+CD45RO− population of adult circulating CD4+ T cells. In contrast, in this study small numbers of Th17 cells originated from human UCB CD4+ T cells after polyclonal stimulation with anti-CD3/CD28 mAb in the presence of IL-1β plus IL-23. Notably, when UCB CD4+ T cells were sorted into CD161− and CD161+ cells, constitutive expression of RORγt, IL-23R, and CCR6 was found, but only in the CD161+ fraction. More importantly, Th17 cells only developed from the CD161+ fraction and their frequency could reach substantial levels (>20%) after a repeated 2-wk stimulation. Because UCB CD4+ T cells consisted not only of CD45RA+CD45RO− cells but also of double-positive CD45RA+CD45RO+ cells, whose functional nature is difficult to define, the possibility that UCB CD161+ cells give rise to IL-17–producing cells, representing the expansion of a small population of contaminating memory T cells present in the UCB as a consequence of fetal immune system stimulation during pregnancy, could be hypothesized. However, this possibility can be reasonably excluded on the basis of three main observations. First, neither CD161+ nor CD161− UCB CD4+ T cells produced any cytokine in response to stimulation with PMA plus ionomycin except IL-2, which is a functional property of naive T cells. Second, both CD161+ and CD161− cells showed comparable TREC levels, which were significantly higher than those of CD4+CD45RA−CD45RO+ (memory) adult circulating T cells. Third, when the CD161+ fraction of UCB CD4+ T cells was completely deprived of CD45RO+ cells, including those coexpressing CD45RA, the remaining CD45RA+CD45RO− population maintained the ability to differentiate into IL-17–producing cells in response to the stimulation with anti-CD3/CD28 mAb in the presence of IL-1β plus IL-23. Finally, and more importantly, completely similar results were obtained when the small population of CD161+ cells present among single-positive CD4+CD8− T cells of postnatal human thymuses was assessed. Obviously, why CD161+ UCB or thymic naive T cells can differentiate into Th17 cells, whereas purified CD45RA+RO− circulating CD4+ T cells from adult subjects do not (see Results) (17), remains unclear. However, the latter finding is in keeping with the observation reported in this paper that CD161+ cells are apparently not present in the CD45RA+CD45RO− fraction of circulating CD4+ T cells from adult subjects. Thus, if adult circulating CD45RA+CD45RO− cells contain some Th17 precursors, they are certainly present in much lower proportions than in UCB. One might speculate that Th17 responses are mainly mounted during the perinatal period. However, this would imply that for an effective Th17 response, all possible bacterial pathogens should present themselves during the perinatal period, which is very unlikely. Thus, a more likely possibility to explain why Th17 precursors are more numerous in UCB than in adult blood may be that they migrate very early into tissues, where they can differentiate into IL-17–producing cells even later in life.
The results of this study also support our previous findings suggesting that human Th1 and Th17 cells may have a close developmental relationship (8). We could indeed confirm that a substantial proportion of IL-17–producing cells, even after their development from naive CD4+ T cells, also possess the ability to produce IFN-γ (Th17/Th1), which may represent an intermediate phenotype between Th1 and Th17 cells. Moreover, we clearly showed that human naive CD4+ T cells can give origin to either Th1 or Th17 cells in the presence of IL-1β plus IL-23, with the presence of IL-12 being determinant for the Th1 choice. Therefore, the origin of human Th17 cells seems to be more similar to that initially reported in mice, when it was thought to depend on the balance between IL-12 and IL-23 production and their interaction with IL-12R or IL-23R, respectively (36).
The mechanisms linking together the expression of CD161 and the ability to develop into IL-17–producing cells in the presence of the appropriate cytokines (IL-1β plus IL-23) are still unclear. Cross-linking of CD161 by a plate-bound anti-CD161 mAb did not exert any effect on the proliferation or cytokine production by CD161+ cells (unpublished data), thus excluding that CD161 ligation may play some role in the expansion or effector functions of these cells. Another more likely explanation is that CD161, present on CD4+ T cells as well as on TCRγδ+ cells, is implicated in their transendothelial migration (37, 38). Although CD161 is expressed on a minority of human circulating CD4+ T cells (30), an abundance of CD161+ cells represented the majority of T cells from the epithelial and lamina propria layers of the human colon (39). Recently, it was found that high numbers of Th17 cells are constitutively present in different compartments of the mouse gut, and their numbers correlate with the bacterial load of the gut segments (thus being preeminent in the lamina propria of the colon), whereas low numbers were found in the spleen and liver (40). One physiological ligand for human CD161 has been identified as the lectin-like transcript 1, which belongs to the C-type lectin domain family 2 (CLEC2) (41). Another member of the CLEC2 family, CLEC2A, has been found to be selectively expressed in the skin (42), where Th17 cells migrate in the course of chronic inflammatory disorders, such as contact dermatitis, psoriasis, and atopic dermatitis (19). The results of our study show that several CD161+ cells are present among T cells infiltrating the skin of psoriatic patients and that virtually all CD4+ T cells able to produce IL-17 in the gut of subjects with CD, or in the skin of those suffering from psoriasis, were contained within the cell population expressing CD161. This finding supports the possibility that this molecule plays an important role in favoring transendothelial migration of Th17 effectors into tissues, where they are recruited in response to the CCR6-binding chemokine CCL20 (7, 8). However, it cannot be excluded, as suggested previously in this paragraph, that this molecule is also critical for the transendothelial migration of Th17 precursors early in life. In conclusion, we have identified CD161 as one of the most important surface markers of human IL-17–producing cells, and we have demonstrated that, at least in humans, IL-17–producing cells can only originate from a naive CD161+CD4+ T cell precursor.
MATERIALS AND METHODS
Subjects.
UCB samples were obtained from 23 donors. Small bowel specimens were obtained from disease-affected gut areas of three subjects with CD, and skin biopsy specimens were obtained from the lesional skin of three patients affected by psoriasis. Normal skin, taken from the abdominal skin of three healthy volunteers who underwent minor surgery (in whom no cutaneous or systemic inflammatory diseases were diagnosed), served as a control. Peripheral blood (PB) samples were obtained from seven healthy volunteers. Three postnatal thymuses were obtained from children (aged between 3 and 15 d) subjected to cardiac surgery. The procedures and all of the experiments of the study were in accordance with the ethical standards of and approved by the Regional Committee on Human Experimentation.
Reagents.
RPMI 1640 medium (Seromed) was used, supplemented with 2 mM l-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, and 2 × 10−5 M 2-ME (all from Invitrogen), and 10% FCS (HyClone; Invitrogen). Unlabeled or fluorochrome-conjugated anti-CD3, -CD4, -CD8, -CD14, -CD16, -CD19, -CD28, -CD31, -CD34, -CD45RA, -CD45RO, -CD56, -CD154, -CD161, -CCR6, –glycophorin A/B, -TCRγδ, –MHC class II (clone Tu39, mouse IgG2a; BD Biosciences), –IFN-γ, –IL-2, –IL-4, and isotype-matched control mAbs were purchased from BD Biosciences. The fluorochrome-conjugated anti–IL-17, –IL-21 and –IL-22 mAbs were obtained from eBioscience. PMA, ionomycin, and brefeldin A were purchased from Sigma-Aldrich. The cytokines IL-1β, IL-6, IL-12, IL-23, and TGF-β were purchased from R&D Systems. IL-21 was purchased from Invitrogen.
CD4+ T cell recovery and expansion.
MNC suspensions were obtained from UCB and PB by centrifugation on Ficoll-Hypaque gradient. Isolation of CD4+ T cells from UCB or PBMCs was performed by using the CD4 isolation kit II (Miltenyi Biotec). For isolation of human single-positive CD4+CD8− thymocytes, thymic tissue fragments were mechanically disrupted with a MEDI machine (BD Biosciences). MNCs were separated from the other cells by Ficoll-Hypaque gradient centrifugation. Thymic MNC suspensions were incubated for 20 min with anti-CD8, -CD14, -CD16, -CD19, -CD34, -CD56, -TCRγδ, and –glycophorin A/B mAbs and extensively washed, and then incubated for an additional 20 min with goat anti–mouse microbead-conjugated polyclonal antibody (Miltenyi Biotec). After washing, cells were separated on a CS+ column. Purified UCB-, PB- and thymus-derived CD4+ cells were further divided into CD161+ and CD161− cells by an anti-CD161–PE or –allophycocyanin (APC) mAb, followed by an anti-PE or -APC microbead mAb (Miltenyi Biotec). Purified UCB, PB, and thymus-derived CD4+CD161+ and CD4+CD161− T cells were (a) analyzed by flow cytometry for surface marker expression, (b) stimulated for intracellular cytokine detection, (c) analyzed by quantitative PCR for gene expression, and (d) stimulated for 7 d with 5 μg/ml anti-CD3 plus 5 μg/ml anti-CD28 mAbs in the absence or presence of 10 ng/ml IL-1β, 2 ng/ml IL-6, 2 ng/ml IL-12, 20 ng/ml IL-23, 5 ng/ml TGF-β, or 50 ng/ml IL-21, or combinations of them. On day 7, T cells were analyzed by quantitative PCR for gene expression and stimulated for intracellular cytokine detection. UCB CD4+CD161+ and CD4+CD161− T cell lines were restimulated under the same conditions described for an additional 7 d and were then analyzed by flow cytometry for intracellular cytokine production. In an additional experiment, PB CD4+CD161+ and CD4+CD161− T cell populations were further divided in CCR6+ and CCR6− cell fractions by a FACSAria (BD Biosciences); each purified T cell population was analyzed by quantitative PCR for IL-23R and RORγt mRNA expression and by flow cytometry for intracellular cytokine production after PMA plus ionomycin stimulation. In additional experiments, PB CD4+ and UCB CD4+CD161+ T cell populations were further purified in the CD45RA+CD45RO− cell fraction by using CD45RO microbead (MACS; Miltenyi Biotec) or CD45RO-APC (FACSAria) mAbs. For isolation of T lymphocytes from the gut and skin biopsies, tissues fragments were mechanically disrupted with a MEDI machine. Cell suspensions were stimulated for 7 d with anti-CD3 plus anti-CD28 mAbs in the presence of 20 IU/ml IL-2. On day 7, T cells were analyzed by flow cytometry for intracellular cytokine production. CD4+CD45RA−CD45RO+ memory T cells used for TREC analysis were obtained by using the CD4 isolation kit II, and CD45RA depletion was performed by using an anti-CD45RA microbead mAb (Miltenyi Biotec). T cell clones were generated from the PBMCs of adult healthy donors and categorized into Th1, Th2, and Th17 subsets, as reported previously (8). In brief, CD4+ cells were seeded under limiting-dilution conditions (0.5 cells per well) in round-bottom microwell plates (Nunc) containing 105 irradiated (6,000 rad) allogeneic PBMCs as feeder cells, 1% PHA (vol/vol), and 50 U/ml rIL-2. Growing microcultures were then supplemented at weekly intervals with 50 U/ml IL-2 and 105 irradiated allogeneic PBMCs as feeder cells. Recovered CD4+ T cell clones were classified into Th1, Th2, or Th17 subsets on the basis of their ability to produce IFN-γ, IL-4, or IL-17, respectively, as assessed by single-cell flow cytometry after polyclonal stimulation.
iNKT cell culture.
PBMCs were expanded in the presence of 100 U/ml rIL-2 and 100 ng/ml α-galactosylceramide (ALEXIS Biochemicals) for 2 wk. The cells were then sorted using anti-Vβ11 and -Vα24 mAbs (Beckman Coulter).
Intracellular cytokine and CD154 detection assay.
Intracellular staining for IFN-γ, IL-17, IL-4, and IL-2 was performed as previously described (8). 105 CD4+CD161+ and CD4+CD161− cells were cultured with or without 0.5 × 105 irradiated (9,000 rad) allogeneic DCs in the presence of 5 μg/ml anti–MHC class II (clone Tu39, mouse IgG2a) mAb or 5 μg/ml of isotype control mAbs for 8 h, with the last 6 h in the presence of brefeldin A. CD154 detection was performed according to a previously described technique (43). In one experiment, a PPD-specific short-term T cell line, obtained as described previously (44), was stimulated for 8 h (the last 6 h in the presence of brefeldin A) with PPD-loaded autologous DCs in the presence of an anti–MHC class II or an isotype control mAb. Cells were stained for CD3, CD4, CD154, intracellular IFN-γ, and IL-17, and analyzed by flow cytometry. PPD was obtained from Novartis.
Confocal microscopy.
Detection of CD161 on T cells present in biopsy specimens of normal skin or in lesional skin from psoriatic patients was performed by confocal microscopy on 5-μm sections of frozen skin tissues by using a laser confocal microscope (LSM 510META; Carl Zeiss, Inc.), as previously described (8). For this staining, unlabeled anti-CD3 mAb (clone UCHT1) was obtained from BD Biosciences, and unlabeled anti-CD161 mAb was obtained from Miltenyi Biotec.
RNA isolation, cDNA synthesis, and real-time quantitative RT-PCR.
Total RNA was extracted by using the RNeasy Micro Kit (QIAGEN) and treated with DNase I to eliminate possible genomic DNA contamination. TaqMan RT-PCR was performed, as previously described (8). Primers and probes used were purchased from Applied Biosystems.
Microarray.
Gene expression profiles on human Th1, Th2, and Th17 clones were assessed by cDNA microarray technique using the Human Genome Survey Microarray (Applied Biosystems). In brief, RNA from different samples was amplified and labeled with Digoxigenin-UTP (DIG-UTP; Applied Biosystems). 10 μg of DIG-labeled cRNA was hybridized to the Human Genome Survey Microarray and read using the 1700 Chemiluminescent Microarray Analyzer (Applied Biosystems). Expression Array System software (Applied Biosystems) was used to analyze the microarrays images. Only microarrays showing a normalized signal intensity >5,000 and a median background <600 were analyzed and normalized using Spotfire and Intergomics software (Spotfire Inc.). Class comparisons expressed as Benjamini-Hochberg false discovery rates were done using parametric tests (LIMMA) after log transformation. Each sample was analyzed three times. Microarray data are available from the Gene Expression Omnibus database under accession no. GSE11553.
TCR analysis.
The TCR repertoire was studied by CDR3 length analysis, a well-established PCR-based technique we refer to as spectratyping or immunoscope (45, 46). cDNAs recovered from the different samples and culture conditions described were amplified under nonsaturating PCR conditions with TCR-BV or TCR-AV family-specific primers (45, 46). Each reaction contained a β-actin–specific primer pair producing a 6-FAM–labeled 230-bp product as an internal control (47). The amount of template cDNA for each sample was assessed by preliminary titration amplifying for total TCR message (46, 47). Each different PCR product was run in a fluorescence-based DNA sequencer (ABI 377; Applied Biosystems) with Rox-labeled size markers (Applied Biosystems), as previously described (46, 47). The data were analyzed using Genescan software (Applied Biosystems) to assign size and peak areas to the different PCR products.
TREC analysis.
TRECs were assessed using a procedure that was previously described (48, 49). DNA was extracted by the different cell populations using a RepliQ-mini kit (QIAGEN). The PCR amplification was performed using an ABI 7500 (Applied Biosystems), and primer cycling and conditions have been previously reported (48, 49). As a standard to calculate the number of copies, we used a cloned Sj TREC fragment inserted in the Hind III site of a pUC-19 vector. The number of Sj copies present in a given cell population was calculated by diluting this standard in each PCR experiment. As a control for memory T cells, we used DNA of CD4+CD45RA-CD45RO+ T cells sorted from the PBMCs of a normal donor.
Statistics.
A standard two-tailed paired t test was used for statistical analysis. P ≤ 0.05 was considered significant.
Online supplemental material.
Fig. S1 shows that CD161 expression by Th17 clones is not affected by in vitro polyclonal activation. Fig. S2 provides an analysis of CD45RA, CD45RO, and CD31 in sorted CD161+ and CD161− circulating CD4+ T cells from adult subjects. Fig. S3 shows the production of IL-21 and IL-22 by CD161− and CD161+ cell fractions from circulating CD4+ T cells of adult subjects. Fig. S4 shows the expression of mRNA for RORγt, IL-23R, IL-17, and CD161 by UCB CD4+ T cells stimulated in the presence of different cytokines or cytokine combinations. Fig. S5 shows the CD161+ expression by sorted CD161+ and CD161− cell fractions. Tables S1–S4 show microarray data of T cell clones. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080397/DC1.
Supplementary Material
Acknowledgments
We thank Chiara Romagnani for critical reading and comments.
This work was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (grant PRIN 2005 prot.2005064784_004), the Italian Ministry of Health, the Italian Spatial Agency (MoMa Project), the FP6 European Union project SENS.IT.IV (grant LSHB-CT 2006-018861), INNOCHEM (grant FP6-LSHB-CT2005-518167), Legge 5 Regione Campania (2005), the Associazione Italiana Ricerca sul Cancro, and Ente Cassa di Risparmio di Firenze.
The authors have no conflicting financial interests.
Abbreviations used: APC, allophycocyanin; CD, Crohn's disease; CLEC2, C-type lectin domain family 2; iNKT, invariant NKT; MNC, mononuclear cell; NKT, natural killer T; PB, peripheral blood; PPD, purified protein derivative; RORγt, retinoic acid–related orphan receptor γt; TREC, TCR excision circle; UCB, umbilical cord blood.
L. Cosmi and R. De Palma contributed equally to this paper.
References
- 1.Abbas, A.K., K.M. Murphy, and A. Sher. 1996. Functional diversity of helper T lymphocytes. Nature. 383:787–793. [DOI] [PubMed] [Google Scholar]
- 2.Romagnani, S. 1997. The Th1/Th2 paradigm. Immunol. Today. 18:263–266. [DOI] [PubMed] [Google Scholar]
- 3.Rengarajan, J., S.J. Szabo, and L.H. Glimcher. 2000. Transcriptional regulation of Th1/Th2 polarization. Immunol. Today. 21:479–483. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal, S., N. Ghilardi, M.H. Xie, F.J. de Sauvage, and A.L. Gurney. 2003. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 278:1910–1914. [DOI] [PubMed] [Google Scholar]
- 5.Harrington, L.E., R.D. Hatton, P.R. Mangan, H. Turner, T.L. Murphy, K.M. Murphy, and C.T. Weaver. 2005. Interleukin 17-producing CD4+ effector T cells develop via lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov, I.I., B.S. McKenzie, L. Zhou, C.E. Tadokoro, A. Lepelley, J.J. Lafaille, D.J. Cua, and D.R. Littman. 2006. The orphan nuclear receptor RORγt directs the differentiation program of pro-inflammatory IL-17+ T helper cells. Cell. 126:1121–1131. [DOI] [PubMed] [Google Scholar]
- 7.Acosta-Rodriguez, E.V., L. Rivino, J. Geginat, D. Jarrossay, M. Gattorno, A. Lanzavecchia, and F. Sallusto. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646. [DOI] [PubMed] [Google Scholar]
- 8.Annunziato, F., L. Cosmi, V. Santarlasci, L. Maggi, F. Liotta, B. Mazzinghi, E. Parente, L. Filì, S. Ferri, F. Frosali, et al. 2007. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettelli, E., Y. Carrier, W. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal development pathways for the generation of pathogenic effector Th17 and regulatory T cells. Nature. 441:235–238. [DOI] [PubMed] [Google Scholar]
- 10.Weaver, C.T., R.D. Hatton, P.R. Mangan, and L.E. Harrington. 2007. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25:821–852. [DOI] [PubMed] [Google Scholar]
- 11.Deenick, E.K., and S.G. Tangve. 2007. Autoimmunity. IL-21: a new player in Th17-cell differentiation. Immunol. Cell Biol. 85:503–505. [DOI] [PubMed] [Google Scholar]
- 12.Acosta-Rodriguez, E.V., G. Napoletani, A. Lanzavecchia, and F. Sallusto. 2007. Interleukins 1β and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 8:942–949. [DOI] [PubMed] [Google Scholar]
- 13.Wilson, N.J., K. Boniface, J.R. Chan, B. McKenzie, W.M. Blumenschein, J.D. Mattson, B. Basham, K. Smith, T. Chen, F. Morel, et al. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957. [DOI] [PubMed] [Google Scholar]
- 14.Evans, H.G., T. Suddason, I. Jackson, L.S. Taams, and G.M. Lord. 2007. Optimal induction of T helper 17 cells in humans requires T cell receptor ligation in the context of Toll-like receptor-activated monocytes. Proc. Natl. Acad. Sci. USA. 104:17034–17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Z., C.M. Tato, L. Muul, A. Laurence, and J.J. O'Shea. 2007. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. 56:2936–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurence, A., and J.J. O'Shea. 2007. Th17 differentiation: of mice and men. Nat. Immunol. 8:903–905. [DOI] [PubMed] [Google Scholar]
- 17.van Beelen, A.J., Z. Zelinkova, E.W. Taanman-Kueter, F.J. Muller, D.W. Hommes, S.A. Zaat, M.L. Kapsenberg, and E.C. de Jong. 2007. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 27:660–669. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, O.H., I. Kirman, N. Rudiger, H. Jebeld, and B. Vainer. 2003. Up-regulation of interleukin-12 and -17 in active inflammatory bowel disease. Scand. J. Gastroenterol. 38:180–185. [DOI] [PubMed] [Google Scholar]
- 19.van Beelen, A.J., M.B.M. Teunissen, M.L. Kapsenberg, and E.C. de Jong. 2007. Interleukin-17 in inflammatory skin disorders. Curr. Opin. Allergy Clin. Immunol. 7:374–381. [DOI] [PubMed] [Google Scholar]
- 20.Godfrey, D.I., H.R. MacDonald, M. Kronenberg, M.J. Smyth, and L. Van Kaer. 2004. NKT cells: what's in a name? Nat. Rev. Immunol. 4:231–237. [DOI] [PubMed] [Google Scholar]
- 21.Onoda, T., M. Rhaman, H. Nara, A. Araki, K. Makabe, K. Tsumoto, I. Kumagai, T. Kudo, N. Ishii, N. Tanaka, et al. 2007. Human CD24+ central and effector memory T cells produce IL-21: effect on cytokine-driven proliferation of CD4+ T cell subsets. Int. Immunol. 19:1191–1199. [DOI] [PubMed] [Google Scholar]
- 22.Liang, S.C., X.Y. Tan, D.P. Luxenburg, R. Karim, K. Dunussi-Joannopoulos, M. Collins, and L.A. Fouser. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanier, L.L., C. Chang, and J.H. Philips. 1994. Human NKR-P1A. A disulphide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J. Immunol. 153:2417–2428. [PubMed] [Google Scholar]
- 24.Bendelac, A., P.B. Savage, and L. Teyton. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336. [DOI] [PubMed] [Google Scholar]
- 25.Godfrey, D.I., and S.P. Berzins. 2007. Control points in NKT-cell development. Nat. Rev. Immunol. 7:505–518. [DOI] [PubMed] [Google Scholar]
- 26.Behar, S.M., T.A. Potrebarac, C.J. Roy, C.R. Wang, and M.B. Brenner. 1999. Diverse TCRs recognize murine CD1. J. Immunol. 162:161–167. [PubMed] [Google Scholar]
- 27.Chiu, Y.H., J. Jayawardena, A. Weiss, D. Lee, S.H. Park, A. Dautry-Varsat, and A. Bendelac. 1999. Distinct subsets of CD1-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 189:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werwitzke, S., A. Tiede, B.E. Drescher, R.E. Schmidt, and T. Witte. 2003. CD8b/CD28 expression defines functionally distinct populations of peripheral blood T lymphocytes. Clin. Exp. Immunol. 133:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi, T., S. Dejbakhsh-Jones, and S. Strober. 2006. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J. Immunol. 176:211–216. [DOI] [PubMed] [Google Scholar]
- 30.Eberl, G., R. Lees, S.T. Smiley, M. Taniguchi, M.J. Grusby, and H.R. MacDonald. 1999. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J. Immunol. 162:6410–6419. [PubMed] [Google Scholar]
- 31.Skold, M., N.N. Faizunessa, C.-R. Wang, and S. Cardell. 2000. CD1d-specific NK1.1+ T cells with a transgenic variant TCR. J. Immunol. 165:168–174. [DOI] [PubMed] [Google Scholar]
- 32.Hammond, K.J.L., D.G. Pellicci, L.D. Poulton, O.V. Naidenko, A.A. Scalzo, A.G. Baxter, and D.I. Godfrey. 2001. CD1d-restricted NKT cells: an inter-strain comparison. J. Immunol. 167:1164–1173. [DOI] [PubMed] [Google Scholar]
- 33.Michel, M.L., A.C. Keller, C. Paget, M. Fujo, F. Trottein, P.B. Savage, C.H. Wong, E. Schneider, M. Dy, and M.C. Leite-de-Moraes. 2007. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J. Exp. Med. 204:995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichavant, M., S. Goya, E.H. Meyer, R.A. Johnston, H.Y. Kim, P. Matangkasombut, M. Zhu, Y. Iwakura, P.B. Savage, R.H. DeKruyff, et al. 2008. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J. Exp. Med. 205:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niemeyer, M., A. Darmoise, H.-J. Mollenkopf, R. Hurwitz, G.S. Besra, U.E. Schaible, and H.E. Kaufmann. 2008. Natural killer T-cell characterization through gene expression profiling: an account of versatility bridging T helper type 1 (Th1), Th2 and Th17 immune responses. Immunology. 123:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bettelli, E., and V.K. Kuchroo. 2005. IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together. J. Exp. Med. 201:169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poggi, A., P. Costa, M.R. Zocchi, and L. Moretta. 1997. NKRP1A molecule is involved in trans-endothelial migration of CD4+ human T lymphocytes. Immunol. Lett. 57:121–123. [DOI] [PubMed] [Google Scholar]
- 38.Poggi, A., M.R. Zocchi, P. Costa, E. Ferrero, G. Borsellino, R. Placido, S. Galgani, M. Salvetti, C. Gasperini, G. Ristori, et al. 1999. IL-12-mediated NKRP1A up-regulation and consequent enhancement of endothelial transmigration of Vδ2 TCRγδ+ T lymphocytes from healthy donors and multiple sclerosis patients. J. Immunol. 162:4349–4354. [PubMed] [Google Scholar]
- 39.O'Keeffe, J., D.G. Doherty, T. Kenna, K. Sheahan, D.P. O'Donoghue, J.M. Hyland, and C. O'Farrelly. 2004. Diverse populations of T cells with NK cell receptors accumulate in the human intestine in health and in colorectal cancer. Eur. J. Immunol. 34:2110–2118. [DOI] [PubMed] [Google Scholar]
- 40.Niess, J.H., F. Leithauser, G. Adler, and J. Reimann. 2008. Commensal flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J. Immunol. 180:559–568. [DOI] [PubMed] [Google Scholar]
- 41.Rosen, D.B., J. Bettadapura, M. Alsharifi, P.A. Mathew, H.S. Warren, and L.L. Lanier. 2005. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J. Immunol. 175:7796–7799. [DOI] [PubMed] [Google Scholar]
- 42.Spreu, J., E.C. Kienle, B. Schrage, and A. Steinle. 2007. CLEC2A: a novel, alternatively spliced and skin-associated member of the NKC-encoded AICL-CD69-LLT1 family. Immunogenetics. 59:903–912. [DOI] [PubMed] [Google Scholar]
- 43.Frentsch, M., O. Arbach, D. Kirchhoff, B. Moewes, M. Worm, M. Rothe, A. Scheffold, and A. Thiel. 2005. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 11:1118–1124. [DOI] [PubMed] [Google Scholar]
- 44.Maggi, L., V. Santarlasci, F. Liotta, F. Frosali, R. Angeli, L. Cosmi, E. Maggi, S. Romagnani, and F. Annunziato. 2007. Demonstration of circulating allergen-specific CD4+CD25highFoxp3+ T-regulatory cells in both nonatopic and atopic individuals. J. Allergy Clin. Immunol. 120:429–436. [DOI] [PubMed] [Google Scholar]
- 45.De Palma, R., and J. Gorski. 1995. Restricted and conserved T-cell repertoires involved in allorecognition of class II major histocompatibility complex. Proc. Natl. Acad. Sci. USA. 92:8836–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pannetier, C., J. Even, and P. Kourilsky. 1995. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today. 16:176–181. [DOI] [PubMed] [Google Scholar]
- 47.Bacsi, S., R. De Palma, G.P. Visentin, J. Gorski, and R.H. Aster. 1999. Complexes of heparin and platelet factor 4 specifically stimulate T cells from patients with heparin-induced thrombocytopenia/thrombosis. Blood. 94:208–215. [PubMed] [Google Scholar]
- 48.Hazenberg, M.D., S.A. Otto, J.W. Cohen Stuart, M.C. Verschuren, J.C. Borleffs, C.A. Boucher, R.A. Coutinho, J.M. Lange, T.F. Rinke de Wit, A. Tsegaye, et al. 2000. Increased cell division but not thymic dysfunction rapidly affects the T-cell receptor excision circle content of the naive T cell population in HIV-1 infection. Nat. Med. 6:1036–1042. [DOI] [PubMed] [Google Scholar]
- 49.Clave, E., V. Rocha, K. Talvensaari, M. Busson, C. Douay, M.L. Appert, C. Rabian, M. Carmagnat, F. Garnier, A. Filion, et al. 2005. Prognostic value of pretransplantation host thymic function in HLA-identical sibling hematopoietic stem cell transplantation. Blood. 105:2608–2613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.