Abstract
In mantle cell lymphoma (MCL) and some cases of multiple myeloma (MM), cyclin D1 expression is deregulated by chromosome translocations involving the immunoglobulin heavy chain (IgH) locus. To evaluate the mechanisms responsible, gene targeting was used to study long-distance gene regulation. Remarkably, these targeted cell lines lost the translocated chromosome (t(11;14)). In these MCL and MM cells, the nonrearranged cyclin D1 (CCND1) locus reverts from CpG hypomethylated to hypermethylated. Reintroduction of the translocated chromosome induced a loss of methylation at the unrearranged CCND1 locus, providing evidence of a transallelic regulatory effect. In these cell lines and primary MCL patient samples, the CCND1 loci are packaged in chromatin-containing CCCTC binding factor (CTCF) and nucleophosmin (NPM) at the nucleolus. We show that CTCF and NPM are bound at the IgH 3′ regulatory elements only in the t(11;14) MCL cell lines. Furthermore, NPM short hairpin RNA produces a specific growth arrest in these cells. Our data demonstrate transvection in human cancer and suggest a functional role for CTCF and NPM.
B cell malignancies such as non-Hodgkin's lymphoma and multiple myeloma (MM) are characterized by 14q32 translocations involving the IgH locus (1). These translocations serve to juxtapose IgH regulatory elements, such as the intronic enhancer (Eμ;1) or 3′ Cα locus control region (LCR) (2, 3), that deregulate transcription of target genes over several hundred kilobases of DNA. The mechanisms involved in long-distance deregulation of target genes by IgH regulatory elements are unknown; however, regulatory elements in the IgH locus are thought to derepress or increase the transcription of target genes such as CMYC (1, 2) and cyclin D1 (CCND1) (1, 4) that are involved in B cell malignancies. Epigenetic changes in chromatin structure involving DNA methylation (5) and/or histone modifications (6) have been implicated in deregulated gene expression (4).
The activation of the CCND1 gene that occurs in mantle cell lymphoma (MCL) and a subset of MM was used as a model system to investigate the mechanisms responsible for long-distance gene deregulation in B cell malignancies (see Fig. 1 A). Cyclin D1 is not expressed in normal lymphocytes, where the unlinked family members cyclin D2 and/or D3 are active (7). In B cell malignancies, cyclin D1 gene expression is activated by the insertion or translocation of IgH regulatory elements, such as the Eμ intronic or 3′ Cα enhancer/LCR, that can be as far as 100–300 kb away from the CCND1 gene (4, 8). The majority of the breakpoints in MCL map to the major translocation cluster (MTC) region located ∼120 kb upstream (centromeric) of the CCND1 gene (8). The nearest gene to CCND1, MyeOV, is located 360 kb centromeric to CCND1 and is expressed in a subset of t(11;14) MM, but not in MCL (9). Although translocations involving the t(11;14) are most common, the MM cell line U266 contains an insertion of IgH regulatory sequences ∼10 kb centromeric of the CCND1 promoter (10).
Figure 1.
Derivative MCL and MM cell lines have lost t(11;14). (A) Maps of the 11q13 and 14q32 regions with the locations of the gene targeting events. TV1 inserts the neoR gene into the MTC region, ∼120 kb upstream of the CCND1 gene on chromosome 11. TV2 inserts a neoR gene downstream of the human IgH 3′α1 LCR region on chromosome 14. (B) Southern blot analysis of U266 parental (U) and TV1-transfected clones (U1–11). Maps of the vector, the endogenous locus, and the targeted clone U7 are shown. (C) Southern blot analysis of MCL parental and mutant cell lines. Genomic DNA from the Granta parental cell line (G) and its targeted derivatives (G4, LG1, and LG2) were analyzed with the Granta breakpoint (P519) probe, and genomic DNA from the parental cell line NCEB1 (N) and its targeted derivatives (N1, N2, and N3) were analyzed with the NCEB1 breakpoint (MTC) probe. Both parental lines have both translocated (Trans.) and nontranslocated (Nor.) CCND1 loci, whereas the derivatives retain only the normal chromosome. The map depicts the locations of the cell line breakpoints. (D) FISH analysis of MCL parental and mutant cell lines. A centromeric probe for chromosome 11 and IgH/CCND1 fusion probes revealed loss of the translocated t(11;14) in the derivative cells lines. A more detailed description is provided in Supplemental results (available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). (E) FISH analyses show the presence of the variant t(11;14) insertion in the U266 MM cell line and its absence in derivative cell lines (LU1 and LU2). Table S1 provides a summary. Bars, 15 μm.
The CCND1 promoter contains a CpG island that can be potentially regulated by DNA methylation (4, 11). We have found that in normal B cells the CCND1 locus is organized into hypomethylated DNA bound by acetylated nucleosomes. Analysis of the CCND1 DNA methylation status in MM and MCL cell lines with t(11;14) indicates that the deregulated as well as the normal, silent CCND1 loci are CpG hypomethylated (4).
Control of DNA methylation in mammalian cells has been shown to involve a cis-acting mechanism (12). However, observations in higher plants (13–15), the fungus Ascobolus immerses (16), and more recently in mice (13, 17) demonstrated that DNA methylation can also be regulated in trans by interactions between homologous chromosomes.
A ubiquitous, complex protein that has emerged as a critical mediator of multiple epigenetic processes is the zinc finger protein CCCTC binding factor (CTCF). CTCF is a highly conserved, 11 zinc finger protein first identified as a c-myc binding factor and subsequently shown to bind to metazoan regulatory elements known as insulators (18). Insulator elements, which act from an intervening position to prevent flanking cis-acting elements from interacting, mediate their function through CTCF (18, 19). Furthermore, the binding of CTCF to insulator elements is blocked by CpG methylation, allowing interactions between distal regulatory elements in the imprinted Igf2/H19 locus (19, 20). Recent evidence has identified the nucleolar protein nucleophosmin (NPM) as a major CTCF-interacting protein that functions to tether promoters and regulatory elements separated by large linear distances together at the nucleolar periphery (21).
To investigate the mechanisms of long-distance activation of the CCND1 gene by the IgH enhancer/LCR, homologous recombination was used to target regulatory regions potentially involved in cyclin D1 deregulation. Recombinants that lost the translocated chromosome also lost the ability to maintain hypomethylation at the normal CCND1 allele. These clones no longer expressed cyclin D1. These findings suggest that the translocated chromosome exerts a long-distance cis DNA hypomethylating effect on the linked CCND1 promoter as well as a transallelic effect on the unlinked CCND1 promoter on the homologous chromosome. Thus, in the absence of the translocated chromosome, the unrearranged CCND1 locus is densely DNA methylated. CTCF and NPM are associated with both the translocated and unrearranged CCND1 loci in cell lines with t(11;14). Interestingly, binding of these proteins at the unrearranged CCND1 locus is dependent on the presence of the translocated allele. These results provide further evidence that long-distance control of DNA methylation in mammalian cells can be communicated in trans, and demonstrate a novel mechanism for chromosome structure and nuclear organization to participate in deregulated gene expression in human malignancy.
RESULTS
B cell lines that have lost the t(11;14) translocation do not express cyclin D1 and have altered DNA methylation patterns at the unrearranged CCND1 locus
A gene targeting strategy was designed to study regulatory regions involved in cyclin D1 deregulation in human B cell malignancies. Our initial approach was to target the potential regulatory regions involved in long-distance cyclin D1 deregulation in B cell malignancies. By analogy, results in the β-globin locus demonstrated that insertion of a selectable marker gene into the β-globin LCR abrogated transcription of downstream genes (22). Two gene targeting vectors (TV1 and 2; Fig. 1 A) were constructed and electroporated into two MCL cell lines, Granta 519 (Granta [G]) and NCEB-1 (NCEB [N]), and the MM cell line U266 (U), all of which express cyclin D1. The initial Southern blot analysis was consistent with successful gene targeting (Fig. 1 B); however, subsequent analyses showed instability and loss of the targeted/translocated chromosomes. Additional clones were isolated using TV1 in NCEB (N1–3) cells, and TV2 in U266 (LU clones) and Granta (LG clones) cells. Southern blot analysis using cell line–specific translocation breakpoint probes (P519 for Granta and MTC for NCEB) demonstrated the absence of translocated chromosome breakpoint sequences in the mutant MCL cell lines (Fig. 1 C). Fluorescent in situ hybridization (FISH) analysis demonstrated that the translocated chromosomes had been lost in all of these clones (Fig. 1, D and E; and Table S1, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1).
We assessed the expression of D-type cyclins in each of the parental and targeted MCL cells to determine whether loss of t(11;14) affected expression of the remaining cyclin D alleles. Parental Granta and NCEB MCL cells expressed cyclin D1 (4). However, the derivative MCL cell lines, which had lost t(11;14), did not express cyclin D1 (Fig. 2, A and B). Instead, up-regulation of cyclin D3 mRNA (not depicted) and protein (Fig. 2 C) levels was observed, presumably to compensate for the loss of cyclin D1 that has been observed in other experimental cell culture and animal systems (23, 24).
Figure 2.
Analysis of D-type cyclin protein levels in parental and mutant clones. (A) IP Western blot analysis of cyclin D1 protein in parental and mutant NCEB and Granta MCL cell lines. Cyclin D1 protein is not detectable in mutant clones. (B) IP Western blot analysis of cyclin D1 protein in control and mutant U266 MM clones. Parental U266 cells express cyclin D1 protein, whereas all the mutant clones express no cyclin D1 protein. (C) Western blot analysis of cyclin D3 protein in parental and cyclin D1− MM and MCL cells. Abundant cyclin D3 protein is observed in cyclin D1− cell lines. Parental MCL and MM lines express minimal amounts of cyclin D3 protein. The white line indicates that intervening lanes have been spliced out. (D) Western blot analysis of cyclin D2 protein in parental and cyclin D1− MCL clones. Cyclin D2 protein is present at low levels in both parental and cyclin D1− clones. Chicken DT40 cells were used as a negative control. Manca (Ma), RD, and DHL-4 cells were used as positive controls. M, marker.
DNA methylation patterns within the CCND1 locus in the variant cyclin D1− MCL cells were analyzed. The CCND1 promoter region and sequences 3 kb upstream were extensively methylated, as shown by Southern blot analysis (Fig. 3 B) and bisulfite sequencing (Fig. 3 C). DNA methylation changes occurred in sequences 90 kb (P519) upstream of the CCND1 gene. Because the translocation breakpoints in the NCEB and Granta cell lines are located at the 3′ end and proximal to the MTC (Fig. 3 A), respectively, most of this region lies on the reciprocal translocation partner. At the MTC, CpG sequences such as at the Hpa II/Msp site remained hypomethylated in both cyclin D1-expressing and nonexpressing clones. Similar results were obtained with independently isolated Granta and NCEB clones (Fig. 3, B and C).
Figure 3.
DNA methylation analysis of the parental and cyclin D1− cell lines. (A) Schematic representation of the 11q13 locus. Breakpoints, probes used, regions studied by bisulfite sequencing, and their positions relative to the CCND1 gene are shown. (B) Analysis of DNA methylation in Granta (G) versus G4, LG1, and LG2 cell lines, and NCEB (N) versus N1, N2, and N3 cell lines. Isolated DNA from each line was cut with Hind III + Msp (M) or Hpa II (H) and Southern blotted. (C) Bisulfite sequencing in G4 and N1 cell lines. Extensive remethylation of the promoter and 3-kb upstream regions compared with parental cell lines Granta and NCEB, and remethylation of some sites in the P519 region located 90 kb upstream of the CCND1 gene are observed. (D) Analysis of DNA methylation in U266 and variant cell lines by Southern blotting. The CCND1 promoter region and upstream region P519 are unmethylated in U266 cells and methylated in nonexpressing LU1 and LU2 cells. CCND1-expressing MM cell lines KMS12 and KMS21BM (t(11;14)+) are unmethylated in all regions examined, similar to U266 cells.
DNA methylation in the cyclin D1− genetic variants was also assayed by methylation-specific PCR (MSP) (25), which complements the Southern blotting and sodium bisulfite sequencing approaches (26). MSP analysis of the CCND1 promoter and the 3-kb upstream regions in Granta, NCEB, G4, and N1 cells revealed that both regions were heavily CpG methylated in the genetic variants (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1).
These data are consistent with a model in which the translocated chromosome exerts a transallelic hypomethylating effect on the nontranslocated chromosome in MCL parental cell lines. This effect disappeared in the targeted variants with the loss of the translocated chromosome, leading to extensive methylation of the CCND1 locus. The transallelic hypomethylating effect extended from the CCND1 promoter at least to the P519 region 90 kb upstream.
To determine if our observations in MCL cells were applicable to cyclin D1–expressing MM cells that contained an insertion of IgH sequences instead of the classical t(11;14), we analyzed similar variant clones from parental U266 cells that contain IgH regulatory sequences inserted ∼10 kb centromeric of the CCND1 promoter (12). Similar to the variant Granta and NCEB clones, U7, LU1, and LU2 clones were isolated where the active chromosome 11 was lost (Fig. 1 E; and Table S1), and no cyclin D1 protein was detected (Fig. 2 B). In addition, two other control MM cell lines that contain t(11;14) and express cyclin D1 (KMS12 and KMS21BM) (27) were analyzed.
Analysis of the DNA methylation status at the CCND1 locus in the parental MM cell lines and the U266 cyclin D1− derivatives revealed that the CCND1 promoter and P519 regions were hypomethylated in all of the cyclin D1+ MM cell lines, including U266, KMS12, and KMS21BM (Fig. 3 D), similar to the results obtained with the parental Granta and NCEB MCL cell lines. In contrast, DNA hypermethylation was observed at the CCND1 locus in the cyclin D1− U266 MM cell lines that had lost the active CCND1 locus with inserted IgH regulatory elements. Thus, transallelic effects at the CCND1 locus are observed with inserted as well as translocated IgH sequences.
Monoallelic histone acetylation patterns at the cyclin D1 loci
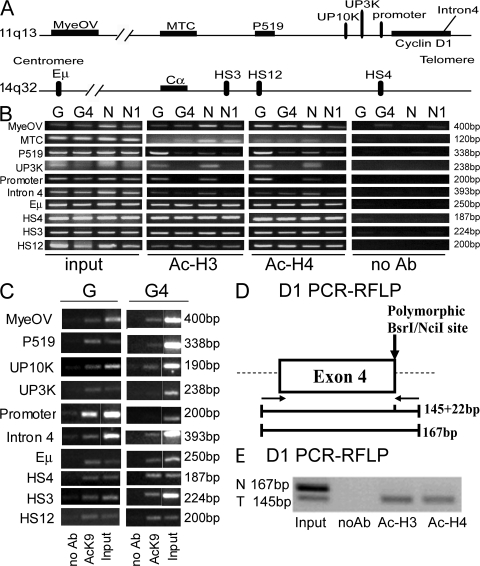
To determine if posttranslational histone modifications such as histone acetylation were coordinately involved with DNA methylation in transallelic effects at the cyclin D1 locus, histone acetylation patterns in the cyclin D1− genetic variants and parental cell lines were studied by chromatin immunoprecipitation (ChIP) assays. In contrast to the parental cell lines, the cyclin D1 coding and promoter regions and sequences extending 3 kb upstream were histone H3 and H4 deacetylated in the mutant cell lines, consistent with the DNA methylation patterns and the lack of transcription of the cyclin D1 gene. However, regions far upstream of the cyclin D1 gene, including the MTC, P519, and the IgH regulatory regions, remained histone H3 and H4 hyperacetylated in the mutant cell lines (Fig. 4 B) (4). Similar results were obtained with specific antibodies to acetyl H3 lysine 9 (AcK9; Fig. 4 C).
Figure 4.
Histone acetylation analyses in parental and cyclin D1− MCL lines. (A) Map of the 11q13 and 14q32 loci, as in Fig. 1 A. Primers used and their positions relative to the CCND1 and Cα genes are shown. (B) ChIP assays with antibodies to acetylated histones H3 and H4 demonstrated hyperacetylation in the parental cell lines Granta (G) and NCEB (N), and hypoacetylation in the mutant cell lines G4 and N1 at the CCND1 promoter and upstream 3-kb (UP3K) regions. The other regions, including upstream regions MyeOV, MTC, P519, the CCND1 gene intron 4, and the IgH regulatory elements showed high levels of histone acetylation. (C) ChIP assays of H3 AcK9 showing a similar pattern as that obtained in B. White lines indicate that intervening lanes have been spliced out. (D) Schematic diagram of the CCND1 gene showing the location of the primers (left and right arrows) used for PCR and the polymorphic BsrI/NciI site, located in the last codon of exon 4 (A870G; reference 30). BsrI digestion of the PCR products distinguishes two alleles: allele A (145 and 22 bp) and allele G (167 bp). (E) ChIP RFLP assay of the CCND1 promoter region in U266 cells. Acetylated histones are only detected on the translocated allele, suggesting no transvection effects as observed with DNA methylation. N, normal allele; T, translocated allele.
Because the changes in histone acetylation observed in the mutant cells were also consistent with the loss of the histone H3 and H4 acetylated, translocated allele, allele-specific histone acetylation assays were performed. Specifically, we wished to determine whether both the translocated and nontranslocated CCND1 alleles in the parental cells contained acetylated histones. Allele-specific patterns of histone acetylation were studied in the U266 MM cell line, whose cyclin D1 alleles can be distinguished by a polymorphism at the cyclin D1 intron/exon 4 junction (Fig. 4 D). Allelic ChIP assays performed on U266 cells indicated that acetylated histones were only present on the translocated allele (Fig. 4 E), suggesting that histone acetylation was not participating in transallelic effects at the CCND1 locus.
Reintroduction of the translocated t(11;14) alters DNA methylation in trans
Somatic cell fusion experiments have been used to study the contribution of individual alleles to both the transcription and DNA methylation status of several genetic loci in human malignancy (28, 29). Thus, cell fusion was used to determine if reintroduction of the translocated chromosome into the mutant cell lines where the CCND1 locus was methylated was capable of effecting transhypomethylation of the methylated allele. The Granta cell line (CCND1 hypomethylated) was fused with the N1 cell line (CCND1 methylated) to enable use of a DNA polymorphism within the CCND1 gene (30) to distinguish the translocated from the normal CCND1 loci (Fig. 5, A and B). All of the hybrid (G/N1) clones isolated contained both the untranslocated CCND1 locus from N1 cells and the translocated CCND1 locus from Granta cells, and expressed cyclin Dl (Fig. 5, C–F). No hybrid clones could be isolated that contained only the nontranslocated CCND1 loci (see Discussion). RFLP analysis demonstrated that cyclin D1 gene expression in these G/N1 hybrid clones originated from the Granta cell–derived translocated CCND1 locus (Fig. 5 E).
Figure 5.
Characterization and analysis of G/N1 hybrid cells. (A) Schematic diagram of the somatic cell fusion strategy used to generate G/N1 hybrid cells. (B) Schematic diagrams showing the location of the primers (arrows) used for PCR (Fig. 4 D) or RT-PCR and the polymorphic BsrI/NciI site (A870G; reference 30). BsrI or NciI digestion of the PCR or RT-PCR products distinguishes two alleles (A and/or G). (C) Allelic CCND1 gene PCR-RFLP analysis. Granta cells had the GG genotype, the mutant cell line N1 had the A genotype, and G/N1 hybrid cells had the AG genotype. (D) RT-PCR analysis demonstrated cyclin D1 expression in the G/N1 hybrid cells. Granta was used as positive control. N1 was used as a negative control and actin was used as an amplification control. (E) Allelic CCND1 RT-PCR analysis. The hybrid cells contain the translocated and transcriptionally active CCND1 locus, as in Granta cells. N1 and N cells were used as controls. (F) Southern blot analysis of the genomic DNA cleaved with BamH1 from Granta (G) and G/N1 hybrid cells shows both the normal and translocated CCND1 loci. (G) Southern blot analysis of DNA methylation in the G/N1 hybrid, Granta, and N1 cell lines. The CCND1 promoter region and upstream regions P519 and MTC are hypomethylated in G/N1 hybrid cells.
DNA methylation in the hybrid G/N1 cells was analyzed using Southern blotting (Fig. 5 G) and sodium bisulfite sequencing (Fig. 6) to determine whether introduction of the translocated CCND1 locus was capable of effecting transhypomethylation of the methylated allele. A 1:1 mix of the two parental DNAs and DNAs from nonhomologous clones were used as controls (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). Analysis of DNA methylation patterns by bisulfite sequencing and MSP (Fig. 6) demonstrated that the translocated CCND1 locus was able to effect DNA hypomethylation at the promoter and 3 kb upstream of the promoter from the unrearranged, methylated CCND1 allele in G/N1 hybrid cells containing both the translocated and nonrearranged CCND1 loci. Extensive hypomethylation of the upstream 3-kb region was observed, but the transhypomethylation at the CCND1 promoter was incomplete in G/N1 hybrids. DNA methylation was reevaluated in these hybrid cells after multiple rounds of DNA replication to determine if the transhypomethylating effect at the promoter could be magnified (Fig. 6 B). In fact, DNA replication was associated with a small increase in CCND1 promoter methylation.
Figure 6.
Transallelic effects are specific to t(11;14). (A) Granta cells demonstrated minimal DNA methylation in the CCND1 gene promoter region. G/N1 hybrid cells showed ∼20% methylation, whereas the 1:1 mixture of Granta and N1 showed ∼50% methylation (∼100% methylation in N1 cells, as shown in Fig. 3 C; and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). Low-level DNA methylation was found in the upstream 3-kb region in both Granta and hybrid (G/N1) cells, whereas N1 cells displayed ∼100% methylation, as shown in Fig. 3 C. (B) Methylation levels in hybrid cells increased to ∼40% with culture (G/N1 late). (C) Hybrid cells treated with 5-azacytidine alone (G/N1 aza) or the combination of 5-azacytidine and trichostatin A (G/N1 TSA + aza) showed decreased methylation levels at the CCND1 gene promoter region compared with untreated cells (B) or cells treated with trichostatin A alone (G/N1 TSA). (D) MSP analysis at the CCND1 3-kb upstream region was performed as in Fig. S1. Methylation was observed in N1, U7, and LCL/N1 hybrid cells but not in parental NCEB, U266, or hybrid G/N1 cells, or in the malignant lymphocytes of a patient with a 11q− deletion and no t(11;14).
Treatment of these hybrid cells with a DNA hypomethylating agent (5-azacytidine) and/or a histone deacetylase inhibitor (trichostatin A) was performed (Fig. 6 C). Treatment with 5-azacytidine alone or in combination with trichostatin A led to almost total DNA hypomethylation at the CCND1 promoter in the hybrid cells, whereas treatment with trichostatin A alone did not change DNA methylation patterns.
Because hybrid clones containing only the nontranslocated CCND1 locus could not be isolated (see Discussion), control fusions were performed using N1 and lymphocyte cell line (LCL) cells to investigate the effects of introduction of a nontranslocated, hypomethylated CCND1 locus (4) on the densely methylated CCND1 locus in N1 cells. MSP analysis of the CCND1 3-kb upstream region demonstrated that in contrast to G/N1 cells, LCL/N1 hybrid cells did not demonstrate hypomethylation in trans at the CCND1 locus (Fig. 6 D).
To further demonstrate that transallelic effects in MCL were specific to t(11;14), malignant lymphoid cells that had deleted one CCND1 allele without the presence of t(11;14) were studied. A variant MCL patient (31) with marked peripheral blood lymphocytosis was identified (expressing cyclin D3 and no cyclin D1 mRNA; not depicted). FISH analysis demonstrated that this patient's malignant B cells contained a deletion of one allele of chromosome 11q, including the cyclin D1 locus (Fig. S3, available at http://www.jem.org/cgi/content/full/jcb.20072102/DC1), and no t(11;14). MSP analysis of this 11q− patient's DNA demonstrated a hypomethylated CCND1 allele (Fig. 6 D). Thus, transallelic effects were specific to the presence of the translocated CCND1 locus.
CTCF, NPM, and the nucleolus are associated with cyclin D1 activation and transvection
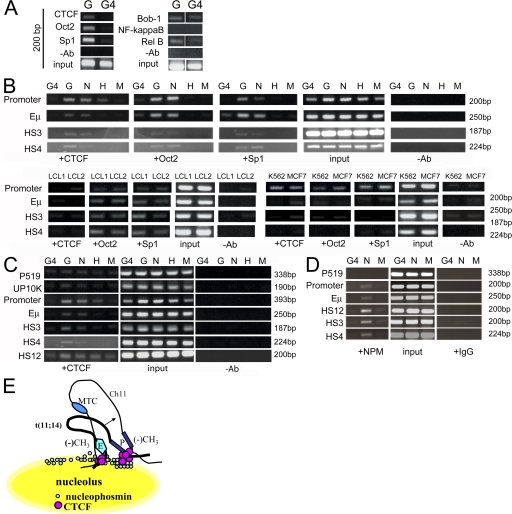
ChIP assays were used to screen parental versus variant cyclin D1− G4 cells for DNA binding proteins that would demonstrate selective binding to the translocated, transcriptionally active allele (Fig. 7). Proteins were assayed that have been reported to bind to the CCND1 promoter in vitro or whose potential consensus binding sites are located in the CCND1 promoter and IgH regulatory regions (32–35). Our results demonstrated that CTCF, Oct2, and Sp1 exhibited selective in vivo binding at the CCND1 promoter region in G versus G4 cells, whereas OCA-B (Bob-1), NF-κB, or a subunit of NF-κB (Rel-B) did not. Additional ChIP assays were performed to further study in vivo DNA–protein interactions of CTCF, Oct2, and Sp1 at the CCND1 promoter, and IgH regulatory regions in cyclin D1–expressing and control cell lines. Oct2, Sp1, and CTCF binding to the CCND1 promoter region correlated with cyclin D1 gene expression in MCL cells (Fig. 7 B). Primers from the IgH Eμ enhancer 3′ Cα LCR regions showed CTCF, Oct2, and Sp1 binding to the Eμ enhancer in both MCL and LCL cells. CTCF but not Oct2 or Sp1 binding to 3′ Cα LCR HS3 and HS4 correlated with cyclin D1 expression (Fig. 7 B).
Figure 7.
CTCF and NPM bind to the CCND1 loci and selectively to 3′ IgH regulatory elements in MCL cells. (A) CTCF, Oct2, and Sp1 showed differential binding in the parental cell line Granta (G) and the mutant cell line G4. OCA-B/Bob-1, NF-κB, and Rel-B showed similar low levels of binding in both cell lines. White lines indicate that intervening lanes have been spliced out. (B) CTCF, Oct2, and Sp1 are bound to the CCND1 promoter and IgH regulatory regions in MCL cell lines. EBV-transformed B lymphocytes LCL1 and LCL2 showed binding of CTCF, Oct2, and Sp1 at the CCND1 promoter region, whereas cyclin D1 mRNA was not expressed in these cells. Oct2 and Sp1 bound to the IgH regulatory elements Eμ and 3′ Cα HS4 and HS3, whereas CTCF was only found bound to the Eμ intronic enhancer in LCL1. CTCF, Oct2, and Sp1 all bound to the CCND1 promoter region. H, HL60 myeloid cells; M, Manca Burkitt's lymphoma cells. (C) CTCF binding was enriched at the CCND1 promoter and IgH regulatory sequences. (D) NPM binding is similar to that of CTCF in Granta MCL cells at the CCND1 locus and IgH regulatory sequences. (E) Model of CTCF tethering of the 3′ Cα IgH enhancers (E) to the CCND1 promoter (P) through interaction at the nucleolar periphery. CTCF functions to juxtapose enhancer and promoter by tethering them to the nucleolus through interaction with NPM. CTCF is also postulated to tether the normal and translocated CCND1 loci, thus facilitating transallelic effects (modified from reference 20).
The binding of CTCF at the CCND1 locus and IgH regulatory regions was further examined using ChIP assays (Fig. 7 C). Although binding at IgH 3′ Cα LCR HS3 and HS4 only occurred in cell lines that expressed CCND1, primers for 3′ Cα HS12 demonstrated binding of CTCF in all cell lines examined (Fig. 7 C). Collectively, these results suggested that 3′ Cα HS3 and HS4 binding by CTCF could be important in deregulated cyclin D1 expression in MCL.
Western blot analyses were performed to assay the level of these proteins in the cell lines used (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). cyclin D1 protein was detected in the MCL cell lines Granta and NCEB, and the breast cancer cell line MCF7, consistent with RT-PCR assays (4). The transcription factors CTCF, Oct2, and Sp1 were detected in all cell lines examined. Thus, the specific binding of CTCF observed was not secondary to variations in the levels of this protein in MCL cells.
CTCF has been shown to interact with the nucleolar protein NPM to tether an insulator sequence to subnuclear sites at the nucleolar periphery (21). ChIP assays demonstrated binding of NPM at the CCND1 promoter and IgH regulatory regions in MCL cells, similar to results obtained with CTCF (Fig. 7 D). These results suggested a potential role for CTCF in juxtaposing 3′ Cα IgH regulatory elements (HS3 and HS4) and the CCND1 promoter by tethering them to the nucleolar periphery with NPM. CTCF and NPM could also play a role in mediating pairing of the translocated and normal CCND1 loci, facilitating transallelic effects (Fig. 7 E).
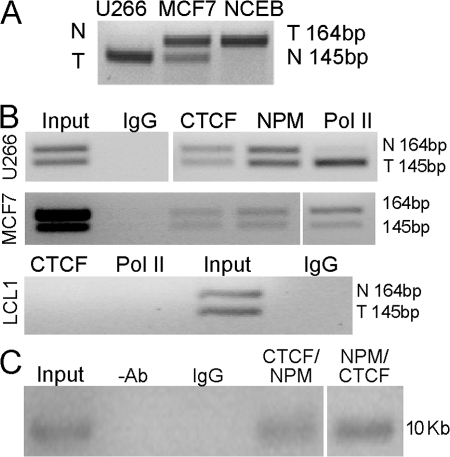
If CTCF and NPM were involved in tethering the CCND1 loci together, evidence of binding of these proteins to both CCND1 alleles would be expected. Allelic ChIP assays were performed with antibodies to CTCF, NPM, and RNA polymerase II (Pol II). MCF7 cells, a cyclin D1–expressing breast cancer line without the t(11;14) translocation, demonstrated biallelic transcription of cyclin D1 (30) as well as biallelic binding of Pol II, NPM, and CTCF (Fig. 8 B). An EBV LCL (LCL1) that does not express cyclin D1 demonstrated no binding of Pol II or CTCF at either allele, whereas the CCND1 loci in this cell line are CpG hypomethylated and histone H3 and H4 are hyperacetylated (4). U266 MM cells only transcribed the CCND1 allele containing the IgH insertion, as shown by RT-PCR analysis and ChIP analysis using Pol II antibodies (Fig. 8, A and B). However, CTCF and NPM ChIP DNA from U266 but not LCL cells contained sequences from both the transcribed and nontranscribed CCND1 alleles (Fig. 8 B). Thus, both the transcribed and nontranscribed CCND1 loci bind CTCF and NPM, consistent with our hypothesis that CTCF and NPM are involved in tethering these loci together.
Figure 8.
CTCF and NPM bind at both the translocated and normal CCND1 alleles. (A) Allelic CCND1 RT-PCR assay. The 145-bp fragment corresponds to the translocated and transcribed CCND1 locus in U266 cells, whereas the 164-bp fragment corresponds to the translocated locus in Granta cells. Both CCND1 loci of MCF7 cells are transcribed. N, normal; T, translocated CCND1 alleles. (B) Allelic ChIP assays. CTCF and NPM binding is seen on both CCND1 loci and Pol II binding only on the translocated and transcribed locus (145-bp band shown, as in Fig. 5 B) in U266. MCF7 cells bind CTCF and Pol II at both CCND1 loci, and an LCL B cell line showed no binding of CTCF and Pol II. (C) ReChIP assay of CTCF and NPM binding to the CCND1 locus in MCL cells. ChIP assays with CTCF and NPM antibodies sequentially in both combinations were performed in HBL-2 MCL cells. PCR analysis with primers from the CCND1 locus (10 kb upstream; Fig. 4) demonstrated that CTCF and NPM were bound together at the cyclin D1 locus in HBL-2 MCL cells. Mouse IgG and no antibody controls showed no binding. White lines indicate that intervening lanes have been spliced out.
To provide further evidence that CTCF and NPM were directly cobinding at the CCND1 locus, sequential ChIP (36) assays were performed on the EBV-negative MCL cell line HBL-2 (37) using sequential IP assays with anti-CTCF and NPM antibodies. These results demonstrated that CTCF and NPM were binding together at the CCND1 locus in MCL cells (Fig. 8 C).
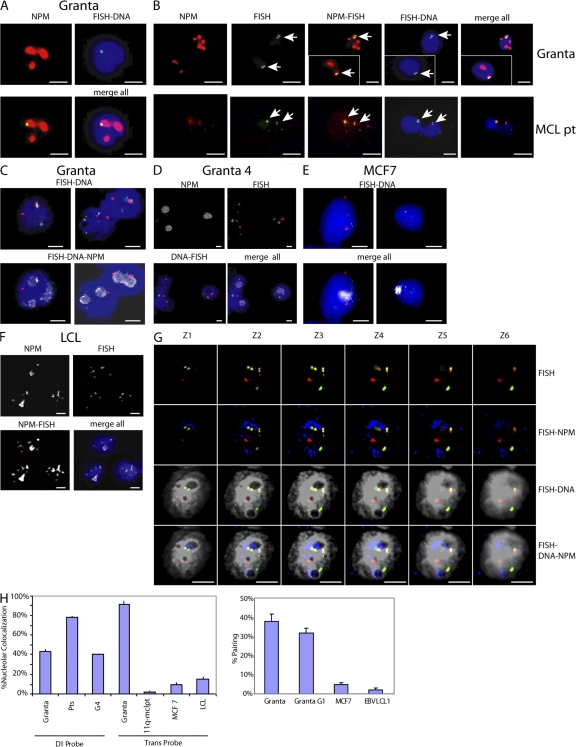
FISH analysis combined with NPM immunofluorescent FISH (immunoFISH) staining was used to visualize the position of the CCND1 loci in relationship to the nucleolus. Fixation protocols using methanol/acetic acid or paraformaldehyde alone, or in combination, yielded similar results. The CCND1 locus colocalized at the nucleolar periphery in CCND1-translocated cell lines (Fig. 9, A and G) and in neoplastic cyclin D1–expressing lymphocytes from MCL patients (Fig. 9 B; and Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). Pairing of the CCND1 loci at the nucleolus was also observed, as indicated by two juxtaposed FISH signals (Fig. 9, B and C; and G and H). The paired CCND1 loci observed were unlikely to be caused by replicated but unseparated sister chromatids, because the same frequency of pairing was observed in flow-sorted Granta nuclei in the G1 cell cycle phase (Fig. 9 H). Paired CCND1 loci were not observed in cyclin D1–expressing MCF7 cells and LCL cells where the CCND1 locus was hypomethylated and histone H3 and H4 were acetylated (4).
Figure 9.
CTCF and NPM participate in tethering of the translocated and nontranslocated CCND1 loci at the nucleolus. (A) immunoFISH assays demonstrate colocalization of CCND1 probe (green) and NPM protein (red) at the nucleolar periphery in Granta MCL cells (merge; yellow). DAPI-stained nuclei are shown in blue. (B) immunoFISH assays (as in A). Paired CCND1 loci as demonstrated by two juxtaposed FISH spots (arrows) colocalize with nucleoli in cells from Granta cells (top) and an MCL patient (bottom). Insets show different fields of view. (C) immunoFISH assays of Granta cells with CCND1 (red) /IgH (green) probes and NPM antibodies (Cy5; gray), demonstrating colocalization of both translocated chromosome partners (yellow) at the nucleolar periphery. (D) G4 cells lacking the translocated CCND1 locus exhibit colocalization of the CCND1 FISH signal with the nucleoli. (E) immunoFISH assay of MCF7 cells (FISH probes as in D) demonstrates no colocalization of the CCND1 locus with the nucleoli. (F) immunoFISH assay of LCL1 B lymphocytes (FISH probes as in D) demonstrates no colocalization of the CCND1 locus with the NPM-stained nucleoli. (G) Z stack images (deconvolution microscopy; FISH probes as in D) show nucleolar colocalization of translocated chromosomes in Granta cells. Nucleoli are blue and DAPI-stained nuclei are gray. The nucleoli are seen as DAPI-poor staining regions (FISH-DNA) that stain with anti-NPM antibodies (FISH–DNA NPM). (H) Quantitative analysis of the percentage of nucleolar colocalization and CCND1 locus pairing in Granta cells, cells from three MCL patients (Pts), and MCF7 cells using CCND1 (D1; see A–C), CCND1/IgH fusion (Trans; probe used in D–G), FISH probes, and NPM antibodies. At least 100 nuclei were scored per sample. G1 cell cycle phase nuclei isolated by flow cytometry were used. Error bars show SEM. Bars, 5 μm.
In MCL cells that lost the translocated CCND1 locus (G4), the nontranslocated chromosome 11 remained associated with the nucleolus, providing further evidence that pairing of the cyclin D1 loci at the nucleolus was occurring in the parental MCL cell lines (Fig. 9, D and H). These observations demonstrated the nucleolar environment is capable of both activation and repression functions at the CCND1 loci in MCL cells (see Discussion). In cyclin D1–expressing MCF7 cells, B lymphocytes (LCLs), and the variant MCL patient (Fig. S4) that do not contain the t(11;14) translocation, colocalization of the CCND1 loci and the nucleolus was not observed (Fig. 9, E and F; and not depicted).
To further investigate the role of the nucleolus and NPM in deregulated cyclin D1 expression in t(11;14) B cell malignancies, we used NPM short hairpin RNA (shRNA) to knock down levels of NPM. Cell growth was specifically inhibited by NPM shRNA in cell lines containing a t(11;14) but not in B lymphocytes without this translocation and MCF7 cells (Fig. 10 A). Western blotting demonstrated significant knockdown of NPM protein levels but no change in cyclin D1 protein levels in either MCF7 or Granta cells (Fig. 10 B). Western analysis showed a marked decrease in NPM levels in cells containing the NPM shRNA (Fig. 10 C). The NCEB MCL cell line demonstrated marked morphological changes that were consistent with apoptosis and were confirmed by Tdt-mediated dUTP-biotin nick-end labeling (TUNEL) assays. Lower levels of apoptosis were observed in Granta cells (Fig. 10 D), and minimal apoptosis was seen in G4 and N1 cells (Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20072102/DC1). MCF7 and K562 erythroleukemia cells treated with the NPM but not control shRNA vectors showed loss of nucleolar NPM staining, and CTCF, cyclin D1, and residual NPM staining in the cytoplasm (Fig. 10 C). These results are consistent with the role of NPM as a shuttle protein (38) and suggest that CTCF and cyclin D1 are among the proteins regulated by NPM. immunoFISH assays revealed no significant change in nucleolar colocalization of the CCND1 loci in NPM knockdown cells (Fig. 10 E), suggesting that additional layers of complexity are involved in the tethering of the CCND1 loci in MCL cells. NPM knockdown cells also did not demonstrate any change in cyclin D1 protein levels (Fig. 10 B), suggesting that NPM binding at the CCND1 locus was not directly involved in regulating expression of the cyclin D1 gene but could be serving a structural role in tethering the CCND1 loci to the nucleolus.
Figure 10.
NPM knockdown in MCL and control cells. (A) Growth of cells containing t(11;14) translocations is inhibited with NPM shRNA. Cells were infected with retroviruses containing either an NPM (red) or a control (blue) shRNA. Cell growth was specifically inhibited in cells with t(11;14) translocations. (B) Western blot analysis of protein levels of cyclins D1, D3, NPM, and α-tubulin genes after infection with NPM shRNA virus. NPM levels are reduced, whereas levels of cyclins D1 (Granta and MCF7) and D3 (K562) are unchanged. (C) Immunofluorescent staining of NCEB, MCF7, and K562 cells after transfection with NPM shRNA vector. NPM shRNA–infected NCEB cells show dramatic morphological changes. MCF7 cells demonstrated cyclin D1 and CTCF cytoplasmic protein staining. K562 erythroleukemia cells also demonstrate cytoplasmic NPM and CTCF staining. Granta NPM shRNA–treated cells do not show cytoplasmic localization of NPM or cyclin D1. (D) TUNEL assays of NPM and control shRNA MCL cells. Fig. S6 (available at http://www.jem.org/cgi/content/full/jem.20072102/DC1) shows control shRNA vector–infected cells. (E) immunoFISH assay of NPM knockdown Granta cells demonstrates no significant change in the percentage of cells showing nucleolar localization. Error bars in A, D, and E show SEM. Bars, 5 μm.
DISCUSSION
Previously, the DNA methylation patterns of the 11q13 region surrounding the CCND1 gene in MCL and MM cells were examined (4). The activation of the cyclin D1 gene by IgH regulatory elements was found to correlate with the absence of DNA methylation at the CCND1 promoter. Interestingly, this domain of hypomethylated DNA appears to be present not only on the translocated but also on the untranslocated allele. Normal B cells and EBV lymphocytes also displayed extensive DNA hypomethylation and hyperacetylation of histones at the CCND1 loci, although the CCND1 gene is not expressed (4). Hypomethylation up to 90 kb upstream of the CCND1 promoter was eliminated with loss of the translocated chromosome initiated through a gene targeting event.
Somatic cell hybrid fusions indicated that reintroduction of the translocated chromosome but not a nontranslocated chromosome was capable of transhypomethylating a densely methylated CCND1 locus. The Granta cells used in the fusion experiments that reintroduced the t(11;14) chromosome into N1 cells also contained a nontranslocated CCND1 locus that possessed the identical cyclin D1 polymorphism as the translocated CCND1 locus. The recipient N1 cell line possessed a different, distinguishable polymorphism in the cyclin D1 gene (Fig. 5). All of the hybrid clones isolated contained the translocated chromosome. We were unable to isolate clones that contained only the nontranslocated CCND1 locus from Granta cells. Thus, the translocated and nontranslocated CCND1 loci could not be segregated. This result provides further, albeit indirect, evidence that the translocated and nontranslocated cyclin D1 loci are tethered together (at the nucleolus), as suggested by our immunoFISH data (Fig. 9). Alternatively, defects in DNA methylation have been associated with chromosomal instability, both in embryogenesis (39) and in colorectal cancer cell lines (40). Therefore, another possibility is that the hypomethylated nontranslocated CCND1 locus was unstable and was lost during the cell fusion process.
Control fusion experiments indicated that fusion of N1 cells with a B lymphocyte line without a translocated chromosome was unable to hypomethylate the densely methylated cyclin D1 locus of N1 cells in trans. Similar results using other systems demonstrated no trans epigenetic effects with nontranslocated chromosomes (28, 29).
Because the CCND1 locus is extensively hypomethylated in normal B cells, at least three DNA methylation states of the CCND1 loci are implied: (a) normal B cells in where both CCND1 loci are unmethylated and independent; (b) B cells containing CCND1 translocations/insertions, where the translocated locus exerts a transhypomethylating effect on the untranslocated locus; and (c) B cells that have lost the translocated chromosome and where the CCND1 locus is CpG methylated. The second and third DNA methylation states are dependent on localization at the nucleolus, whereas the second DNA methylation state appears to be associated with pairing of the CCND1 loci. Thus, the association of genetic loci with the nucleolus can result in either active (state 2) or inactive (state 3) chromatin, consistent with the results obtained in other systems (21, 41). The mechanisms involved in gene expression or repression by association with the nucleolus remain obscure, but could be related to the ability of the translocated allele to recruit activator molecules that protect the tethered cyclin D1 loci from the general repressive effects of the nucleolus. There may also be subtle differences in nucleolar localization that may be important in epigenetic activation versus repression.
The change in DNA methylation patterns observed in the normal CCND1 locus after loss of the translocated chromosome are consistent with transallele sensing effects (42), which are well studied in Drosophila (transvection; references 42–44) and plants (paramutation) (13–15). Trans effects in both endogenous and transgenic mouse loci have been reported (45–49). Such effects often involve DNA methylation and occur over long distances (45, 46, 48). Transactivation of the endogenous Igf2 gene by trangenes has been reported (49), but given the results reported in this paper, transallelic effects should be considered.
Recent observations in transgenic mice have described paramutation-like effects at the Igf2 (45), U2af1rs-1 (46), c-kit (47), and Rosa26 (48) loci. Paramutation/transvection in human genetic disease associated with the susceptibility to type I diabetes has been suggested at the insulin minisatellite variable number tandem repeat element (50). However, our observed transallelic effects are similar to transvection but differ from paramutation because the DNA transhypomethylation effects are not heritable in the absence of the inducer (translocated) allele (14).
Physical evidence of transcommunication has recently been observed between loci on nonhomologous chromosomes regulated by the T cell enhancers/LCRs (51), and the Igf2/H19 and Wsb1/Nf1 loci (52). CTCF has been demonstrated to be involved in long-distance cis and trans effects at the Igf2/H19 locus (52, 53). Thus, transcommunication may be an important means of regulation in mammalian cells.
At least two nonexclusive mechanisms are implicated in the homology-based transfer of DNA methylation patterns (14). One mechanism involves direct communication between homologous chromosomes, as has been demonstrated in imprinted loci (54) or in mouse embryonic stem cells at the onset of X inactivation (55). Recent work has shown that CTCF is required for this transient X chromosome pairing (56). The nucleolus has also been implicated in silencing of the inactive X chromosome using immunoFISH assays similar to those performed in this study (57). Our FISH and allelic ChIP data provide evidence for direct communication of the CCND1 alleles in MCL cells that we suggest is mediated via CTCF, NPM, and the nucleolus.
Alternatively, small RNA molecules have been shown to direct chromatin modification in trans in plants and Drosophila (13, 14, 58–61). This epigenetic silencing mechanism involves CpG and non-CpG DNA methylation in plants. Tandem and inverted repeats have been shown to be involved in this type of RNA-mediated repression (58–61). More recently, a paramutation-like effect mediated by small RNAs has been described at the mouse c-kit locus (47).
Our allelic ChIP and immunoFISH data suggest that CTCF and NPM participate in transallelic interactions by tethering the cyclin D1 loci from both translocated and nontranslocated cyclin D1 loci together at the nucleolar periphery, as depicted in Fig. 7 E. Our data demonstrate that the 3′ Cα IgH LCR region is sufficient to mediate this effect and that CTCF binding to 3′ Cα LCR HS3 and HS4 correlates with cyclin D1 activation in MCL cells.
Our finding in cyclin D1–deregulated B cell malignancies represents the first report of the association of transvection with human cancer. The critical role of NPM in t(11;14) B cell malignancies is demonstrated by growth arrest and apoptosis selectively in t(11;14) cell lines with NPM shRNA. Our data also suggest a novel approach to therapy of t(11;14) malignancies using agents targeting NPM.
MATERIALS AND METHODS
Cell culture, electroporation, and retroviral infection.
Cell lines were obtained and maintained as previously described (4). The HBL-2 MCL cell line (37) was provided by S. Dave (National Cancer Institute, Bethesda, MD). Fig. 1 shows details of gene targeting constructs. Electroporations were performed, as previously described (22), using 10–20 μg of linearized targeting vector. Retroviral infections were performed as previously described (62).
FISH/immunoFISH.
FISH (63) and immunoFISH (64) were performed as previously described. For immunoFISH, cells were fixed using 4% paraformaldehyde, methanol/acetic acid (3:1), and methanol/acidic acid, followed by paraformaldehyde with similar results. For two-color immunoFISH, a combination of CCND1 probes from chromosome 11 (9) was labeled by nick translation (63). Three-color immunoFISH used a t(11;14) fusion probe (Vysis; Fig. 1). Cy5-conjugated anti–mouse secondary antibody (Invitrogen) was used with anti-NPM antibody after FISH, according to the supplied protocol. Deconvolution microscopy (Deltavision) was performed using the OHSU core facility. Granta cells were stained with Hoechst dye, and G1 cells were isolated by flow cytometry for immunoFISH analysis. At least 100 cells were assayed per cell line.
Peripheral blood or bone marrow was obtained from MCL patients giving informed consent under an OHSU institutional review board–approved Southwest Oncology Group study. Buffy coat was purified using Ficoll, as previously described (4).
Somatic cell fusions.
Somatic cell fusions were performed using polyethylene glycol, as previously described (65). Hybrids were selected using combined resistance to neomycin and hygromycin or hygromycin and puromycin.
Southern blotting and bisulfite sequencing.
Southern blotting and bisulfite sequencing were performed as previously described. At least 10 clones were sequenced for each bisulfite reaction (4). Hybridization probes and PCR primer sequences have been previously described (4).
ChIP.
ChIP assays were performed and PCR primers were used as previously described (4). Supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20072102/DC1) describes primers and antibodies.
PCR-RFLP.
The CCND1 G/A polymorphism was detected by the PCR- RFLP method. A 167-bp fragment of the CCND1 gene at the junction of exon 4/intron 4 was amplified by PCR using 0.1 μg of genomic DNA at an annealing temperature of 64°C. For RFLP analyses, each PCR product was purified by gel extraction and digested with BsrI (ACTGGN/) at 50°C before electrophoresis. The DNA fragments were separated using a 3% 2:1 Nusieve/SeaKem agarose gel. The allele types were determined as shown in Fig. 4. Primer sequences are provided in Supplemental materials and methods.
TUNEL assays.
TUNEL assays were done according to the manufacturer's protocol (Roche). 300–500 nuclei were assayed per sample.
Online supplemental material.
Table S1 summarizes the FISH results demonstrating the absence of t(11;14) in the targeted clones. Fig. S1 shows the results of MSP analysis of parental and cyclin D1− clones. Fig. S2 shows the results of bisulfite sequencing mixing control experiments. Fig. S3 demonstrates FISH analysis of a cyclin D1− 11q− MCL patient. Fig. S4 shows Western blot analysis of proteins assayed by ChIP analysis in the cell lines used. Fig. S5 shows the results of staining of blood from an MCL patient with antibodies to cyclin D1 and NPM. Fig. S6 shows the results of TUNEL assays on control and NPM shRNA–treated cells. Supplemental results describes Fig. 1 D in detail. Supplemental materials and methods provides details about antibodies and PCR primers. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20072102/DC1.
Supplementary Material
Acknowledgments
We thank Mark Groudine, William Forrester, Matt Lorincz, Richard Maziarz, Janine Lasalle, and Vicki Chandler for discussions and comments. Rich Fisher, Tom Miller, and the Southwest Oncology Lymphoma Committee provided MCL patient samples.
This work was supported by grants from the National Institutes of Health (HL069133 and HL077818 to W.H. Fleming, and DK56798 to E. Epner), the International Myeloma Foundation (to E. Epner), and the Lymphoma Research Foundation (to E. Epner).
The authors declare no financial conflicts of interest.
Abbreviations used: CCND1, cyclin D1; ChIP, chromatin immunoprecipitation; CTCF, CCCTC binding factor; FISH, fluorescent in situ hybridization; immunoFISH, immunofluorescent FISH; LCL, lymphocyte cell line; LCR, locus control region; MCL, mantle cell lymphoma; MM, multiple myeloma; MSP, methylation-specific PCR; MTC, major translocation cluster; NPM, nucleophosmin; Pol II, RNA polymerase II; shRNA, short hairpin RNA; TUNEL, Tdt-mediated dUTP-biotin nick-end labeling.
H. Liu and J. Huang contributed equally to this paper.
H. Liu's present address is Nimblegen Systems, Madison, WI 53711.
References
- 1.Willis, T.G., and M.J. Dyer. 2000. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 96:808–822. [PubMed] [Google Scholar]
- 2.Madisen, L., and M. Groudine. 1994. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. 8:2212–2226. [DOI] [PubMed] [Google Scholar]
- 3.Mills, F.C., N. Harindranath, M. Mitchell, and E.E. Max. 1997. Enhancer complexes located downstream of both human immunoglobulin Calpha genes. J. Exp. Med. 186:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu, H., J. Wang, and E.M. Epner. 2004. Cyclin D1 activation in B-cell malignancy: association with changes in histone acetylation, DNA methylation, and RNA polymerase II binding to both promoter and distal sequences. Blood. 104:2505–2513. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein, M., G. Keini, H. Cedar, and Y. Bergman. 1994. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 76:913–923. [DOI] [PubMed] [Google Scholar]
- 6.Madisen, L., A. Krumm, T.R. Hebbes, and M. Groudine. 1998. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol. Cell. Biol. 18:6281–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajchenbaum, F., K. Ando, J.A. DeCaprio, and J.D. Griffin. 1993. Independent regulation of human D-type cyclin gene expression during G1 phase in primary human T lymphocytes. J. Biol. Chem. 268:4113–4119. [PubMed] [Google Scholar]
- 8.Rimokh, R., F. Berger, G. Delsol, I. Digonnet, J.P. Rouault, J.D. Tigaud, M. Gadoux, B. Coiffier, P.A. Bryon, and J.P. Magaud. 1994. Detection of the chromosomal translocation t(11;14) by polymerase chain reaction in mantle cell lymphomas. Blood. 83:1871–1875. [PubMed] [Google Scholar]
- 9.Janssen, J.W., J.W. Vaandrager, T. Heuser, A. Jauch, P.M. Kluin, E. Geelen, P.L. Bergsagel, W.M. Kuehl, H.G. Drexler, T. Otsuki, et al. 2000. Concurrent activation of a novel putative transforming gene, myeov, and cyclin D1 in a subset of multiple myeloma cell lines with t(11;14)(q13;q32). Blood. 95:2691–2698. [PubMed] [Google Scholar]
- 10.Gabrea, A., P.L. Bergsagel, M. Chesi, Y. Shou, and W.M. Kuehl. 1999. Insertion of excised IgH switch sequences causes overexpression of cyclin D1 in a myeloma tumor cell. Mol. Cell. 3:119–123. [DOI] [PubMed] [Google Scholar]
- 11.Kitazawa, S., R. Kitazawa, and S. Maeda. 1999. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J. Biol. Chem. 274:28787–28793. [DOI] [PubMed] [Google Scholar]
- 12.Ng, H.H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158–163. [DOI] [PubMed] [Google Scholar]
- 13.Chandler, V.L. 2007. Paramutation: from maize to mice. Cell. 128:641–645. [DOI] [PubMed] [Google Scholar]
- 14.Chandler, V.L., and M. Stam. 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5:532–544. [DOI] [PubMed] [Google Scholar]
- 15.Walker, E.L. 1998. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics. 148:1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colot, V., L. Maloisel, and J.L. Rossignol. 1996. Interchromosomal transfer of epigenetic states in Ascobolus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell. 86:855–864. [DOI] [PubMed] [Google Scholar]
- 17.Herman, H., M. Lu, M. Anggraini, A. Sikora, Y. Chang, B.J. Yoon, and P.D. Soloway. 2003. Trans allele methylation and paramutation-like effects in mice. Nat. Genet. 34:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell, A.C., A.G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic. Science. 291:447–450. [DOI] [PubMed] [Google Scholar]
- 19.Bell, A.C., A.G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 98:387–396. [DOI] [PubMed] [Google Scholar]
- 20.Bell, A.C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 405:482–485. [DOI] [PubMed] [Google Scholar]
- 21.Yusufzai, T.M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 13:291–298. [DOI] [PubMed] [Google Scholar]
- 22.Kim, C.G., E.M. Epner, W.C. Forrester, and M. Groudine. 1992. Inactivation of the human beta-globin gene by targeted insertion into the beta-globin locus control region. Genes Dev. 6:928–938. [DOI] [PubMed] [Google Scholar]
- 23.Ciemerych, M.A., A.M. Kenney, E. Sicinska, I. Kalaszczynska, R.T. Bronson, D.H. Rowitch, H. Gardner, and P. Sicinski. 2002. Development of mice expressing a single D-type cyclin. Genes Dev. 16:3277–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam, E.W., J. Glassford, L. Banerji, N.S. Thomas, P. Sicinski, and G.G. Klaus. 2000. Cyclin D3 compensates for loss of cyclin D2 in mouse B-lymphocytes activated via the antigen receptor and CD40. J. Biol. Chem. 275:3479–3484. [DOI] [PubMed] [Google Scholar]
- 25.Herman, J.G., J.R. Graff, S. Myohanen, B.D. Nelkin, and S.B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 93:9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warnecke, P.M., C. Stirzaker, J.R. Melki, D.S. Millar, C.L. Paul, and S.J. Clark. 1997. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 25:4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaandrager, J.W., P. Kluin, and E. Schuuring. 1997. The t(11;14) (q13;q32) in multiple myeloma cell line KMS12 has its 11q13 breakpoint 330 kb centromeric from the cyclin D1 gene. Blood. 89:349–350. [PubMed] [Google Scholar]
- 28.Kuzmin, I., L. Geil, H. Ge, U. Bengtsson, F.M. Duh, E.J. Stanbridge, and M.I. Lerman. 1999. Analysis of aberrant methylation of the VHL gene by transgenes, monochromosome transfer, and cell fusion. Oncogene. 18:5672–5679. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., L. Myeroff, L. Kasturi, L. Krumroy, S. Schwartz, J.K. Willson, E. Stanbridge, G. Casey, and S. Markowitz. 2002. Chromosomal autonomy of hMLH1 methylation in colon cancer. Oncogene. 21:1443–1449. [DOI] [PubMed] [Google Scholar]
- 30.Hosokawa, Y., and A. Arnold. 1998. Mechanism of cyclin D1 (CCND1, PRAD1) overexpression in human cancer cells: analysis of allele-specific expression. Genes Chromosomes Cancer. 22:66–71. [DOI] [PubMed] [Google Scholar]
- 31.Fu, K., D.D. Weisenburger, T.C. Greiner, S. Dave, G. Wright, A. Rosenwald, M. Chiorazzi, J. Iqbal, S. Gesk, R. Siebert, et al. 2005. Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood. 106:4315–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boulon, S., J.C. Dantonel, V. Binet, A. Vie, J.M. Blanchard, R.A. Hipskind, and A. Philips. 2002. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol. Cell. Biol. 22:7769–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrett, F.E., A.V. Emelyanov, M.A. Sepulveda, P. Flanagan, S. Volpi, F. Li, D. Loukinov, L.A. Eckhardt, V.V. Lobanenkov, and B.K. Birshtein. 2005. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol. Cell. Biol. 25:1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joyce, D., C. Albanese, J. Steer, M. Fu, B. Bouzahzah, and R.G. Pestell. 2001. NF-kappaB and cell-cycle regulation: the cyclin connection. Cytokine Growth Factor Rev. 12:73–90. [DOI] [PubMed] [Google Scholar]
- 35.Nagata, D., E. Suzuki, H. Nishimatsu, H. Satonaka, A. Goto, M. Omata, and Y. Hirata. 2001. Transcriptional activation of the cyclin D1 gene is mediated by multiple cis-elements, including SP1 sites and a cAMP-responsive element in vascular endothelial cells. J. Biol. Chem. 276:662–669. [DOI] [PubMed] [Google Scholar]
- 36.Rincon-Arano, H., V. Valadez-Graham, G. Guerrero, M. Escamilla-Del-Arenal, and F. Recillas-Targa. 2005. YY1 and GATA-1 interaction modulate the chicken 3′-side alpha-globin enhancer activity. J. Mol. Biol. 349:961–975. [DOI] [PubMed] [Google Scholar]
- 37.Abe, M., Y. Nozawa, H. Wakasa, H. Ohno, and S. Fukuhara. 1988. Characterization and comparison of two newly established Epstein-Barr virus-negative lymphoma B-cell lines. Surface markers, growth characteristics, cytogenetics, and transplantability. Cancer. 61:483–490. [DOI] [PubMed] [Google Scholar]
- 38.Borer, R.A., C.F. Lehner, H.M. Eppenberger, and E.A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 56:379–390. [DOI] [PubMed] [Google Scholar]
- 39.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 300:455. [DOI] [PubMed] [Google Scholar]
- 40.Lengauer, C., K.W. Kinzler, and B. Vogelstein. 1997. DNA methylation and genetic instability in colorectal cancer cells. Proc. Natl. Acad. Sci. USA. 94:2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, L., R.A. Haeusler, P.D. Good, M. Thompson, S. Nagar, and D.R. Engelke. 2005. Silencing near tRNA genes requires nucleolar localization. J. Biol. Chem. 280:8637–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff, S., and L. Comai. 1998. Trans-sensing effects: the ups and downs of being together. Cell. 93:329–332. [DOI] [PubMed] [Google Scholar]
- 43.Duncan, I.W. 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36:521–556. [DOI] [PubMed] [Google Scholar]
- 44.Southworth, J.W., and J.A. Kennison. 2002. Transvection and silencing of the Scr homeotic gene of Drosophila melanogaster. Genetics. 161:733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forne, T., J. Oswald, W. Dean, J.R. Saam, B. Bailleul, L. Dandolo, S.M. Tilghman, J. Walter, and W. Reik. 1997. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl. Acad. Sci. USA. 94:10243–10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatada, I., A. Nabetani, Y. Arai, S. Ohishi, M. Suzuki, S. Miyabara, Y. Nishimune, and T. Mukai. 1997. Aberrant methylation of an imprinted gene U2af1-rs1(SP2) caused by its own transgene. J. Biol. Chem. 272:9120–9122. [DOI] [PubMed] [Google Scholar]
- 47.Rassoulzadegan, M., V. Grandjean, P. Gounon, S. Vincent, I. Gillot, and F. Cuzin. 2006. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 441:469–474. [DOI] [PubMed] [Google Scholar]
- 48.Rassoulzadegan, M., M. Magliano, and F. Cuzin. 2002. Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J. 21:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, F.L., W.L. Dean, G. Kelsey, N.D. Allen, and W. Reik. 1997. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 389:809–815. [DOI] [PubMed] [Google Scholar]
- 50.Bennett, S.T., A.J. Wilson, L. Esposito, N. Bouzekri, D.E. Undlien, F. Cucca, L. Nistico, R. Buzzetti, E. Bosi, F. Pociot, et al. 1997. Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. The IMDIAB Group. Nat. Genet. 17:350–352. [DOI] [PubMed] [Google Scholar]
- 51.Spilianakis, C.G., M.D. Lalioti, T. Town, G.R. Lee, and R.A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645. [DOI] [PubMed] [Google Scholar]
- 52.Ling, J.Q., T. Li, J.F. Hu, T.H. Vu, H.L. Chen, X.W. Qiu, A.M. Cherry, and A.R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 312:269–272. [DOI] [PubMed] [Google Scholar]
- 53.Kurukuti, S., V.K. Tiwari, G. Tavoosidana, E. Pugacheva, A. Murrell, Z. Zhao, V. Lobanenkov, W. Reik, and R. Ohlsson. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA. 103:10684–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaSalle, J.M., and M. Lalande. 1996. Homologous association of oppositely imprinted chromosomal domains. Science. 272:725–728. [DOI] [PubMed] [Google Scholar]
- 55.Xu, N., C.L. Tsai, and J.T. Lee. 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 311:1149–1152. [DOI] [PubMed] [Google Scholar]
- 56.Xu, N., M.E. Donohoe, S.S. Silva, and J.T. Lee. 2007. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 39:1390–1396. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, L.F., K.D. Huynh, and J.T. Lee. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 129:693–706. [DOI] [PubMed] [Google Scholar]
- 58.Martienssen, R.A. 2003. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 35:213–214. [DOI] [PubMed] [Google Scholar]
- 59.Pal-Bhadra, M., U. Bhadra, and J.A. Birchler. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 9:315–327. [DOI] [PubMed] [Google Scholar]
- 60.Stam, M., A. Viterbo, J.N. Mol, and J.M. Kooter. 1998. Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol. Cell. Biol. 18:6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zilberman, D., X. Cao, and S.E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 299:716–719. [DOI] [PubMed] [Google Scholar]
- 62.Welcker, M., J. Singer, K.R. Loeb, J. Grim, A. Bloecher, M. Gurien-West, B.E. Clurman, and J.M. Roberts. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 12:381–392. [DOI] [PubMed] [Google Scholar]
- 63.Aladjem, M.I., M. Groudine, L.L. Brody, E.S. Dieken, R.E. Fournier, G.M. Wahl, and E.M. Epner. 1995. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 270:815–819. [DOI] [PubMed] [Google Scholar]
- 64.Jiang, S., L. Walker, M. Afentoulis, D.A. Anderson, L. Jauron-Mills, C.L. Corless, and W.H. Fleming. 2004. Transplanted human bone marrow contributes to vascular endothelium. Proc. Natl. Acad. Sci. USA. 101:16891–16896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forrester, W.C., E. Epner, M.C. Driscoll, T. Enver, M. Brice, T. Papayannopoulou, and M. Groudine. 1990. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 4:1637–1649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.