Abstract
We have investigated the variables that influence DNA coverage on gold nanoparticles. The effects of salt concentration, spacer composition, nanoparticle size, and degree of sonication have been evaluated. Maximum loading was obtained by salt aging the nanoparticles to ~0.7 M NaCl in the presence of DNA containing a poly(ethylene glycol) (PEG) spacer. In addition, DNA loading was substantially increased by sonicating the nanoparticles during the surface loading process. Lastly, nanoparticles up to 250 nm in diameter were found have ~2 orders of magnitude higher DNA loading than smaller (13–30 nm) nanoparticles, a consequence of their larger surface area. Stable large particles are attractive for a variety of biodiagnostic assays.
Keywords: DNA, DNA density, gold nanoparticles, bio-detection, sonication
INTRODUCTION
Gold nanoparticles exhibit several interesting physical and chemical properties which have made them an integral part of research in nanoscience.1 In addition to their striking optical properties, gold nanoparticles are important because they can be stabilized with a wide variety of molecules by taking advantage of well known chemistry involving alkyl thiol adsorption on gold.2 In particular, thiol modified oligonucleotides (short synthetic DNA sequences) can be loaded onto the surface of Au nanoparticles. The resulting DNA functionalized Au nanoparticles have become widely used as nanoscale building blocks in assembly strategies,3, 4 as antisense agents in nanotherapeutics for gene regulation,5 and as probes in many biodiagnostic systems.6 In all of these applications, it is essential to understand the coverage of the DNA on the nanoparticle, and, in many cases, it is favorable to have higher DNA loadings.
The advantages provided by higher DNA loading have the potential to dramatically impact biodetection and nanotherapeutics. The first biodetection assays using DNA functionalized gold nanoparticles involved colorimetric readout strategies.7–9 These procedures were inspired by the aggregation induced red-to-blue color transition which is due to a dampening and red shifting of the nanoparticle surface plasmon resonance (SPR) band. Since the development of the initial colorimetric assays, DNA functionalized Au nanoparticles have become a central component in a wide variety of schemes that use readout strategies including fluorescence,10, 11 radioactivity,12 quartz crystal microbalance,13 Raman spectroscopy,14, 15 light scattering,16 and electrical signal.17
In addition, the bio-bar-code method is a strategy that has made a marked impact on the field of gold nanoparticle-based biodiagnostics, by providing a protocol to detect proteins,18, 19 DNA,20, 21 and other biomolecules22, 23 at remarkably low concentrations both serially18, 20 and, in some cases, in a multiplexed format.19, 21 The high sensitivity of this assay stems from the indirect amplification of the target sequence by the sizable number of DNA strands which can be loaded on a single gold nanoparticle. Therefore, the amount of DNA on each nanoparticle directly correlates to the amount of amplification possible and therefore the sensitivity attainable in this system.
Moreover, recent work in the area of nanotherapeutics has demonstrated the use of DNA functionalized gold nanoparticles as antisense agents for intracellular gene regulation.5 The nanoparticles act as a non-toxic and highly efficient antisense agent that by virtue of cooperative binding properties can very effectively scavenge mRNA within the cell.5, 24 In addition, the tight packing of the DNA on the surface of the nanoparticle likely plays a role in the inhibition of its degradation by nucleases. This opens the door for the use of functionalized gold nanoparticles in several very efficient gene regulation therapies.
The diagnostic and therapeutic applications of oligonucleotide-modified nanoparticles benefit from the ability to maximize and tailor the amount of DNA on the gold nanoparticle surface. Herein, we fully quantify the loading of DNA on a range of gold nanoparticle sizes while examining the effects of several different parameters. We determine the dependence of DNA loading on: 1) salt concentrations from 0 to 1.0 M NaCl, 2) adenine (A), thymine (T), and non-DNA base (poly(ethylene glycol) (PEG)) spacers (the region of the oligonucleotide between the recognition sequence and the thiol functionality), and 3) sonication. Importantly, we determine the DNA loading obtained from these parameters on several sizes of gold nanoparticles: 15, 30, 50, 80, 150 and 250 nm in diameter. Through these studies, two new parameters that are significant in achieving particles with the highest DNA loading have been determined. They are 1) the use of PEG as a spacer and 2) the use of sonication. We have discovered that even 250 nm particles can be heavily loaded with DNA and made indefinitely stable. This is a significant advance since these large nanoparticles have the potential to be loaded with several orders of magnitude more DNA strands than the smaller particles (13–30 nm) that are often used in biodiagnostic assays that rely on gold clusters as probes.
PROCEDURE
Materials
Gold nanoparticles were purchased from Ted Pella (Redding, CA). Oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA) (5′HS-spacer-ATC CTT TAC AAT ATT 6′FAM 3′, where spacer=A10, T10, or 3[(CH2CH2O)6-phosphoramidite]. Dithiothreitol (DTT) was purchased from Pierce Biotechnology, Inc. (Rockford, IL). NAP-5 columns (Sephadex G-25 DNA grade) were purchased from G. E. Healthcare (Piscatiway, NJ). Carbowax 20 M was purchased from Supelco, Inc. (Bellefonte, PA). All other salts and reagents, unless specified, were purchased from Sigma-Aldrich (St. Louis, MO). Clear 96-well plates (Costar 3696) and black shell, clear bottom 96-well plates (Costar 3603) were purchased from Corning, Inc. (Corning, NY). NANOpure® H2O (>18.0 MΩ), purified using a Barnstead NANOpure® Ultrapure water system, was used for all experiments.
Instrumentation
Absorbance measurements of oligonucleotides and gold nanoparticles were collected using a Bio-Tek Synergy HT Microplate Spectrophotometer. Fluorescence measurements were performed on a Molecular Devices Gemini EM Microplate Spectrofluorometer. All sonication was performed using a Branson 2510 sonicator.
Preparation of Alkanethiol Oligonucleotide-Modified Gold Nanoparticles
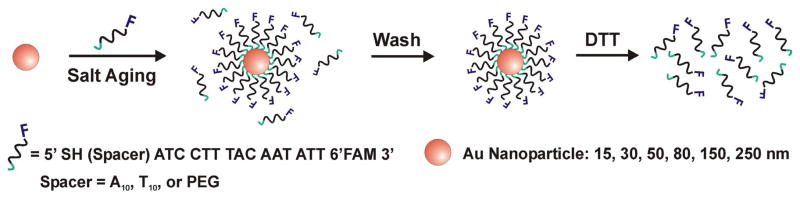
Gold nanoparticles were functionalized with fluorophore (fluorescein, 6′FAM) modified alkanethiol oligonucleotides. Prior to use, the disulfide functionality on the oligonucleotides was cleaved by addition of DTT to lyophilized DNA and incubated at room temperature for 1 hour (0.1 M DTT, 0.18 M phosphate buffer (PB), pH 8.0). The cleaved oligonucleotides were purified using a NAP-5 column. Freshly cleaved oligonucleotides were added to gold nanoparticles (1 OD/1 mL) and the concentration of PB and sodium dodecyl sulfate (SDS) were brought to 0.01 M and 0.01 % respectively. The oligonucleotide/gold nanoparticle solution was allowed to incubate at room temperature for 20 min. The concentration of NaCl was increased to 0.05 M using 2 M NaCl, 0.01 M PBS while maintaining an SDS concentration of 0.01 %. The oligonucleotide/gold nanoparticle solution was then sonicated for approximately 10 s followed by a 20 min incubation period at room temperature. This process was repeated at one more increment of 0.05 M NaCl and for every 0.1 M NaCl increment thereafter until a concentration of 1.0 M NaCl was reached. The salting process was followed by incubation overnight at room temperature. To remove excess oligonucleotides, the gold nanoparticles were centrifuged and the supernatant was removed, leaving a pellet of gold nanoparticles at the bottom. The particles then were resuspended in 0.01 % SDS. This washing process was repeated for a total of 5 supernatant removals.
Quantification of Alkanethiol Oligonucleotides Loaded on Gold Nanoparticles
To determine the number of oligonucleotides loaded on each particle, the concentration of nanoparticles and the concentration of fluorescent DNA in each sample were measured. The concentration of gold nanoparticles in each aliquot was determined by performing UV-visible spectroscopy measurements. These absorbance values were then related to the nanoparticle concentration via Beer’s Law (A=εbc). The wavelength of the absorbance maxima (λ) and extinction coefficients (ε) used for each particle size are as follows: 15 nm, λ=524 nm, ε=2.4 × 108 L/(mol*cm); 30 nm, λ=526 nm, ε=3.0 × 109 L/(mol*cm); 50 nm, ε=531 nm, ε=1.5 × 1010 L/(mol*cm); 80 nm, λ=545 nm, ε=6.85 × 1010 L/(mol*cm); 150 nm, λ=622 nm, ε=2.19 × 1011 L/(mol*cm); 250 nm, λ=600 nm, ε=5.07 × 1011 L/(mol*cm).
In order to determine the concentration of fluorescent oligonucleotides in each aliquot, the DNA was chemically displaced from the nanoparticle surface using DTT. The displacement was achieved by adding equal volumes of oligonucleotide-functionalized gold nanoparticles and 1.0 M DTT in 0.18 M PB, pH 8.0. The oligonucleotides were released into solution during an overnight incubation and the gold precipitate was removed by centrifugation. To determine oligonucleotide concentration, 100 μL of supernatant was placed in a 96-well plate and the fluorescence was compared to a standard curve. Because the 6′FAM fluorophore is sensitive to pH,25 the oligonucleotide samples for the standard curve were prepared with the same 1.0 M DTT buffer solution. During the fluorescence measurement, the fluorophore was excited at 495 nm and the emission was collected from 530 to 560 nm.
The number of oligonucleotides per particle for each aliquot was calculated by dividing the concentration of fluorescent oligonucleotides by the concentration of nanoparticles. All experiments were repeated three times using fresh samples to obtain reliable error bars.
RESULTS AND DISCUSSION
Gold nanoparticles (15, 30, 50, 80, 150, and 250 nm) were functionalized with fluorophore labeled, alkanethiol modified oligonucleotides. Nanoparticle concentration was determined using UV-vis spectroscopy. Nanoparticle-bound oligonucleotides were liberated into solution by addition of DTT,23, 26, 27 and quantified using fluorescence spectroscopy, Scheme 1.
Scheme 1.
DNA loading on Au nanoparticles and quantification of surface coverage.
Since the initial functionalization of gold nanoparticles with modified DNA in 1996,3 modifications have been made to the procedure. Recent publications have described a “fast” salt aging process where the final salt concentration of the solution is reached within several hours21 as opposed to 40 hours, as originally described. This “fast” salt aging is made possible by the addition of surfactant molecules prior to salt aging. These molecules decrease the tendency of the nanoparticles to aggregate and coalesce, particularly at high salt concentrations. Surfactants also increase the stability of larger nanoparticles (up to 250 nm) during the salt aging process.
The initial step was to ensure that these modifications did not detrimentally affect the loading of DNA on Au nanoparticles. Control experiments found DNA loading to be independent of salting rate (see Supporting Information). The presence of surfactant molecules improves reproducibility and slightly increases the DNA loading (~39 % more strands/particle at 1.0 M NaCl) since these molecules reduce sticking of the nanoparticles either to each other or to the surface of the glass vials. The type of surfactant used (SDS, Tween 20, or Carbowax) did not affect DNA loading (See Supporting Information). In addition, no change in loading was observed when varying the salt cation (Li+, Na+, K+) while keeping the Cl− anion constant (See Supporting Information). Experiments also were performed which compared DNA loading in a phosphate buffer to that in a Tris buffer, which contains molecules with a positively charged amine group. The DNA loading was found to be slightly lower (~16 % less strands/particle at 1.0 M NaCl) in the Tris buffer than in the phosphate buffer. We attribute this result to the electrostatic interactions between the positively charged Tris molecules and the negatively charged DNA backbone.28 Because of this interaction, each DNA strand occupies a larger effective volume at the surface of the nanoparticle. This added bulk decreases the amount of DNA on the particle surface since fewer DNA strands can pack into a given space (See Supporting Information). Since the goal of this study is to maximize DNA loading, all studies reported herein are performed using phosphate buffer.
Effect of Salt Concentration
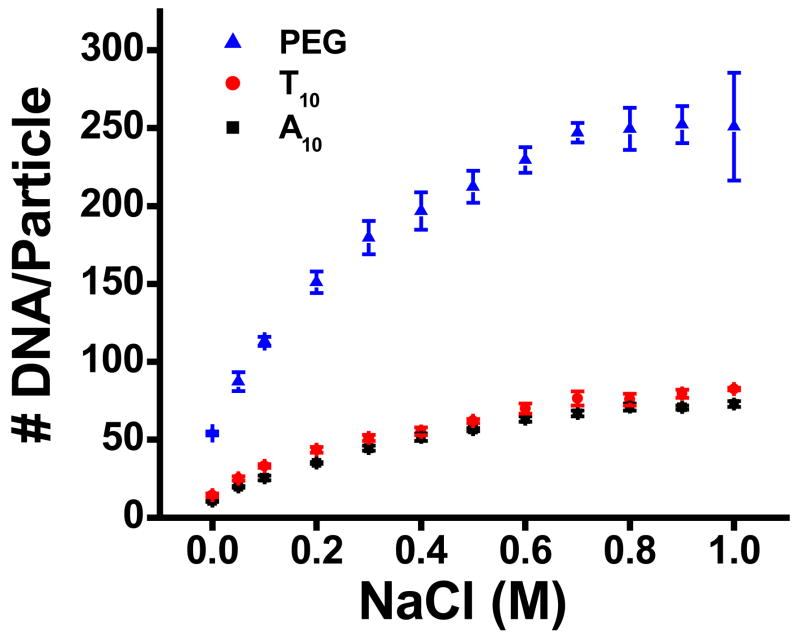
Initial loading experiments were performed to investigate DNA loading as a function of NaCl concentration (0–1.0 M), Figure 1 (15 nm nanoparticles). While higher salt concentrations were previously reported to result in higher DNA loading,29 we have extended this study to determine the salt concentration at which maximum loading is achieved. The number of DNA strands was found to increase with addition of salt and plateau between 0.7 and 1.0 M NaCl. The initial increase is attributed to the reduction of repulsive forces between the DNA strands as the concentration of counter ions is increased. At high salt concentrations (~0.7 M), maximum screening is achieved between neighboring DNA strands and the loading remains relatively constant. These results indicate that gold nanoparticle probes should be salt aged to at least 0.7 M NaCl to obtain maximum loading.
Figure 1.
DNA loading (# DNA strands/nanoparticle) vs. NaCl concentration for A10, T10, and PEG spacers on 15 nm gold nanoparticles.
Effect of Spacer Composition
We investigated the effect of three different spacers on DNA loading. Typically, probe DNA sequences are designed with a spacer region between the alkanethiol and the recognition sequence. The spacer serves the purpose of moving the recognition sequence further from the particle surface, thereby reducing steric crowding of this region during hybridization steps. The loading of DNA sequences containing an A10, T10, or PEG spacer (having roughly the same length as 10 DNA bases) was measured as a function of NaCl concentration between 0 and 1.0 M, Figure 1. To the best of our knowledge, this is the first study looking at DNA loading using a PEG spacer. Interestingly, a dramatic increase in loading was observed with DNA containing the PEG spacer (~250 DNA strands/particle) compared to the A10 and T10 spacers (~70 and ~80 DNA strands/particle, respectively). The data pertaining to DNA loading with the nucleobase spacers agrees with previous studies which found DNA containing a poly-T spacer to give higher loadings compared to DNA with a poly-A spacer.29 Similar effects from salt aging and spacer type were observed for all other nanoparticle sizes. The effect of size on DNA loading will be discussed below.
The influence of the spacer type on DNA loading can be explained by considering the interactions between neighboring spacer regions and the spacer region with the gold surface. Spacer regions composed of nucleotide bases will exhibit interstrand repulsion due to the negatively charged phosphate/sugar backbone, thus restricting DNA loading. In addition, the tendency of the DNA bases to interact with the gold will cause the DNA to partially lie on the gold surface, further reducing DNA loading. The adenine nucleobase has a much stronger relative affinity for gold than the thymine nucleobase. 30–36 As a result, DNA strands containing the A10 spacer are more likely to lie on the Au nanoparticle surface compared to DNA containing the T10 spacer. The spacer region containing the PEG chain behaves quite differently. For instance, molecules with thiolated-PEG moieties have been shown to form self-assembled monolayers (SAMs) on gold surfaces.37 Both the lack of intermolecular repulsions between neighboring PEG moieties and the decreased interactions between the PEG and the gold surface allow higher packing densities to be achieved.38, 39 Here, these factors translate to substantially higher DNA loading (~3 times more) on 15 nm gold nanoparticles for strands containing a PEG spacer compared to the nucleobase spacers, Figure 1.
Effect of Sonication
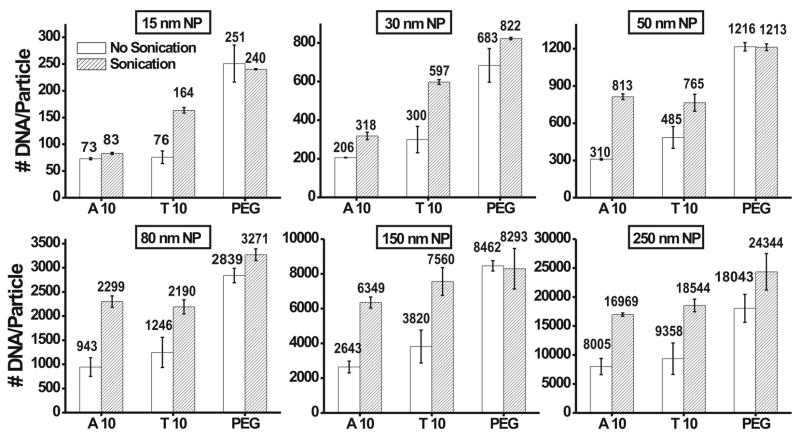
The DNA loading was further increased when the gold nanoparticles were sonicated during the salt aging process, Figure 2. On average, the loading was found to nearly double for the A10 and T10 nucleobase spacers. This increase in loading was slightly less for the A10 spacer on the smaller particles (15 and 30 nm). Very little effect was seen on the DNA loading using the PEG spacer. Similar effects of sonication were observed for all nanoparticles sizes. We believe that sonication disrupts the interactions between the DNA bases and the gold surface. This creates room for additional thiolated DNA to attach to the exposed gold nanoparticle surface, thereby increasing the DNA loading. This effect is not observed with DNA containing the PEG spacer due to the reduced affinity of PEG for gold. Control experiments revealed that increasing the duration of sonication does not substantially increase loading (see Supporting Information). In addition, nanoparticles salt aged at elevated temperatures (55 °C for 10 min after each salt addition) showed increased loading, with results comparable to those obtained with sonication (See Supporting Information).
Figure 2.
DNA loading values at 1.0 M NaCl, with and without sonication, as a function of spacer for 15, 30, 50, 80, 150, and 250 nm gold nanoparticles.
Control experiments determined that the DNA strands were not altered due to this level and duration of sonication. HPLC analysis determined that DNA strands were not chemically degraded after sonication (see Supporting Information). In addition, identical melting temperatures (Tm) were found when comparing the melting transition of the sonicated and unsonicated DNA (See Supporting Information). Also, control experiments determined that additional sonication, after the removal of excess DNA, did not cause nanoparticle bound DNA to detach (see Supporting Information). Transmission electron microscopy (TEM) reveals that the nanoparticles remain highly monodisperse after sonication and washing steps (see Supporting Information).
Effect of Nanoparticle Size
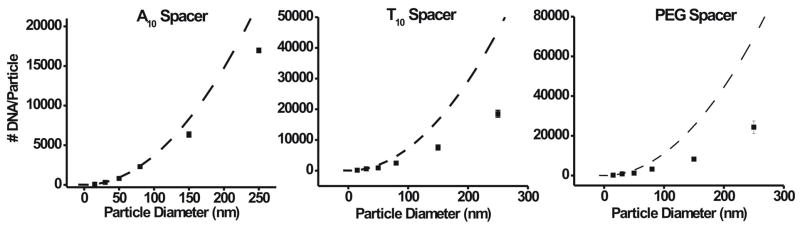
We explored the relationship between nanoparticle size and DNA loading. To the best of our knowledge, this is the first example where large gold particles150 and 250 nm in diameter have been successfully stabilized with alkanethiol modified DNA. As the nanoparticle diameter increases from 15 to 250 nm, the DNA loading increases by two orders of magnitude for all spacers, Figure 3. For example, in the case of the PEG spacer, the DNA loading on a single 15 nm nanoparticle is ~250 strands, while that on a 250 nm nanoparticle is ~25,000 strands. However, the DNA density is largest for the 15 nm particles and decreases as the particle size increases, Table 1. The dashed lines in Figure 3 represent the theoretical loading for each spacer assuming a fixed density equal to that of a 15 nm nanoparticle. The divergence from the theoretical loading density becomes more pronounced as particle size increases. This effect is due to the decrease in the curvature of the nanoparticle surface as the particle size increases. This causes the DNA strands to be closer together and intensifies interstrand repulsion. A decrease in DNA density was previously observed when comparing a high-curvature nanoparticle surface to a flat Au substrate.29
Figure 3.
DNA loading as a function of nanoparticle size at 1.0 M NaCl for DNA containing an A10, T10, or PEG spacer. The dashed line shows the theoretical values of DNA loading assuming a density fixed to that of a 15 nm nanoparticle.
Table 1.
Surface coverage of DNA on gold nanoparticles from 15–250 nm at 1.0 M NaCl after sonication for all spacers.
| A10 Spacer pmol/cm2 | T10 Spacer pmol/cm2 | PEG Spacer pmol/cm2 | |
|---|---|---|---|
| 15 nm | 19 | 38 | 56 |
| 30 nm | 19 | 35 | 48 |
| 50 nm | 17 | 19 | 26 |
| 80 nm | 19 | 20 | 27 |
| 150 nm | 15 | 18 | 19 |
| 250 nm | 14 | 16 | 21 |
The deviation of the actual loading from the theoretical loading is dependent on the type of spacer. For the 250 nm nanoparticles, the loading values for the A10, T10, and PEG spacer are 65, 60, and 26 % of the theoretical loading. When the curvature of the surface is high (smaller particles), the effective footprint of the DNA dictates the maximum density at which the DNA strands can pack. Therefore, DNA containing the PEG spacer can pack very densely. This results in a much higher theoretical loading for the PEG spacer. However, as the curvature decreases (larger particles) the repulsion between the DNA strands plays a more substantial role, and the density is less affected by spacer type.
The larger gold particles, 150 nm and above, behave differently during the salt aging possess compared to the smaller particles. Typically, when ~0.6 M NaCl is reached during the salt aging process, the larger particles precipitate out of solution. At this time, the solution turns clear, and small aggregates can be seen at the bottom of the vial. These aggregates do not redisperse with heating (up to 80 °C) but will resuspend (returning to original color) only after gentle sonication (<5 seconds). However, when the particles are resuspended in NANOpure® water or a salt buffer below ~0.3 M NaCl, sonication is no longer necessary. We attribute this reversible aggregation to an increase in screening between the particles (higher salt concentration) that allows surface interactions to dominate. However, because the particles are protected by the surface bound DNA, the particles do not coalesce to form gold aggregates and the aggregation is reversible with sonication. When the 1.0 M NaCl PBS buffer is replaced with NANOpure® water, the screening between the particles decreases and the particles redisperse.
CONCLUSION
This study investigated the effects of several parameters on the loading of thiolated-DNA on 15, 30, 50, 80, 150 and 250 nm gold nanoparticles. The intent of this investigation was to determine which parameters maximize DNA loading. In general, DNA loading can be increased by salt aging up to ~0.7 M NaCl and by using a PEG moiety as a spacer chain in place of the more common nucleobase (A or T) spacers. This is the first quantification of DNA on a nanoparticle surface wherein a PEG spacer is utilized. In most cases, the loading can be further increased by sonicating the DNA/nanoparticle solution during the salt aging process. Finally, we have shown for the first time a general method to stabilize gold nanoparticles (up to 250 nm) with DNA. These larger particles carry a substantially greater amount (~2 orders of magnitude) of DNA compared to smaller nanoparticles (13–30 nm). This study can be used as a guideline for tailoring the DNA loading on Au nanoparticles as a function of size. These findings are important for researchers developing Au nanoparticle probes for biodetection assays, nanoparticle-based therapeutics, and building blocks for novel synthetically programmable assembly strategies.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the HSARPA, NSF, AFOSR, and NIH for support of this work. CAM is grateful to the NIH for a Director’s Pioneer Award. We thank Dr. Dimitra G. Georganopolou and Aeryn R. Mayer for helpful discussions and Dr. Gabriella Métraux and Jill E. Millstone for TEM images.
References
- 1.Burda C, Chen X, Narayanan R, El-Sayed MA. Chem Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- 2.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 4.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Jr, MPB, Schultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 5.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 6.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 7.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;227:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 8.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 9.Reynolds RA, Mirkin CA, Letsinger RL. J Am Chem Soc. 2000;122:3795–3796. [Google Scholar]
- 10.Zhao X, Tapec-Dytioco R, Tan W. J Am Chem Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Wu H, Huang F, Song S, Li W, Cao Y, Fan C. Nucleic Acids Res. 2005;33:e83. doi: 10.1093/nar/gni084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weizmann Y, Patolsky F, Willner I. Analyst. 2001;126:1502–1504. doi: 10.1039/b106613g. [DOI] [PubMed] [Google Scholar]
- 13.Patolsky F, Lichtenstein A, Willner I. J Am Chem Soc. 2000;122:418–419. doi: 10.1021/ja0036256. [DOI] [PubMed] [Google Scholar]
- 14.Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536–1539. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 15.Cao YC, Jin R, Nam JM, Thaxton CS, Mirkin CA. J Am Chem Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 16.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 17.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 18.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 19.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J Am Chem Soc. 2006 doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 20.Nam JM, Stoeva SI, Mirkin CA. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 21.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Angew Chem Int Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 22.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. P Natl Acad Sci USA. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaxton CS, Hill HD, Georganopoulou DG, Stoeva SI, Mirkin CA. Anal Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 24.Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Roitberg A, Meuse C, Gaigalas AK. Spectrochim Acta, Part A. 2001;57:1781–1791. doi: 10.1016/s1386-1425(01)00408-5. [DOI] [PubMed] [Google Scholar]
- 26.Letsinger RL, Elghanian R, Viswanadham G, Mirkin CA. Bioconjugate Chem. 2000;11:289–291. doi: 10.1021/bc990152n. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Jin R, Mirkin CA, Letsinger RL. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazur S, Chen C, Allison SA. J Phys Chem B. 2001;105:1100–1108. [Google Scholar]
- 29.Demers LM, Mirkin CA, Mucic RC, Robert A, Reynolds I, Letsinger RL, Elghanian R, Viswanadham G. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 30.Demers LM, Ostblom M, Zhang H, Jang NH, Liedberg B, Mirkin CA. J Am Chem Soc. 2002;124:11248–11249. doi: 10.1021/ja0265355. [DOI] [PubMed] [Google Scholar]
- 31.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. J Am Chem Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 32.Parak WJ, Pellegrino T, Micheel CM, Gerion D, Williams SC, Alivisatos AP. Nano Lett. 2003;3:33–36. [Google Scholar]
- 33.Kryachko ES, Remacle F. J Phys Chem B. 2005;109:22746–22757. doi: 10.1021/jp054708h. [DOI] [PubMed] [Google Scholar]
- 34.Oestblom M, Liedberg B, Demers LM, Mirkin CA. J Phys Chem B. 2005;109:15150–15160. doi: 10.1021/jp051617b. [DOI] [PubMed] [Google Scholar]
- 35.Petrovyhk DY, Perez-Dieste V, Opdahl A, Kimura-Suda H, Sullivan JM, Tarlov MJ, Himpsel FJ, Whitman LJ. J Am Chem Soc. 2006;128:2–3. doi: 10.1021/ja052443e. [DOI] [PubMed] [Google Scholar]
- 36.Storhoff JJ, Elghanian R, Mirkin CA, Letsinger RL. Langmuir. 2002:18. [Google Scholar]
- 37.Pale-Grosdemange C, Simon ES, Prime KL, Whitesides GM. J Am Chem Soc. 1991;113:12–20. [Google Scholar]
- 38.Levin CS, Bishnoi SW, Grady NK, Halas NJ. Anal Chem. 2006;78:3277–3281. doi: 10.1021/ac060041z. [DOI] [PubMed] [Google Scholar]
- 39.Latham AH, Williams ME. Langmuir. 2006;22:4319–4326. doi: 10.1021/la053523z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.