Abstract
We have designed a chip-based assay, using microarray technology, for determining the relative binding affinities of duplex and triplex DNA binders. This assay combines the high discrimination capabilities afforded by DNA-modified Au nanoparticles with the high-throughput capabilities of DNA microarrays. The detection and screening of duplex DNA binders are important because these molecules, in many cases, are potential anticancer agents as well as toxins. Triplex DNA binders are also promising drug candidates. These molecules, in conjunction with triplex forming oligonucleotides, could potentially be used to achieve control of gene expression by interfering with transcription factors that bind to DNA. Therefore, the ability to screen for these molecules in a high-throughput fashion could dramatically improve the drug screening process. The assay reported here provides excellent discrimination between strong, intermediate, and weak duplex and triplex DNA binders in a high-throughput fashion.
Keywords: DNA binding molecules, colorimetric screening, high-throughput, DNA-modified gold nanoparticles
JUSTIFICATION
Small molecules that can bind to duplex and triplex DNA are important for a variety of reasons. Typically, the association of either duplex or triplex DNA with a binding molecule results in a more stabile complex relative to the duplex or triplex alone. When binding molecules are introduced into biological systems, they form DNA/small molecule complexes which exhibit increased stability compared to free DNA, and can induce cell death. The toxicity is a direct result of the small molecules interfering with the cells ability to interact with the DNA. As a consequence, small molecules with DNA binding properties are also potential anti-cancer drugs and gene regulation agents. Therefore, the ability to screen for duplex and triplex DNA binding molecules is an important step in the drug discovery process. In this manuscript, we present a high-throughput assay which is designed to screen for DNA binding molecules. The design of this assay allows for duplex and triplex binders to be easily distinguished. Importantly, this assay combines the unique properties of DNA-modified Au nanoparticles, which provide excellent discrimination between weak, intermediate, and strong DNA binders, with the high-throughput capabilities of microarray technology. This method provides benefits over fluorescence-based methods because it does not suffer from signal interference and provides improved discrimination between binders of different strengths.
INTRODUCTION
Since the initial development of Au nanoparticle probes through the covalent attachment of thiol-capped oligonucleotides to citrate stabilized gold nanoparticles in 19961, these composite nanomaterials have become the central component in several PCR-less DNA2–9 and protein detection assays10–12. These assays benefit from the unique properties of these materials which are not displayed by either free DNA, Au nanoparticles without DNA, or Au nanoparticles that have been modified with oligonucleotides using different coupling schemes that lead to less efficient oligonucleotide loading. These properties include: 1) enhanced binding properties which directly lead to increased sensitivity13 and 2) sharp DNA melting transitions which dramatically increase the level of selectivity of DNA binding2. In addition, the intense optical properties of the Au nanoparticles provide a strong signal by which to monitor the melting behavior of DNA, which has a relatively weaker spectroscopic signature. The combination of these properties has led to assays with impressive analytical capabilities with respect to sensitivity, selectivity, and assay readout for both nucleic acid and protein analytes.
Recently, DNA-modified Au nanoparticles have been used for the colorimetric detection of duplex14 and triplex15 DNA binding molecules.16, 17 Researchers are interested in probing the interactions between small molecules and DNA in order to obtain leads for new anti-cancer and gene regulation agents. Many duplex DNA binders such as doxorubicine, daunorubicin and amsacrine have been identified and used as anticancer drugs.18 They are known to reversibly interact with duplex DNA to form a drug/DNA complex, and by doing so disrupt cellular replication. Triple helix specific DNA binding molecules, or triplex binders, are also promising drug candidates. These molecules, in conjunction with triplex forming oligonucleotides, could potentially be used to achieve control of gene expression by interfering with transcription factors that bind to DNA.19–21
The recently developed DNA-modified Au nanoparticle-based colorimetric screening methods display excellent discrimination between weak, intermediate and strong duplex and triplex DNA binders16, 17. However, with the development of combinatorial chemistry where many compounds can be synthesized within a short time period,22, 23 high-throughput screening techniques are necessary to screen large libraries for potential drug candidates.24–26 In the past, typical screening processes have included UV-vis melting experiments, competitive dialysis, and mass spectroscopy.21, 27–29 Unfortunately, these methods are not applicable to high-throughput screening. Only recently have protocols been developed which are compatible with high-throughput screening techniques.16, 17, 30, 31 Herein, we present a new high-throughput technique, based on microarray technology, which incorporates the high discrimination capabilities of DNA-modified Au nanoparticles for the screening of duplex and triplex DNA binding molecules.
These assays are based on a perturbation of the DNA melting temperature (Tm) induced by the presence of DNA binders. Strong DNA binders result in a greater increase in Tm whereas weaker DNA binders have very little effect on the Tm. Since DNA functionalized Au nanoparticles have such an intense optical signature, one can work at higher ratios of potential binder:probe DNA, which leads to larger absolute changes in Tm as compared with conventional UV-vis approaches. This increased perturbation expands the temperature window that can be used to screen for duplex and triplex DNA binders. Generally, the strength of binding between the drug and target molecule correlates with the drug’s biological activity and, therefore, it is important to obtain a quantitative measure of the difference in binding capabilities of the molecules that make up a particular library.32 The authors note that there are a few cases where strong DNA binding molecules do not increase the Tm associated with the bound DNA.33 However, this is an uncommon characteristic, and the high-throughput screening assay described here is directed towards screening the more general DNA binders which make up the majority of these molecules and not special cases.
The assay described herein relies on scanometric detection and the use of a commercial reader, the Verigene ID.4 In a typical experiment, synthetic alkyl amine functionalized oligonucleotides are arrayed on a glass chip via literature procedures (see Methods). Gold nanoparticles, functionalized with a complementary DNA sequence, are hybridized to the chip. A sample holder with 10 different wells that reside over the microarrays is attached to the microscope slide with the arrays. Each well is filled with an aqueous solution containing a different potential DNA binder. After hybridization, the probes are developed with a silver enhancement solution, which increases the size and light scattering of the Au nanoparticle probes, Scheme 1 and 2. The amount of light scattering from each spot within the microarrays is measured using a Verigene ID scanner. To elucidate strong, intermediate and weak DNA binders, the incubation process with the DNA binder is performed under more stringent conditions by increasing the temperature or decreasing the salt concentration of the solution. As conditions become more stringent, the DNA interconnects require DNA binders to increase the stability of the linking DNA in order for the Au nanoparticles to remain intact on the chip surface. The dehybridization of the Au nanoparticles on the chip surface directly correlates with a reduction of signal post silver enhancement, thereby allowing one to differentiate DNA binders of different strengths. To demonstrate this technique, we chose seven known duplex DNA binders (4′,6-diamidino-2-phenylindole (DAPI), ellipticine (EIPT), amsacrine (AMSA), daunorubicin (DNR), anthraquinone-2-carboxylic acid (AQ2A), ethidium bromide (EB), and 9-aminoacridine (9-AA)). The chosen duplex binders represent a range of binding strengths and include both groove binders (DAPI, DNR) and intercalators (EIPT, AMSA, AQ2A, EB, and 9-AA). Since only a few triplex binders are known, our choices of triplex binders were limited and include coralyne (CORA), a weak binder, and benzo[e]pyridoindole (BePI), a strong binder.
Scheme 1.
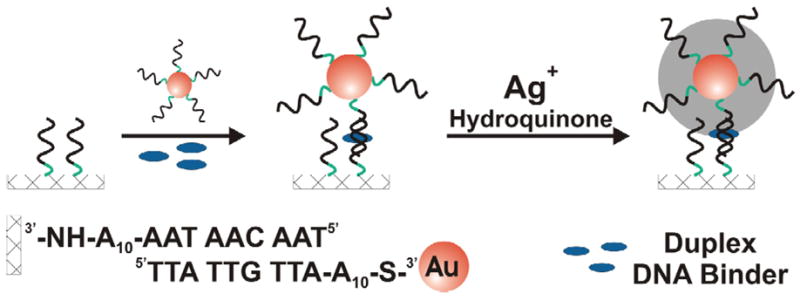
scanometric detection of duplex DNA binders on a microarray.
Scheme 2.
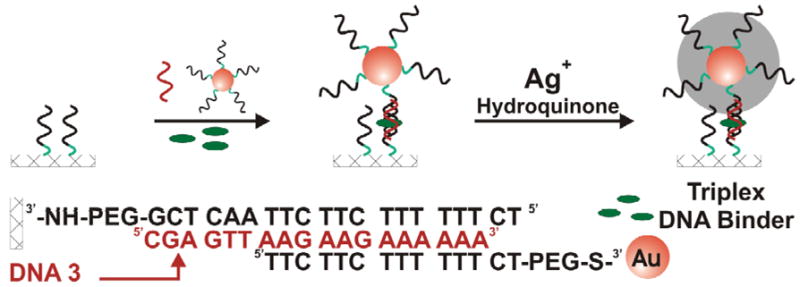
Scanometric detection of triplex DNA binders on a microarray.
EXPERIMENTAL SECTION
All oligonucleotides used in this paper were purchased from Integrated DNA Technologies. Ellipticine was purchased from Biomol (Plymoth Meeting, PA). Coralyne was purchased from Acros Organics (Geel, Belgium). All other chemicals and buffer solutions were prepared using reagents purchased from Sigma-Alrdich (St. Louis, MO).
DNA Modification of Gold Nanoparticles
Au nanoparticles (13 nm diameter), prepared by citrate reduction of HAuCl4 were used to prepare all probes.34 Gold nanoparticles were functionalized by derivatizing aqueous Au colloid with 3′-alkylthiol-modified oligonucleotides (final concentration of oligonucleotides ~4 μM). After 12 hours, the phosphate concentration was increased from 0 to 10 mM by adding 1.0 M pH 7 phosphate buffer (NaH2PO4/Na2HPO4). This was followed by a series of salt additions in order to slowly increase the loading of the oligonucleotides. Initially, 2 M NaCl was added dropwise to increase the salt concentration to 0.05 M NaCl. The solution was allowed to stand for 6–8 h. This process was repeated to increase the salt concentration to 0.1, 0.2 and 0.3 M NaCl. To remove excess thiol-DNA, the solution was centrifuged using 1.5 mL Eppendorf tubes (Fisher Scientific) (30 min at 15 000 rpm). Following removal of the supernatant, the precipitate was suspended in NANOpure® water. This process was repeated five times with a final resuspension in 0.5 M NaCl buffer (10 mM PBS, 0.001 % NaN3).
Determination of Melting Temperature of Control DNA (No Nanoparticles)
To determine the Tm of the duplex DNA in the presence of DNA binders, duplex DNA and a DNA binder were combined in 0.3 M NaCl 10 mM PBS to final concentrations of 2 μM and 5 μM, respectively. To determine the Tm of the triplex DNA in the presence of DNA binders, triplex DNA (1:1:1 each DNA strand) and a DNA binder were combined in 0.5 M NaCl 10 mM PBS to final concentrations of 1 μM and 6 μM, respectively. UV-vis melting curves were measured on a CARY 500 spectrophotometer equipped with a circulating bath. The melting process was monitored by measuring the change in extinction at 260 nm. The solutions were stirred continuously with a magnetic stir bar as the solution temperature was increased at a rate of 0.5 °C/min. The first derivative generated from the melting curve was used to determine the Tm.
Preparation of DNA Chip Array
N-Hydroxysuccinimide-activated Codelink glass microarray slides (Amersham, G. E. Healthcare) were spotted with alkylamine-modified oligonucleotides (Duplex Array:5′-TAA CAA TAA-A10-NH2-3′, Triplex Array: 5′-TC TTT TTT CTT CTT AAC TCG-PEG-NH2-3′, Integrated DNA Technologies, Inc.) according to the slide manufacturer’s protocol. The oligonucleotides were printed in triplicate using a GME 418 robotic pin-and-ring microarrayer (Affymetrix).
Scanometric Detection of Duplex and Triplex DNA Binding Molecules
Glass slides containing the microarrays were fitted with sample chambers (Nanosphere, Inc.), creating 10 individually addressable wells. Gold nanoparticles were added to each well (1nM) in 10 mM PBS with varying concentrations of NaCl (0.1–0.3 M). For detection of triplex binders, 150 nM DNA-3 was added. DNA binding molecules (5 μM) were added to the sample wells. Duplex DNA binders included 4′,6-diamidino-2-phenylindole (DAPI), ellipticine (EIPT), amsacrine (AMSA), daunorubicin (DNR), anthraquinone-2-carboxylic acid (AQ2A), ethidium bromide (EB), and 9-aminoacridine (9-AA). Triplex DNA binders were coralyne (CORA) and benzo[e]pyridoindole (BePI). Incubation with the DNA binders was allowed to proceed for 30 min for duplex DNA binders at a specified temperature and 1 hour for triplex DNA binders at 25 °C. After hybridization, the slides were washed three times with 0.5 M NaNO3 containing 0.02% Tween 20. The microarrays were dried using a benchtop spinner. Gold probes were developed with silver enhancement solution (Ag+/Hydroquinone, Nanosphere, Inc.) (1 mL/chip, 1.5-min development time for duplex DNA binders and 3 min for triplex binders). The Au nanoparticles catalyze the reduction of silver, resulting in silver deposition around the nanoparticles. The reaction was terminated by washing the arrays with NANOpure® water followed by spin-drying. The light scattering from the silver enhancement is then measured by a high-resolution Verigene ID scanner (Nanosphere, Inc.).
RESULTS
Scanometric detection of duplex DNA binders was performed at different temperatures, Figure 1. At 25 °C, all seven binders and the control displayed signal after silver development. Performing the assay at 30 °C resulted in signal from only arrays incubated with DAPI (strong binder), DNR and EB (intermediate binders). Increasing the assay temperature further to 35 °C resulted in signal from only DAPI, a strong duplex binder. Scanometric detection of triplex DNA binders was performed at varying salt concentrations, Figure 2. At 0.3 M NaCl, signal was obtained from both triplex binders, BePI and CORA, while no discernable signal was obtained from the duplex DNA binders. Reducing the salt concentration to 0.2 and 0.1 M NaCl resulted in signal from only BePI, a strong triplex binder.
Figure 1.
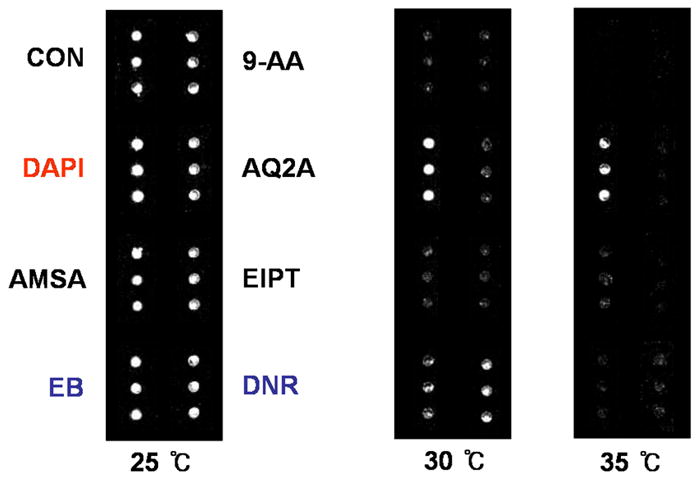
Light scattering image of a microarray post silver development. The spots are scattered light signifying duplex DNA binding molecules.
Figure 2.
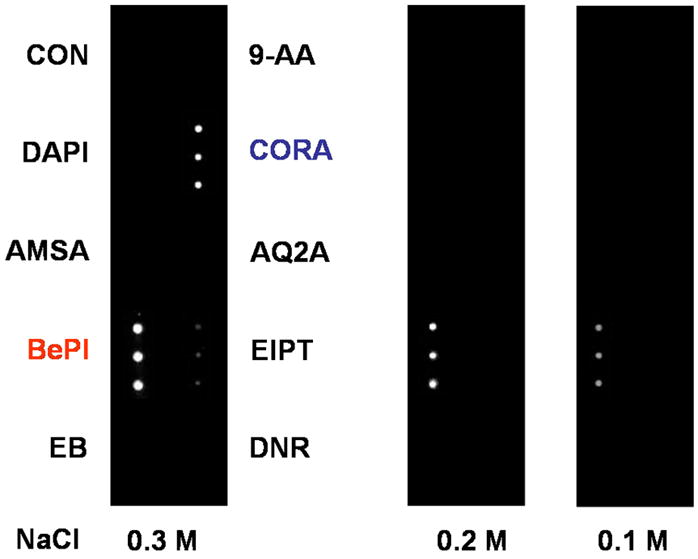
Light scattering image of a microarray post silver development. The spots are scattered light signifying triplex DNA binding molecules. Experiments were performed at 25 °C.
Control experiments with free DNA (no nanoparticles) were performed to verify the trend of DNA binding strength found using the scanometric method. Melting temperatures are shown for the duplex and triplex binders in Table 1 and Table 2, respectively.
Table 1.
Melting temperatures of solution-phase unmodified DNA in the presence of duplex DNA binders: 4′,6-diamidino-2-phenylindole (DAPI), ellipticine (EIPT), Amsacrine (AMSA), daunorubicin (DNR), anthraquinone-2-carboxylic acid (AQ2A), ethidium bromide (EB), and 9-aminoacridine (9-AA)).
| Duplex Binders | Tm (°C) |
|---|---|
| Control (No DNA Binder) | 19.1 |
| DAPI | 27.8 |
| DNR | 25.4 |
| EB | 21.8 |
| EIPT | 20.5 |
| AMSA | 19.5 |
| AQ2A | 19.4 |
| 9-AA | 19.3 |
Table 2.
Melting temperatures of solution-phase unmodified DNA in the presence of triplex DNA binders: coralyne (CORA) and benzo[e]pyridoindole (BePI) and duplex DNA binders: 4′,6-diamidino-2-phenylindole (DAPI), ellipticine (EIPT), Amsacrine (AMSA), daunorubicin (DNR), anthraquinone-2-carboxylic acid (AQ2A), ethidium bromide (EB), and 9-aminoacridine (9-AA). Tm3-2 indicates a melting transition from a triplex to a duplex. Tm2-1 indicates a melting transition from a duplex to single stranded DNA.
| Compounds | Tm3-2 (°C) | Tm 2-1 (°C) |
|---|---|---|
| Control | ND | 58.9 |
| 9-AA | ND | 59.1 |
| AQ2A | ND | 59.1 |
| AMSA | ND | 59.2 |
| EIPT | ND | 59.3 |
| EB | ND | 59.8 |
| CORA | 17.1 | 60.1 |
| BePI | 34.8 | 61.4 |
| DNR | ND | 61.7 |
| DAPI | ND | 66.3 |
DISCUSSION
Detection of Duplex DNA Binders
Capture strands of alkylamine-modified oligonucleotides were spotted on a glass chip using a microarrayer. The microarray was then exposed to DNA-modified Au nanoparticles (complementary to the capture sequence) followed by addition of a duplex DNA binder. The nanoparticle-bound DNA hybridizes to the capture strand and forms a duplex, simultaneously immobilizing the nanoparticle on the array and providing a binding site for the duplex DNA binder, Scheme 1. The array was washed with buffer and the gold probes are developed with silver enhancement solution and the signal is measured with a Verigene ID.
The hybridization of complementary DNA is a reversible process and the stability of the resulting duplex is affected by several factors such as temperature and salt concentration. In addition, the presence of duplex DNA binders increases the stability of the DNA duplex and is reflected by an increase in the duplex Tm. The strength of binding of the duplex DNA binder is represented by the magnitude of increased Tm.32 Therefore, by performing the nanoparticle hybridization process in the presence of different duplex DNA binders as a function of temperature, the relative binding strength of duplex DNA binders can be determined, Figure 1.
Analysis of seven known duplex DNA binders was performed to demonstrate the ability of this method to discriminate between strong, intermediate and weak binders. At 25 °C, all seven duplex DNA binders and a control showed positive signal after silver development. Increasing the temperature to 30 °C during the hybridization process eliminates signal from the control and the weak duplex DNA binders, leaving the intermediate (EB and DNR) and strong (DAPI) DNA binders. Further increasing the temperature eliminates the signal from the intermediate strength binders, leaving only the strong duplex DNA binder, DAPI.
These results correlate well with control experiments which measure the Tm of free solution-phase DNA in the presence of the seven duplex DNA binders, Table 1. The relative trend in binding strength determined from the control experiments is DAPI > DNR > EB > other intercalators. This is in agreement with the trend in relative binding strengths determined by the chip-based scanometric method.
Detection of Triplex DNA Binders
Capture strands of alkylamine-modified oligonucleotides are patterned on a glass chip using a microarrayer. The microarray is then exposed to a solution containing three components: 1) complementary DNA (no nanoparticles), 2) DNA functionalized Au nanoparticles (DNA is non-complementary to the arrayed DNA) and 3) DNA binders, Scheme 2. The complementary solution-phase DNA, called DNA-3 in Scheme 2, hybridizes to the microarray and forms a duplex. The Au nanoparticle-bound DNA has the correct sequence to form a triplex with the surface bound duplex through sequence-specific Hoogsteen base pairing.35 The triplex structure is relatively unstable at room temperature and will only form in the presence of triplex DNA binding molecules. The presence of the triplex DNA binding molecules increases the stability of the triplex, immobilizing the Au nanoparticles onto the microarray. The array was then washed with buffer and the gold probes were developed with silver enhancement solution. The light scattering from the microarray was measured using a Verigene ID scanner.
The assay was performed in the presence of two triplex binders, seven duplex DNA binders, and a control (no DNA-3). Similar to the DNA duplex, the formation of the DNA triplex is a reversible process which is affected by both temperature and salt concentration. However, compared to the duplex, the stability of the triplex is much more sensitive to salt concentration.36 Therefore, to determine the relative binding strength of the triplex DNA binders, the assay was performed at room temperature at varying salt concentrations, Figure 2. At 0.3 M NaCl, both the strong (BePI) and weaker (CORA) triplex binders show signal. At decreased salt concentrations (0.2 and 0.1 M NaCl), the stability of the triplex is decreased and signal is only seen in the presence of the stronger triplex binder (BePI). No signal is seen in the presence of the duplex DNA binders indicating that the assay is specific for identifying triplex DNA binders.
These results correlate well with control experiments which measure the Tm of free solution-phase DNA in the presence of the two triplex DNA binders and the seven duplex DNA binders, Table 2. A melting transition associated with the triple helix was found only for experiments performed in the presence of BePI (34.8 °C) and CORA (17.1 °C) where BePI is determined to be the stronger binder. This is in agreement with the results determined by the scanometric method described above.
In conclusion, we have designed a microarray assay for screening the relative binding discrimination capabilities afforded by the DNA modified Au nanoparticles with the affinities of duplex and triplex DNA binders. This assay combines the high high-throughput capabilities of DNA microarrays. The detection and screening of duplex DNA binders are important because these molecules, in many cases, are potential anticancer agents as well as toxins. Triplex DNA binders are also promising drug candidates. These molecules, in conjunction with triplex forming oligonucleotides, could potentially be used to achieve control of gene expression by interfering with transcription factors that bind to DNA. Therefore, the ability to screen for these molecules in a high-throughput fashion could dramatically improve the drug screening process.
Acknowledgments
This work was supported by the NCI through a CCNE, the NSF, and the AFOSR. CAM is grateful for a Director’s Pioneer Award. MSH is grateful for a fellowship from the Korea Research Foundation.
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 3.Nam JM, Stoeva SI, Mirkin CA. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 4.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, Hosokawa K, Maeda M. J Am Chem Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- 6.He L, Musick MD, Nicewarner SR, Salinas FG, Stephen J, Benkovic, Natan MJ, Keating CD. J Am Chem Soc. 2000;122:9071–9077. [Google Scholar]
- 7.Maxwell DJ, Taylor JR, Nie S. J Am Chem Soc. 2002;124:9609–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 8.Su M, Li S, Dravid VP. Appl Phys Lett. 2003;82:3562–3564. [Google Scholar]
- 9.Niazov T, Pavlov V, Xiao Y, Gill R, Willner I. Nano Lett. 2004;4:1683–1687. [Google Scholar]
- 10.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 11.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 12.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 13.Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 14.Ihmels H, Otto D. Supermolecular Dye Chemistry. 2005;58:161–204. [Google Scholar]
- 15.Strekowski L, Say M, Zegrocka O, Tanious FA, Wilson WD, Manzel L, Macfarlane DE. Bioorganic & Medicinal Chemistry. 2003;11:1079–1085. doi: 10.1016/s0968-0896(02)00525-4. [DOI] [PubMed] [Google Scholar]
- 16.Han MS, Lytton–Jean AKR, Oh B-K, Heo J, Mirkin CA. Angew Chem Int Ed. 2006;45:1807–1810. doi: 10.1002/anie.200504277. [DOI] [PubMed] [Google Scholar]
- 17.Han MS, Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2006;128:4954–4955. doi: 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XL, Wang AHJ. Pharm Therap. 1999;83:181–215. doi: 10.1016/s0163-7258(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 19.Praseuth D, Guieysse AL, Hélène C. Biochim Biophys Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 20.Haq I, Ladbury J. J Mol Recognit. 2000;13:188–197. doi: 10.1002/1099-1352(200007/08)13:4<188::AID-JMR503>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Mergny JL, Duval-Valentin G, Nguyen CH, Perrouault L, Faucon B, Rougée M, Montenay-Garestier T, Bisagni E, Hélène C. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 22.Thompson LA, Ellman JA. Chem Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 23.Balkenhohl F, von dem Bussche-Hünnefeld C, Lansky A, Zechel C. Angew Chem Int Ed. 1996;35:2288–2337. [Google Scholar]
- 24.Johnston PA, Johnston PA. Drug Discovery Today. 2002;7:353–363. doi: 10.1016/s1359-6446(01)02140-7. [DOI] [PubMed] [Google Scholar]
- 25.Boger DL, Desharnais J, Capps K. Angew Chem Int Ed Engl. 2004;42:4138–4176. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Sim TB, Kim YS, Chang YT. Curr Opin Chem Biol. 2004;8:371–377. doi: 10.1016/j.cbpa.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Silver GC, Sun J-S, Nguyen CH, Boutorine AS, Bisagni E, Hélène C. J Am Chem Soc. 1997;119:263–268. [Google Scholar]
- 28.Rosu F, De Pauw E, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou JF, Mergny JL. Biochemistry. 2003;42:10361–10371. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- 29.Chaires JB, Ren J, Hamelberg D, Kumar A, Pandya V, Boykin DW, Wilson WD. J Med Chem. 2004;47:5729–5742. doi: 10.1021/jm049491e. [DOI] [PubMed] [Google Scholar]
- 30.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J Am Chem Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 31.Darby RAJ, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonini I, Polucci P, Jenkins TC, Kelland LR, Menta E, Pescalli N, Stefanska B, Mazerski J, Martelli S. J Med Chem. 1997;40:3749–3755. doi: 10.1021/jm970114u. [DOI] [PubMed] [Google Scholar]
- 33.Willis B, Arya DP. Biochemistry. 2006;45:10217–10232. doi: 10.1021/bi0609265. [DOI] [PubMed] [Google Scholar]
- 34.Freeman RG, Hommer MB, Grabar KC, Jackson MA, Natan MJ. J Phys Chem. 1996;100:718–724. [Google Scholar]
- 35.Fox KR. Curr Med Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 36.Rougée M, Faucon B, Mergny JL, Barcelo F, Giovannangeli C, Garestier T, Hélène C. Biochemistry. 1992;31:9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]


