Abstract
High macromolecular concentrations, or crowded conditions, have been shown to affect a wide variety of molecular processes, including diffusion, association and dissociation, and protein folding and stability. Here, we model the effect of macromolecular crowding on the internal dynamics of a protein, HIV-1 protease, using Brownian dynamics simulations. HIV-1 protease possesses a pair of flaps which are postulated to open in the early stages of its catalytic mechanism. Compared to low concentrations, close packed concentrations of repulsive crowding agents are found to significantly reduce the fraction of time that the protease is open. Macromolecular crowding is likely to have a major effect on in vivo enzyme activity, and may play an important regulatory role in the viral life cycle.
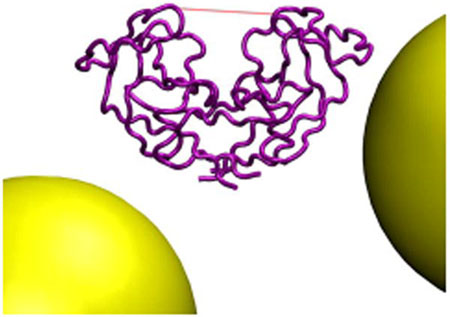
The aspartic protease of human immunodeficiency virus type 1 (HIVpr) possesses a pair of flaps which are postulated to open in the early stages of its catalytic mechanism (Figure 1). Here, we study the effect of high macromolecular concentrations, or crowded conditions, on the internal dynamics of HIVpr. Compared to low concentrations, high concentrations of repulsive crowding agents are found to significantly reduce the fraction of time that the protease flaps are open. Since flap dynamics are likely to have a major effect on in vivo enzyme activity, macromolecular crowding may play an important regulatory role in the viral life cycle.
Figure 1.
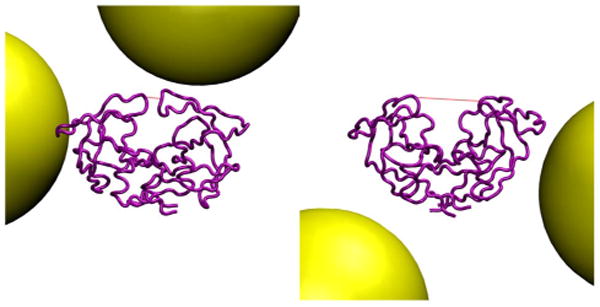
HIVpr in closed and open conformations, in the presence of crowder molecules at 5% concentration by volume. The lines indicate flap tip distances of 5.2 Å (left) and 21.9 Å (right).
Macromolecular crowding has been seen to affect the equilibria and rates of many molecular processes, including diffusion,1 association and dissociation,2 and protein folding3 and stability.4 These effects can be qualitatively studied through polymer chain models,5 with the understanding that nonspecific interactions between macromolecules reduce the available volume of a particle in solution.4 Using a mean-field approximation, macromolecular crowding can be modeled by confinement;6 the qualitative effects are assumed to be the same.
Computer simulations can add realism to the theory of macromolecular crowding.7 Crowding effects on the free energy of protein escape from a chaperonin cage have been studied using Brownian dynamics simulations.8 Coarse-grained molecular dynamics simulations have been used to study crowding effects on the protein native state stability and folding kinetics.9 Here, we report the first, to our knowledge, simulation of crowding effects on internal protein dynamics.
We study HIVpr, an essential viral enzyme that is the target of many antiretroviral drugs. HIVpr is a homodimer which possesses a pair of β-hairpin flaps, one from each monomer, that cover the enzyme active site in most extant crystal structures. The opening and closing of the flaps is thought to be involved in substrate/inhibitor binding and the enzymatic mechanism.
Nuclear magnetic resonance measurements indicate that the flaps are very mobile.10 Flap motions have been studied by atomistically detailed molecular dynamics (MD) simulations11 and accelerated MD12 simulations. Our coarse-grained model has shown bunches of opening and closing events on the nanosecond timescale separated by longer closing times.13
Starting with coordinates from protein data bank entry 1HHP, we treat amino acid interactions by a statistically parameterized coarse grained potential in which each amino acid residue is represented by a single bead.13 Crowder agents are modeled as spheres which interact with each other and with amino acids by the pairwise potential E = ε(σij/deff)12, where σij = (σi + σj), the van der Waals radii of the respective species, ε is 0.6 kcal/mol, and deff is the effective distance between them.
Crowding agents have been modeled as “spheres of spheres” - the surface of a large sphere, covered with many evenly spaced smaller spheres.8 As we consider “spheres of spheres” to be a more realistic model for amino acids on the surface of a globular protein than a single repulsive sphere, we mimic this effect by using the following potential for crowders: deff = d − (rcwd-σcwd), where d is the distance between species, rcwd is the radius of the crowding agent (chosen to be 30 Å, which is slightly larger than the longest axis of HIVpr), and σcwd is the radius of the crowding agent surface particle (7 Å, the average size of an amino acid bead in our coarse-grained model). This model effectively places a surface particle directly between a crowding agent and its interaction partner (Figure 2).
Figure 2.
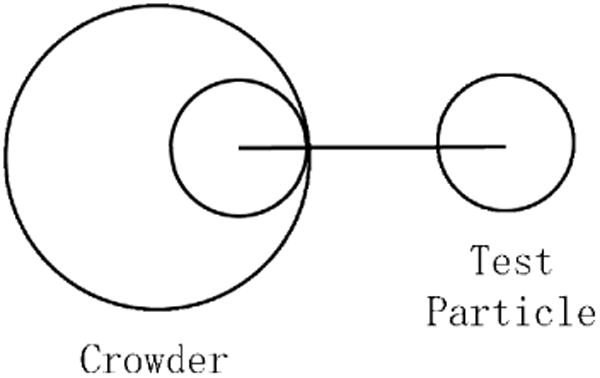
Schematic of crowder potential.
One HIVpr molecule is placed in the center of a periodic cube with a side length of 170 Å. Simulations are run at three crowder concentrations: 0%, 5%, and 68% by volume. At 5% crowding, the agents are randomly added in a serial fashion while avoiding steric clashes with particles in solution. For 68% crowding, a face-centered cubic lattice was generated and particles in contact with the central HIVpr molecule were removed. Brownian dynamics simulations were performed using a modified version of UHBD,14 with a time step of 0.05 picoseconds. Diffusion coefficients were assigned to individual amino acid beads and crowding agents by the Einstein-Stokes equation, D = kbT/6πηrh, where kb is Boltzmann's constant, T is the temperature, η is the viscosity (1 cP for water), and rh is the hydrodynamic radius. The hydrodynamic radius is defined as the particle radius plus a 1.4 Å water solvation shell. After 1 microsecond of equilibration, eight simulations were run under each concentration, spanning 20, 25, and 20 microseconds, respectively.
Flap tip distances – the distances between glycine 51 of each monomer – were output every 200 picoseconds. Flaps were considered open when this distance was beyond 10 Å. Our simulations show that flap opening is significantly suppressed at high crowder concentrations (Figure 3). At 0%, 5%, and 68% crowding, the fraction of open conformations was 0.078±0.018, 0.087±0.025, and 0.019±0.006, respectively.
Figure 3.
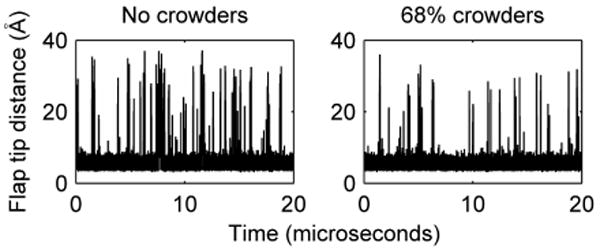
Representative plots of flap tip distance versus simulation time.
A metric for the stability of a conformational state is the decay coefficient. In a first-order decay process, S(Δt)=e-kΔt, where Δt is the elapsed time, S(Δt) is the survival probability, and k is the decay coefficient. The survival probability can be estimated from flap tip distance trajectories by a constructing a histogram of open state dwell times. Using a least squares linear regression of Δt vs. log(S(Δt)), first-order decay coefficients of the HIVpr open state from our trajectories at 0%, 5%, and 68% crowder concentrations were calculated to be 0.046, 0.039, and 0.1029 ns-1, respectively. The increase in the decay coefficient at 68% concentration indicates that the open state is less stable.
The decrease in open state fraction and stability is correlated with an increase in collision frequency. A collision was defined a reduction in the distance between a crowder and an amino acid bead to within 34 Å. At 5% and 68% crowder concentrations, simulated collision frequencies were 0.0013±0.0002 and 0.1893±0.0015 ns-1. The increased collision frequency between the molecule and surrounding crowding agents are the likely cause of changes in HIVpr internal dynamics.
With a significant change in the fraction of open conformations, our results suggest that macromolecular crowding plays an important role in the activity of HIVpr. Under physiological conditions, the concentration of crowding species can be expected to be very high.15 Our recent theoretical analysis suggests that HIVpr ligand binding kinetics depend on the flap opening fraction; computed gated association rates yield good agreement with experimental results.16 The effects of macromolecular crowding on suppressing the protease open state and reducing enzyme-ligand diffusional encounter rates likely combine to slow in vivo enzymatic activity.
The effect of macromolecular crowding on HIVpr flap dynamics, and thereby enzymatic activity, is a potentially important regulatory mechanism. The function of HIVpr in the viral life cycle is to cleave the Gag and Gag-pol polyproteins,17 and wild-type HIVpr is also subject to autoproteolysis. Premature cleavage of polyprotein products in the cellular cytoplasm may preclude subcellular localization of viral species necessary for budding. In the budding virion, however, crowding effects should be offset by the high concentration of substrate species among the surrounding molecules.18
We hope that our study will inspire experimental work along the same vein and anticipate that our methods will be applied to other systems to help elucidate the role of crowding in many processes.
Acknowledgments
DDLM thanks L. Makowski and D. Rodi for discussions about crowding experiments, and R. Konecny for assistance with computing resources, and an anonymous reviewer for suggesting the collision frequency calculation. Support has been provided by the NSF (MCB-0506593 for JAM), NIH (GM31749 for JAM), HHMI, CTBP, NBCR, W.M. Keck Foundation, and Accelrys, Inc. DDLM was supported by the NIH Molecular Biophysics Training Grant at UCSD. JT was supported by the Ministry of Education and Science (115/E-343/ICM/BST-1076/2005) and by European CoE MAMBA.
References
- 1.(a) Maramatsu N, Minton AP. Proc Natl Acad Sci USA. 1988;85:2984–2988. doi: 10.1073/pnas.85.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Weiss M, Elsner M, Kartberg F, Nilsson T. Biophys J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. J Am Chem Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 3.Martin J, Hartl F. Proc Natl Acad Sci USA. 1997;94:1107–1112. doi: 10.1073/pnas.94.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tokuriki N, Kinjo M, Negi S, Hoshino M, Goto Y, Urabe I, Yomo T. Prot Sci. 2004;13:125–133. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ai X, Zhou Z, Bai Y, Choy W. J Am Chem Soc. 2006;128:3916–3917. doi: 10.1021/ja057832n. [DOI] [PubMed] [Google Scholar]
- 4.Minton AP. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 5.Dill KA, Bromberg S. Molecular Driving Forces. Garland Science; New York: 2003. pp. 629–642. [Google Scholar]
- 6.Thirumalai D, Klimov DK, Lorimer GH. Proc Natl Acad Sci USA. 2003;100:11195–11197. doi: 10.1073/pnas.2035072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Hall D, Minton AP. Biochim Biophys Acta. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]; (b) Zhou H. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 8.Elcock A. Proc Natl Acad Sci USA. 2003;100:2340–2344. doi: 10.1073/pnas.0535055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Friedel M, Sheeler DJ, Shea JJ. Chem Phys. 2003;118:8106–8113. [Google Scholar]; (b) Cheung MS, Klimov D, Thirumalai D. Proc Natl Acad Sci USA. 2005;102:4653–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich E, Louis JM, Torchia DA. Prot Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Scott WR, Schiffer CA. Struct Fold Des. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]; (b) Perryman AL, Lin JH, McCammon JA. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hornak V, Okur A, Rizzo RC, Simmerling C. Proc Natl Acad Sci USA. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamelberg D, McCammon JA. J Am Chem Soc. 2005;127:13778–13779. doi: 10.1021/ja054338a. [DOI] [PubMed] [Google Scholar]
- 13.Tozzini V, McCammon JA. Chem Phys Lett. 2005;413:123–128. [Google Scholar]
- 14.Davis ME, Madura JD, Luty BA, McCammon JA. Comp Phys Comm. 1991;62:187–197. [Google Scholar]
- 15.Fulton A. Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 16.Chang C, Shen T, Trylska J, Tozzini V, McCammon JA. Biophys J. in press. [Google Scholar]
- 17.Oroszlan S, Luftig RB. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 18.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen G. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]


