Abstract
The motor protein kinesin is implicated in the intracellular transport of organelles along microtubules. Kinesin light chains (KLCs) have been suggested to mediate the selective binding of kinesin to its cargo. To test this hypothesis, we isolated KLC cDNA clones from a CHO-K1 expression library. Using sequence analysis, they were found to encode five distinct isoforms of KLCs. The primary region of variability lies at the carboxyl termini, which were identical or highly homologous to carboxyl-terminal regions of rat KLC B and C, human KLCs, sea urchin KLC isoforms 1–3, and squid KLCs. To examine whether the KLC isoforms associate with different cytoplasmic organelles, we made an antibody specific for a 10-amino acid sequence unique to B and C isoforms. In an indirect immunofluorescence assay, this antibody specifically labeled mitochondria in cultured CV-1 cells and human skin fibroblasts. On Western blots of total cell homogenates, it recognized a single KLC isoform, which copurified with mitochondria. Taken together, these data indicate a specific association of a particular KLC (B type) with mitochondria, revealing that different KLC isoforms can target kinesin to different cargoes.
INTRODUCTION
Kinesins are motor proteins that utilize ATP hydrolysis to drive the transport of macromolecular structures along microtubules (for review, see Bloom and Endow, 1995). Among the members of the superfamily, conventional kinesin (reviewed in Scholey, 1996) is the most ubiquitous motor, which is found in a variety of cells and tissues (Hollenbeck, 1989) and functions during both interphase and mitosis (Wright et al., 1991). The kinesin molecule is an elongated heterotetramer containing two heavy (KHC) and two light (KLC) chains (Bloom et al., 1988; Kuznetsov et al., 1988). Molecular masses vary in different species from 110 to 130 kDa for the KHCs and from 51 to 76 kDa for KLCs (Bloom and Endow, 1994). The globular, amino-terminal domain of the KHC forms the motor head containing the ATPase and microtubule-binding activities (Scholey et al., 1989; Yang et al., 1989). Two motor domains are connected with the globular tail domain through a coiled-coil stalk (Hirokawa et al., 1989).
Kinesin is implicated in the transport of various membrane-bound organelles (for review, see Goodson et al., 1997). However, mechanisms regulating kinesin-dependent transport and particular kinesin–cargo interactions still remain vague. Since the KLCs have been localized to the cargo-binding tail domain of the molecule, they have been suggested to be involved with cargo interaction (Hirokawa et al., 1989). This hypothesis is very attractive, particularly because each organism usually contains multiple KLC isoforms. Two distinct KLCs were found in kinesin purified from bovine brain (Pfister et al., 1989; Wagner et al., 1989) and unfertilized sea urchin eggs (Johnson et al., 1990). KLCs isolated from various cDNA libraries also show evidence of a family of isoforms for this protein (Cyr et al., 1991; Beushausen et al., 1993; Wedaman et al., 1993).
KLCs, cloned from rat (Cyr et al., 1991), Drosophila (Gauger and Goldstein, 1993), human (Cabeza-Arvelaiz et al., 1993), sea urchin (Wedaman et al., 1993), squid (Beushausen et al., 1993), and Caenorhabditis elegans (Fan and Amos, 1994), are highly conserved for most of their sequence. The amino-terminal polypeptide regions contain multiple heptad repeats that are responsible for binding to KHC, presumably through a coiled-coil mechanism (Gauger and Goldstein, 1993). The central region of the molecule contains several long imperfect repeats which suggest protein–protein interactions (Cabeza-Arvelaiz et al., 1993; Gindhart and Goldstein, 1996; Stenoien and Brady, 1997). Both the extreme amino and carboxyl termini of KLCs reveal notable sequence variability. Multiple methionines at the amino terminus might give rise to isoforms differing at their amino-terminal sequences, whereas alternative splicing likely generates different carboxyl terminus ends (Cyr et al., 1991; Beushausen et al., 1993; Cabeza-Arvelaiz et al., 1993; Fan and Amos, 1994; Wedaman et al., 1993). Such variability has led to the suggestion that the amino-terminal splicing of KLCs leads to formation of isoforms with different affinity to KHCs, whereas differential cargo trafficking is accommodated through the heterogeneity of the carboxyl termini (Beushausen et al., 1993).
In this article, we address the question whether unique sequences at the carboxyl terminus of KLCs determine the association of the motor with particular cytoplasmic organelles. By screening an expression library from CHO-K1 cells, we found cDNA molecules encoding at least five different variants of KLC. We made a monospecific antibody against one of these unique sequences and showed that it specifically associates with mitochondria. Thus, among multiple KLCs expressed in a distinct cell type, there is at least one that specifically targets kinesin to mitochondria.
MATERIALS AND METHODS
Library Screening and Sequence Analysis
A commercial CHO-K1 Uni-Zap cDNA expression library (Stratagene, La Jolla, CA) was used for screening. Immunoscreening was performed with the α-KLC antibody (see below) following standard methods (Sambrook et al., 1989). Purified phage was excised in vivo according to the manufacturer’s instructions (Stratagene). Resulting pBluescript SK− plasmids carrying KLC inserts were sequenced using the Sequenase II kit (United States Biochemical, Cleveland, OH).
Antibody Production and Purification
Antibody α-KLC was raised in a rabbit against the recombinant rat KLC-A. The pBluescript KS+ plasmid carrying the corresponding gene was kindly provided by Dr. Janet Cyr and Dr. Scott Brady. The cDNA insert was subcloned into the pET21b vector (Novagen, Madison, WI) and has been expressed in Escherichia coli strain BL21(DE3). The recombinant KLC-A was purified from bacterial lysate by 30% ammonium sulfate precipitation. The pellet was dialyzed against phosphate-buffered saline (PBS) and used for immunization. For the affinity column preparation, KLC-A was recloned into the pGEX-3X vector (Pharmacia, Piscataway, NJ) modified by adding an XhoI site and expressed as the fusion protein with glutathione S-transferase (GST) in E. coli strain JM109. The GST-KLC-A fusion protein was affinity purified on glutathione-agarose (Sigma Chemical Co., St. Louis, MO) and bound to BrCN-Sepharose (Sigma Chemical Co.). Affinity purification of the monospecific antibodies was performed as described elsewhere (Harlow and Lane, 1988).
To generate the antibody against the unique sequence of hamster KLC-B (α-B), a PstI–XhoI DNA fragment, encoding a 78-amino acid carboxyl-terminal region of this KLC isoform was recloned into modified pGEX-3X vector. Fusion protein was expressed in E. coli strain JM109, affinity purified on glutathione-agarose, and used as an immunogene. To purify the specific antibody, the IgG fraction was isolated from the immune serum using 50% ammonium sulfate precipitation. To remove the antibodies against GST and the conserved KLC sequence, the IgG fractions were thoroughly depleted on a column of immobilized recombinant rat KLC-A fused with GST. Finally, α-B antibody was affinity purified on BrCN-Sepharose containing the immobilized carboxyl-terminal portion of KLC-B fused with GST.
To obtain the antibody α-HCT, an antiserum was raised in a rabbit against a synthetic peptide, corresponding to the last 57-carboxyl-terminal amino acid residues of the human KHC, which was conjugated with hemocyanin. The specific antibody was affinity purified on the same peptide conjugated with bovine serum albumin (both conjugates were kindly provided by Dr. Sergei Axenovich, University of Illinois, Chicago, IL).
Cell Culture
Green monkey kidney epithelial cells CV-1 (American Tissue Culture Collection, Rockville, MD), and human skin fibroblasts, line 1029 (kind gift from Dr. V. Kukharenko, Institute of Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia), were maintained in DMEM (Flow Labs, Woodcock Hill, England) supplemented with 10% heat-treated fetal calf serum (Hyclone, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin. CHO-K1 cells (American Tissue Culture Collection) were maintained in Ham’s F12 medium (Flow Labs) supplemented with 10% fetal calf serum and antibiotics. All cultures were grown at 37°C in a 5% CO2 atmosphere.
Indirect Immunofluorescence Microscopy
Cells grown on glass coverslips were rinsed in warm PBS and fixed with methanol at −20°C. For immunolabeling, PBS was supplemented with 1% bovine serum albumin and 0.1% Triton X-100. Fixed cells were blocked with 5% normal goat serum in this solution and incubated with 50 μg/ml primary antibody for 30 min at room temperature followed by TRITC-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Labs, West Grove, PA) diluted 1:100. Immunofluorescence was observed and photographed using a Zeiss Photomicroscope III.
Mitochondria Labeling in Live Cells
A 1 mM stock solution of MitoTracker Green FM (M-7514, Molecular Probes, Eugene, OR) in DMSO was stored at −20°C and diluted in DMEM before labeling. Live cells on coverslips were rinsed in warm DMEM and transferred to a Petri dish containing a fresh portion of DMEM with 0.5 μM MitoTracker. The cells were labeled for 40 min at 37°C, rinsed in DMEM and then with warm PBS, and fixed with methanol at −20°C. The green fluorescence of MitoTracker was observed in the FITC channel.
Mitochondria Isolation from CV-1 Cells
Mitochondria were isolated from cultured CV-1 cells following the method described for the purification of mitochondrial membranes from bovine brain (Leopold et al., 1992). CV-1 cells were grown to confluence on 90-mm cell culture plates. For each experiment, 25 plates were used. Cells were rinsed three times with warm PBS and collected with a rubber policeman in ice-cold buffer A containing 100 mM Tris-HCl (pH 7.4), 250 mM sucrose, 1 mM potassium-EDTA, and protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, and 1 μg/ml N2-p-tosyl-l-arginine methyl ester (TAME) (1 ml of the buffer per each plate). All succeeding procedures were performed at +4°C. Cells were homogenized with a tight-fitting Dounce homogenizer and spun at 2000 × g for 5 min to remove nuclei and cell debris. The supernatant was recentrifuged at 12,500 × g for 8 min to obtain a crude fraction mitochondrial fraction and postmitochondrial supernatant. The crude mitochondrial fraction was then resuspended in 100 μl of buffer B: 3% Ficoll 400 (Pharmacia), 120 mM mannitol, 30 mM sucrose, 50 μM potassium-EDTA, 10 mM Tris-HCl (pH 7.4) and layered onto 500 μl of buffer 2×B (6% Ficoll 400, 240 mM mannitol, 60 mM sucrose, 50 μM potassium-EDTA, 10 mM Tris-HCl, pH 7.4). Mitochondria were centrifuged through the cushion in a SW55 Beckman rotor at 10,000 rpm for 30 min. The pellet was resuspended in 1 ml of buffer A and centrifuged in an Eppendorf centrifuge. The final pellet was resuspended in 100 μl of buffer A and considered as the mitochondrial fraction.
The postmitochondrial supernatant (see above) was centrifuged for 40 min at 150,000 × g to isolate cytosol and microsomes.
Protein concentration was determined with bicinchoninic acid (Pierce Chemical Co., Rockford, IL) using bovine serum albumin as a standard.
During mitochondria isolation, the activity of the mitochondrial enzyme succinate dehydrogenase, determined as described in Vinogradov (1979), increased 11.6 fold, as compared with the initial level in the cell homogenate.
Immunoprecipitation of Mitochondria-associated KLC Isoform
Mitochondria, isolated from CV-1 cells, were resuspended in 1 ml of ice-cold buffer containing 0.2 M KI, 1% Nonidet P-40, 50 mM Tris-HCl (pH 8.0), and protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptine, 1 μg/ml pepstatin A, 1 μg/ml TAME, and 1 μg/ml aprotinin. After a 15-min incubation on ice, mitochondria were centrifuged at 25,000 × g at 0°C for 15 min. The supernatant was divided equally and supplemented with either 20 μg of α-B antibody or 20 μg of α-B* antibody preadsorbed on 20 μl of protein A beads (Sigma Chemical Co.). Reactions were incubated for 1 h at +4°C, then the beads were rinsed three times with buffer (KI + Nonidet) and three times with 50 mM Tris-HCl (pH 8.0), and resuspended in SDS sample buffer.
SDS-PAGE and Western Blotting
Samples were analyzed by SDS-PAGE in 4–12% linear gradient gels according to the method of Laemmli (1970) and electroblotted onto a nitrocellulose membrane (Schleicher & Schuell, Keene, NH). After staining with Ponceau S, blots were blocked in 5% goat serum in TTBS (0.05% Tween 20, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5), incubated with 1–5 μg/ml primary antibodies in the blocking solution for 1 h at room temperature, followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Labs) at 1:5,000–10,000 dilution. For detection, diaminobenzidine or SuperSignal reagents (Pierce Chemical Co.) were used.
RESULTS
Pan Antibody against KLCs
We raised a polyclonal antibody against the light chain A of rat brain kinesin (the plasmid was kindly provided by J. L. Cyr and S. T. Brady). This polypeptide is the shortest isoform among three KLCs cloned from rat brain and contains only one unique carboxyl-terminal amino acid residue (Cyr et al., 1991). Therefore, we reasoned that the antibody would recognize mostly the conserved amino acid sequence of KLCs.
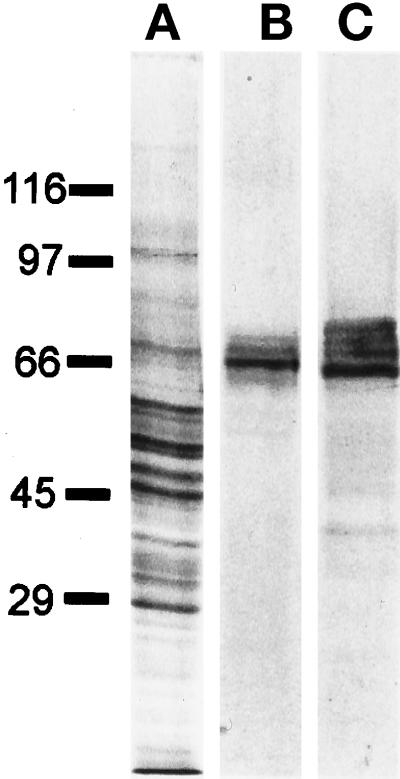
Affinity-purified antibody (α-KLC) recognized several polypeptides of the appropriate molecular mass for KLCs (62–70 kDa; Cyr et al., 1991; Cabeza-Arvelaiz et al., 1993; Beushausen et al., 1993) in Western blots of total rat brain homogenate and in purified bovine brain kinesin preparations (Figure 1). Similar staining patterns were obtained in Western blots of total cell homogenates from CHO-K1, CV-1 cells, or human skin fibroblasts (see below).
Figure 1.
Characterization of the α-KLC antibody by immunoblotting. (A) Rat brain homogenate Coomassie R-250-stained gel; (B) corresponding immunoblot, probed with α-KLC antibody; and (C) immunoblot of purified bovine brain kinesin probed with α-KLC antibody. The positions of the molecular mass markers are shown on the left.
Library Screening
We used the α-KLC antibody to screen a CHO-K1 cDNA expression library (Stratagene). After three rounds of phage selection and in vivo excision into pBluescript (SK−) vector, 13 immunoreactive clones were isolated. The molecular masses of the expressed in E. coli polypeptides correlated with the length of cDNA inserts (1200–200 bp). To determine whether the isolated clones represented KLCs, the longest cDNA was sequenced (the sequence data have been submitted to the EMBL database under accession number Y14586).
Deduced amino acid sequence revealed a high level of homology to the rat KLC-A (overall identity of >80%, Figure 2). Throughout the entire sequence length the rat KLC-A differed from the CHO KLC only by substitutions of 13-amino acid residues that all were conservative and randomly distributed, and a short deletion of 9-amino acid residues (positions 496–503 of rat KLC-A sequence, Figure 2).
Figure 2.
Alignment of the deduced amino acid sequences of rat KLC-A and hamster KLC-D. Asterisks indicate identical amino acids.
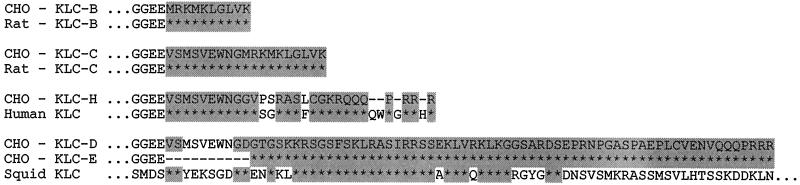
Restriction analysis showed that, although the 5′ regions of 13 isolated DNA clones were very similar, the 3′ ends were different. To characterize this variability, we sequenced 3′ segments of all of the KLC cDNAs (starting approximately from the amino acid 510 of rat KLC-A). Figure 3A shows the carboxyl-terminal amino acid sequences deduced from the nucleotide sequences of the isolated cDNAs. Beginning from the position corresponding to the last amino acid residue of rat KLC-A and running upstream to the ends of the hamster KLC cDNAs, there was no heterogeneity between 13 isolated clones. However, on the basis of diversity of the extreme 3′ ends, the 13 clones fell into 5 different groups. A BLASTP sequence homology search using carboxyl-terminal regions of hamster KLCs as query sequences revealed significant local similarity to known mammalian KLCs (Figure 3). Two hamster KLC polypeptides, designated as KLC-B and C, were identical to those of rat KLC isoforms B and C (Cyr et al., 1991). The carboxyl terminus of a third hamster polypeptide, KLC-H, was highly homologous to the human KLC (Cabeza-Arvelaiz et al., 1993). Two other isoforms, KLC-D and KLC-E, were homologous to sea urchin and squid KLCs (Beushausen et al., 1993; Wedaman et al., 1993). Thus, by screening one CHO-K1 expression library we found five variants of KLC polypeptides. This suggests that at least five KLC isoforms can be expressed simultaneously in a single cell type.
Figure 3.
Deduced carboxyl-terminal amino acid sequences of different CHO KLC isoforms compared with the unique regions of the rat KLC-B and -C, the human, and the squid KLCs.
Generation of Anti-B Isoform-specific Antibodies
To test whether a particular KLC was associated with a specific class of cargoes, we raised an antibody against the unique sequence MRKMKLGLVK of the hamster KLC. This sequence was present in the B and C isoforms but not in the other three KLCs (Figure 3). After failing to obtain an antiserum against a synthetic oligopeptide MRKMKLGLVK, we immunized a rabbit with a recombinant 7-kDa carboxyl-terminal portion of the B isoform fused with GST. To obtain the specific antibody to the unique region, we then used a purification scheme based on the thorough removal of undesirable antibodies against the conservative sequence. For this purpose, the antiserum was exposed to rat KLC-A-GST fusion protein bound to BrCN-Sepharose. We repeated the chromatography until the flow-through fraction was completely devoid of reactivity to the rat KLC-A-GST fusion protein in Western blots. Antibodies were then affinity purified from the depleted antiserum on the 7-kDa carboxyl-terminal fragment of CHO KLC-B isoform expressed as GST fusion protein. We designated the resulting antibody as α-B, although it should recognize both B and C isoforms. To prove that the reactions demonstrated by the antibody were specific to the unique sequence of B and C isoforms, we preadsorbed the α-B antibody on the synthetic oligopeptide MRKMKLGLVK bound to an Affigel 10 column and used the flow-through fraction (designated as antibody α-B*) as a negative control.
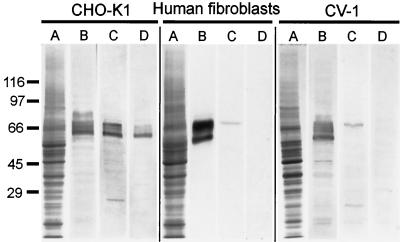
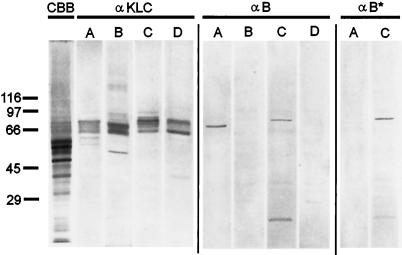
We tested antibody α-B by Western blotting of whole-cell homogenates from several different cell types (Figure 4). In CHO-K1 antibody α-B reacted with a set of polypeptides similar to those stained with a pan KLC antibody, α-KLC (Figure 4, B and C). A similar staining pattern was obtained with α-B* antibody (Figure 4D), and therefore we had to conclude that our purification scheme was not rigorous enough to produce a monospecific reagent for CHO-K1 cells.
Figure 4.
Characterization of α-B antibody by immunoblotting with total cell homogenates of CHO-K1, human skin fibroblasts, and CV-1 cells. (A) Coomassie R-250 stained gel; (B) immunoblot probed with α-KLC antibody; (C) immunoblot probed with α-B antibody; and (D) immunoblot, probed with α-B* antibody. The positions of the molecular mass markers are shown on the left.
In contrast, antibody α-B selectively reacted with a single polypeptide in both human skin fibroblasts and CV-1 cells (Figure 4, lanes C). The estimated molecular mass of this immunoreactive polypeptide was the same for both cell types (68 kDa). Preadsorption of antibody α-B on the MRKMKLGLVK oligopeptide (α-B* antibody, Figure 4, lanes D) completely eliminates the staining. Therefore, we conclude that in these cell lines, the α-B antibody selectively recognizes an endogenous light chain that contains the unique MRKMKLGLVK sequence.
Immunolocalization of 68-kDa KLC Isoform in Cultured Cells
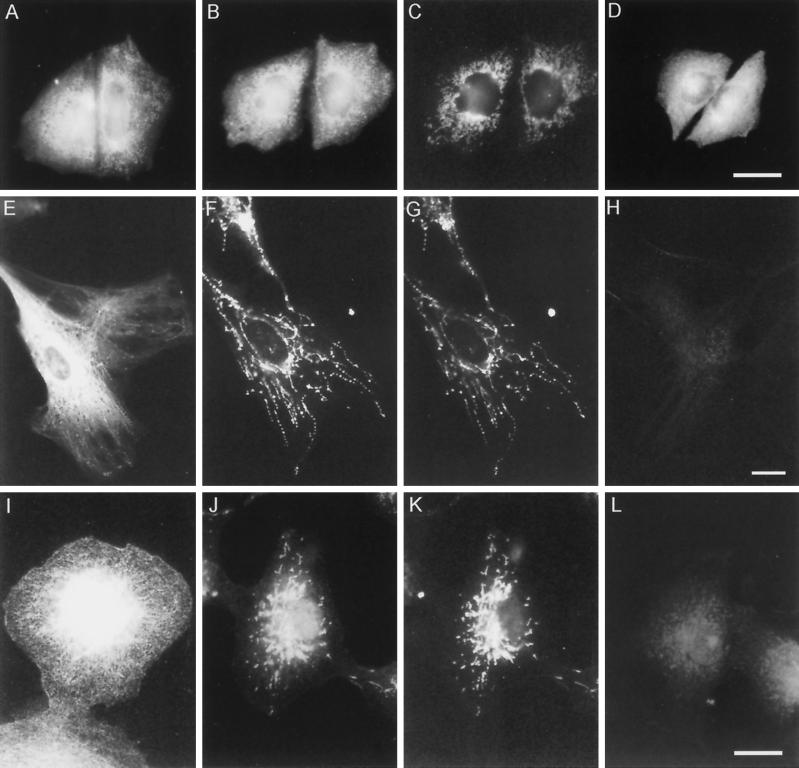
To establish whether the 68-kDa KLC isoform was specifically associated with cytoplasmic structures, we stained CHO-K1 cells, CV-1 cells, and human fibroblasts with α-KLC, α-B, and α-B* antibodies (Figure 5). As expected from immunoblotting results (see above), all three antibodies stained similar patterns in CHO-K1 cells (Figure 5, A, B, and D). However, in the CV-1 cells and human fibroblasts, the α-KLC and α-B staining patterns were dramatically different. Antibody α-KLC revealed a dense vesicular staining pattern (Figure 5, A, E, and I). A similar pattern of fluorescence was obtained with the antibody specific to the motor domain of KHC (antibody HD; Rodionov et al., 1991) in methanol-fixed cells (not shown). In contrast, organelles labeled with the α-B antibody were homogenous and, by their appearance and distribution, resembled mitochondria (Figure 5, F and J). To confirm this, we used the fluorescent dye MitoTracker to label mitochondria in live cells and then fixed cells with cold methanol and stained them with the α-B antibody. The fluorescent patterns obtained in the cells double stained with α-B antibody and MitoTracker were identical (Figure 5, compare F with G and J with K). No specific staining was obtained in CV-1 and human fibroblasts treated with the α-B antibody preadsorbed with the synthetic oligopeptide MRKMKLGLVK (α-B* antibody, Figure 5, H and L) or the fraction of IgG isolated from the preimmune serum. Thus, our immunofluorescence microscopy indicates the specific association of the 68-kDa KLC-isoform with mitochondria.
Figure 5.
Immunolocalization of different KLC isoforms in CHO-K1 cells (A–D), human skin fibroblasts (E–H), and CV-1 cells (I–L). (A, E, and I) α-KLC staining; (B, F, and J) α-B staining; (C, G, and K) MitoTracker staining of the corresponding cells presented in B, F, and J. (D, H, and L) α-B* staining. Bars, 10 μm.
Detection of the 68-kDa KLC in Mitochondrial Fraction
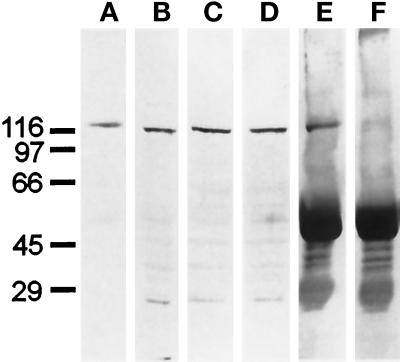
To independently confirm the immunolocalization data, we isolated fractions of mitochondria, microsomes, and cytosol from cultured CV-1 cells and compared them in Western blots with α-KLC and α-B antibodies. Both membrane fractions as well as cytosol contain multiple KLC polypeptides recognized by the α-KLC antibody (Figure 6, left panel). The molecular masses of the immunoreactive polypeptides varied slightly in the fractions, suggesting a selectivity in their distribution. In Western blots with the α-B antibody, the 68-kDa KLC was found only in the mitochondrial fraction; it was not detected in microsomal or cytosol fractions (Figure 6, middle panel). Preadsorbed antibody α-B* failed to recognize this KLC isoform in immunoblotting (Figure 6, lane A in the right panel). Thus, we concluded that the 68-kDa isoform of KLC copurifies with mitochondria from CV-1 cells.
Figure 6.
Western blot analysis of KLCs in fractions of mitochondria (A), postmitochondrial supernatant (B), microsomes (C), and cytosol (D) of CV-1 cells. Immunoblots probed with α-KLC, α-B, and α-B* antibodies. The positions of the molecular mass markers and the Coomassie R-250 stained gel of the mitochondrial fraction (CBB) are shown on the left.
Interestingly, another polypeptide with the molecular mass of approximately 72 kDa was detected with α-B antibody in the fraction of microsomes (Figure 6, middle panel, lane C). However, this polypeptide could also be recognized with preadsorbed antibody α-B* (α-B*, Figure 6, lane C), and was not detected in the total cell homogenate (CV-1 cells, Figure 4, lane C) and postmitochondrial supernatant (α-B, Figure 6, lane B). Moreover, the residual staining pattern observed in CV-1 cells and human fibroblasts probed with α-B* antibody (Figure 5, H and L) was not affected by preadsorption of the α-B* with microsomal fraction (not shown).
The 68-kDa KLC in the Mitochondrial Fraction Is Associated with KHC
To verify that the 68-kDa polypeptide really corresponds to a KLC isoform bound to KHC, and thus represents a full kinesin molecule, we tested whether KHC was also present in the mitochondrial fraction and whether the α-B antibody could immunoprecipitate a complex of the light and heavy chains.
For these experiments, a polyclonal antibody against the last 57-carboxyl-terminal amino acid residues of the human KHC was raised in a rabbit and affinity-purified (antibody α-HCT). On Western blots, this antibody recognized the heavy chain of purified bovine brain kinesin (Figure 7A) and a single polypeptide corresponding to the mass of KHC in the total homogenate of CV-1 cells (Figure 7B). The polypeptide of the same molecular mass was also detected in the mitochondrial fraction isolated from CV-1 cells (Figure 7C). This result is consistent with previous studies where KHC was found in mitochondria isolated from mammalian brain (Leopold et al., 1992; Jellali et al., 1994).
Figure 7.
Immunoblot probed with anti-kinesin heavy chain antibody. (A) Purified bovine brain kinesin. (B) CV-1 whole-cell homogenate. (C) Mitochondrial fraction of CV-1 cells. (D) KI extract of CV-1 mitochondrial fraction. (E) Immunoprecipitate of KI extract with α-B antibody. (F) Immunoprecipitate of KI extract with α-B* antibody.
To perform immunoprecipitation, we first solubilized mitochondria-bound kinesin with 0.2 M KI (Jellali et al., 1994) and 1% Nonidet P-40. Under these conditions both the 68-kDa KLC (not shown) and KHC (Figure 7D; see also Jellali et al., 1994) can be detected in the extract. We then immunoprecipitated KLC from the KI extract using either α-B or α-B* antibody and probed the precipitates with α-HCT antibody, recognizing KHC. As shown in Figure 7E, KHC clearly coprecipitated with KLC when antibody α-B was used, but not with antibody a-B*. We therefore conclude that the 68-kDa KLC isoform in the mitochondrial fraction is associated with a KHC, and thus is a part of the full kinesin molecule, capable of motor activity.
DISCUSSION
Hamster KLC Isoforms: Identification of Conserved Sequence Motifs
In this study, we isolated five different isoforms of KLC from an expression library of CHO cells. Sequence comparisons showed that the hamster polypeptides were identical or highly homologous to other KLCs; however, distinct variability was revealed at the extreme carboxyl-terminal ends. The variability found among these clones corresponds to carboxyl-terminal regions of rat KLC isoforms B and C (Cyr et al., 1991), human KLC (Cabeza-Arvelaiz et al., 1993), sea urchin KLC isoforms 1, 2, and 3 (Wedaman et al., 1993), and squid KLC (Beushausen et al., 1993). As such, the clones group together to form a family of five related but distinct polypeptides. A BLASTP search of the National Center for Biotechnology Information (Bethesda, MD) peptide-sequence database using as query sequences the unique regions MRKMKLGLVK and VSMSVEWNG, which hamster polypeptides share with rat and human isoforms (Figure 3), failed to find any other related proteins. The same result was obtained for the short unique sequence QQQPRRR ending three of the hamster isoforms.
Previously, multiple isoforms of KLC have been isolated from the expression libraries derived from heterogeneous cell populations: rat brain (Cyr et al., 1991), squid optic lobe (Beushausen et al., 1993), and the whole nematode Caenorhabditis elegans (Fan and Amos, 1994). The only exception was the library obtained from sea urchin eggs, where four KLC isoforms were found (Wedaman et al., 1993). However, these cells are known to store inactive mRNAs needed for the following early development (Alberts et al., 1994). Our data provide the first evidence that several KLCs isoforms can be simultaneously expressed in a single cell type.
Isoform-specific Antibody
To directly test the hypothesis that KLC diversity provides specific interaction with different cargoes, we raised an antiserum against the recombinant carboxyl-terminal portion of hamster KLC-B and purified the antibody against the unique sequence MRKMKLGLVK. Since this motif was found in two of five hamster KLCs, we expected the antibody to distinguish the B and C isoforms from the other three. Contrary to our expectations, the α-B antibody did not show absolute specificity in the reaction with hamster KLCs. The reason for this is unclear but probably indicates that our purification scheme was not rigorous enough to remove all undesired antibodies cross-reacting with other KLC isoforms in CHO-K1 cells. There are several amino acid substitutions in the conserved part of the KLC sequences specific for the CHO cells (Figure 2). These amino acids can be recognized by particular antibodies in the immune α-B serum. Since we used rat KLC-A isoform to remove antibodies against conserved sequences from original antiserum, these species-specific anti-KLC antibodies possibly contaminated the purified α-B antibody. This can also explain why we observed similar staining patterns in CHO-K1 cells stained with the antibodies α-B and α-B*.
The α-B antibody did prove to be highly specific in two other cell types, CV-1 and human fibroblasts, for a 68-kDa polypeptide, which likely represents the KLC homologue of B or C hamster isoforms. This conclusion is based on several different types of evidence: 1) the molecular mass of the 68-kDa polypeptide was appropriate for KLC; 2) it was not recognized with the α-B antibody after preadsorption with the MRKMKLGLVK synthetic peptide; and 3) it was precipitated by the antibody α-B in the complex with KHC.
Taken together, these data indicate that we obtained an antibody specific to a distinct KLC isoform that can be used for its localization in situ and in subcellular fractions.
Mitochondria-associated KLC
The role of kinesin in the transport of mitochondria has been a subject of intense study for several years. Upon microinjection, an antibody raised against the motor domain of KHC (antibody HD) causes mitochondria to collapse into the perinuclear region of cultured fibroblasts (Rodionov et al., 1993). Since this antibody was raised against the highly homologous motor domain, it could affect not only conventional kinesin but also kinesin-related motors. Indeed, two such motors, mouse KIF1B (Nangaku et al., 1994) and KLP67A from Drosophila (Pereira et al., 1997), were recently implicated in mitochondria transport. Moreover, the distribution of mitochondria did not change upon the antisense oligonucleotide suppression of KHC in rat neurons and astrocytes (Feiguin et al., 1994) or upon overexpression of the dominant negative KHC in mouse fibroblasts (Nakata and Hirokawa, 1995). Taken together, these data suggest that there are multiple mitochondria-associated motor proteins, which is consistent with the general redundancy of motor proteins in essential cellular activities (Goldstein, 1991). Nevertheless, association of the conventional kinesin with mitochondria has been directly shown by immunoblotting of isolated organelles with monoclonal (Leopold et al., 1992) and polyclonal (Jellali et al., 1994) antibodies against KHC. In both cases, a single immunoreactive polypeptide with the molecular mass of KHC was detected in mitochondrial fraction. In our study, the antibody directed against the carboxyl-terminal portion of the heavy chain, and thus specific to conventional kinesin, also detected a single polypeptide in the homogenate and mitochondrial fraction of CV-1 cells.
Analysis of the light chain distribution revealed that several KLC polypeptides were always present in mitochondrial as well as microsomal and cytosolic fractions of CV-1 cells. However, the particular set of KLCs associated with the mitochondrial fraction differed from those found in cytosol and microsomal fraction. This is consistent with the observation showing an existence of two different pools of kinesin: soluble and membrane associated (Hollenbeck, 1989; Schmitz et al., 1994). The presence of multiple KLCs with different molecular masses has been demonstrated for the soluble kinesin isolated from mammalian brain (Wagner et al., 1989; Matthies et al., 1993). The cytosolic pool has been suggested to contain enzymatically inactive forms of the motor that require some posttranslational modification; for instance, phosphorylation, to become competent for both ATP hydrolysis and cargo binding (Hollenbeck, 1993). Contrary to cytosolic fractions, multiple light chain isoforms in the membrane fractions were totally unexpected.
Despite the observation that each subcellular fraction contains more than one KLC isoform, our data reveal selective association of particular KLC isoforms with membranes. Indeed, the 68-kDa B-type KLC isoform was found only in the mitochondrial and not in any other subcellular fraction. This conclusion was supported by the immunolocalization of the 68-kDa KLC isoform in cultured cells by immunofluorescent staining. In two different cell lines it proved to be associated with mitochondria, verifying that this KLC was bound to mitochondria prior to homogenization of the CV-1 cells.
In previous studies, the L2 monoclonal antibody against bovine brain KLC has been shown to label structures in primary rat brain cells with morphologies suggestive of mitochondria (Pfister et al., 1989). Another experiment using immunogold staining of isolated brain mitochondria showed that kinesin was restricted to the large clusters on the surface of mitochondria (Leopold et al., 1992). Our data confirm the association of kinesin with mitochondria, although the lower resolution of immunofluorescent microscopy could not resolve patches on mitochondrial surface even if they occurred.
Interestingly, α-B antibody reacted with another polypeptide with the molecular mass of approximately 72 kDa in the microsomal fraction. However, this band also reacted with the α-B* antibody and thus did not contain the unique MRKMKLGLVK sequence. Whether this polypeptide is a KLC isoform specific for a microsomal fraction or represents some other protein not related to kinesin remains unknown.
Since the KLCs have been localized to the tail domain of the molecule, the region opposite from the motor domain, they have been suggested to mediate kinesin to cargo binding (Hirokawa et al., 1989). However, evidence to support this hypothesis has been indirect.
A monoclonal antibody (KLC-All) that recognizes a highly conserved epitope in the tandem repeat domain of KLC can inhibit directional vesicles movement in isolated squid axoplasm and release kinesin from the membrane surfaces (Stenoin and Brady, 1997). This implies that KLC is directly involved in kinesin–membrane interaction and that this interaction is likely to be mediated by sequences that are present in all different KLC isoforms. On the other hand, there is evidence that KHC alone is sufficient for binding microsomes in vitro (Skoufias et al., 1994) and exocytotic vesicles in vivo (Bi et al., 1997). Moreover, there are members of the kinesin superfamily which do not appear to have light chains or sequences homologous to KLCs (e.g., Steinberg and Schliwa, 1995; Seiler et al., 1997).
This discrepancy in data indicates that kinesin–cargo interactions are complex. Kinesin appears to be involved in trafficking at least several different types of cargo, e.g., endoplasmic reticulum (Henson et al., 1992), lysosomes (Hollenbeck and Swanson, 1990), synaptic vesicles, mitochondria, coated vesicles (Leopold et al, 1992), etc. This implies that there should exist a mechanism that specifically targets kinesin molecules to the different cargoes. It has been proposed that kinesin may be targeted to a particular type of cargo depending on a particular light chain associated with the heavy chains (Cyr et al., 1991, Wedaman et al., 1993, Skoufias et al., 1994). Two facts observed in our study, simultaneous expression of multiple KLC isoforms in a single cell type and association of one specific KLC isoform with a particular type of cargo (mitochondria), bring some additional support to this hypothesis. However, this does not imply that specific KLCs involved in targeting function are also directly involved in kinesin to cargo binding. The binding per se may be mediated directly by KHC or through a cooperative action of KHC and some ubiquitous isoforms of KLCs. The fact that we always observed multiple KLCs associated with different membrane fractions may indicate that a specific KLC isoform that targets kinesin to the specific cargo is not sufficient for binding.
At present, the particular mechanism of kinesin–cargo interaction remains unclear. Nevertheless, our data establish for the first time that there is a specific membrane fraction (mitochondria) that carries a distinct light chain of conventional kinesin (B/C KLC isoforms). This finding constrains the involvement of conventional kinesin in mitochondria transport and supports the hypothesis that different KLC isoforms can target kinesin molecules to different types of cargoes.
ACKNOWLEDGMENTS
The authors thank Dr. Scott Brady for his generous gift of the rat KLC-A plasmid, Dr. Leah Haimo for the recloning of rat KLC-A into pET-21b vector, and Dr. Sergei Axenovich for providing the carboxyl-terminal portion of the human KHC. We thank Dr. Conly Rieder for continuing support and encouragement. We appreciate the excellent technical assistance provided by Natasha Minina and Olya Deryabina. We are particularly grateful to Dr. Vladimir Gelfand for helpful comments and discussion of the manuscript. Peptides and oligonucleotides used in this work were synthesized in Peptide Synthesis Core and Molecular Genetics Core facilities of Wadsworth Center. This work was supported by Howard Hughes Medical Institute grant 75195–544704 (to F.K.G.), Russian Basic Science Foundation grant 96–04-50517 (to F.K.G.), and National Institutes of Health grant GM-51532 (to M.P.K.)
REFERENCES
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 3rd ed. New York: Garland Publishing; 1994. [Google Scholar]
- Beushausen S, Kladakis A, Jaffe H. Kinesin light chains: identification and characterization of a family of proteins from the optic lobe of the squid Loligo pealii. DNA Cell Biol. 1993;12:901–910. doi: 10.1089/dna.1993.12.901. [DOI] [PubMed] [Google Scholar]
- Bi G-Q, Morris RL, Liao G, Alderton JM, Scholey JM. Kinesin- and myosin-driven steps of vesicle recruitment for Ca2+-regulated exocytosis. J Cell Biol. 1997;138:999–1008. doi: 10.1083/jcb.138.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS, Endow SA. Motor proteins 1: kinesins. Protein Profile. 1995;1:1059–1088. [PubMed] [Google Scholar]
- Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Cabeza-Arvelaiz Y, Shih L-CN, Hardman N, Asselbergs F, Bilbe G, Schmitz A, White B, Siciliano MJ, Lachman LB. Cloning and genetic characterization of the human kinesin light chain (KLC) gene. DNA Cell Biol. 1993;12:881–892. doi: 10.1089/dna.1993.12.881. [DOI] [PubMed] [Google Scholar]
- Cyr JL, Pfister KK, Bloom GS, Slaughter CA, Brady ST. Molecular genetics of KLC: generation of isoforms by alternative splicing. Proc Natl Acad Sci USA. 1991;88:10114–10118. doi: 10.1073/pnas.88.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Amos LA. Kinesin light chain isoforms in Caenorhabditis elegans. J Mol Biol. 1994;240:507–512. doi: 10.1006/jmbi.1994.1465. [DOI] [PubMed] [Google Scholar]
- Feiguin F, Ferreira A, Kosik KS, Caceres A. Kinesin-mediated organelle translocation revealed by specific cellular manipulations. J Cell Biol. 1994;127:1021–1039. doi: 10.1083/jcb.127.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger AK, Goldstein LSB. The Drosophila kinesin light chain. Primary structure and interaction with kinesin heavy chain. J Biol Chem. 1993;268:13657–13666. [PubMed] [Google Scholar]
- Gindhart JG, Goldstein LSB. Tetratrico peptide repeats are present in the kinesin light chain. Trends Biochem Sci. 1996;21:52–53. [PubMed] [Google Scholar]
- Goldstein LSB. The kinesin superfamily: tails of functional redundancy. Trends Cell Biol. 1991;1:93–98. doi: 10.1016/0962-8924(91)90036-9. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Valetti C, Kreis TE. Motors and membrane traffic. Curr Opin Cell Biol. 1997;9:18–28. doi: 10.1016/s0955-0674(97)80147-0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Henson JH, Nesbitt D, Wright BD, Scholey JM. Immunolocalization of kinesin in sea urchin coelomocytes. Association of kinesin with intracellular organelles. J Cell Sci. 1992;103:309–320. doi: 10.1242/jcs.103.2.309. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. The distribution, abundance and subcellular localization of kinesin. J Cell Biol. 1989;108:2335–2342. doi: 10.1083/jcb.108.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ. Phosphorylation of neuronal kinesin heavy and light chains in vivo. J Neurochem. 1993;60:265–275. doi: 10.1111/j.1471-4159.1993.tb03513.x. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ, Swanson JA. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature. 1990;346:864–866. doi: 10.1038/346864a0. [DOI] [PubMed] [Google Scholar]
- Jellali A, Metz-Boutigue M-H, Surgucheva I, Jancsik V, Schwartz C, Filliol D, Gelfand VI, Rendon A. Structural and biochemical properties of kinesin heavy chain associated with rat brain mitochondria. Cell Motil Cytoskeleton. 1994;28:79–93. doi: 10.1002/cm.970280108. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Buster D, Scholey JM. KLC of sea urchin kinesin identified by immunoadsorption. Cell Motil Cytoskeleton. 1990;16:204–213. doi: 10.1002/cm.970160307. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Vaisberg EA, Shanina NA, Magretova NN, Chernyak VY, Gelfand VI. The quaternary structure of bovine brain kinesin. EMBO J. 1988;7:353–357. doi: 10.1002/j.1460-2075.1988.tb02820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–695. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leopold PL, McDowall AW, Pfister KK, Bloom GS, Brady ST. Association of kinesin with characterized membrane-bounded organelles. Cell Motil Cytoskeleton. 1992;23:19–33. doi: 10.1002/cm.970230104. [DOI] [PubMed] [Google Scholar]
- Matthies HJG, Miller RJ, Palfrey HC. Calmodulin binding to and cAMP-dependent phosphorylation of kinesin light chain modulate kinesin ATPase activity. J Biol Chem. 1993;268:11176–11187. [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anteriograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxey SJ, Goldstein LSB. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol. 1997;136:1081–1091. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Wagner MC, Stenoien DL, Brady ST, Bloom GS. Monoclonal antibodies to kinesin heavy and KLC stain vesicle-like structures, but mot microtubules, in cultured cells. J Cell Biol. 1989;108:1453–1463. doi: 10.1083/jcb.108.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Gyoeva FK, Tanaka E, Bershadsky AD, Vasiliev JM, Gelfand VI. Microtubule-dependent control of cell shape and pseudopodial activity is inhibited by the antibody to kinesin motor domain. J Cell Biol. 1993;123:1811–1820. doi: 10.1083/jcb.123.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitz F, Wallis KT, Pho M, Drenckhahn D, Murphy DB. Intracellular distribution of kinesin in chromaffin cells. Eur J Cell Biol. 1994;63:77–83. [PubMed] [Google Scholar]
- Scholey JM. Kinesin-II, a membrane traffic motor in axons, axonemes, and spindles. J Cell Biol. 1996;133:1–4. doi: 10.1083/jcb.133.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Heuser J, Yang JT, Goldstein LSB. Identification of globular mechanochemical heads of kinesin. Nature. 1989;338:355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- Seiler S, Nargang FE, Steinberg G, Schliwa M. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 1997;16:3025–3034. doi: 10.1093/emboj/16.11.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Cole DG, Wedaman KP, Scholey JM. The carboxyl-terminal domain of kinesin heavy chain is important for membrane binding. J Biol Chem. 1994;269:1477–1485. [PubMed] [Google Scholar]
- Steinberg G, Schliwa M. The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol Biol Cell. 1995;6:1605–1618. doi: 10.1091/mbc.6.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Brady ST. Immunochemical analysis of kinesin light chain function. Mol Biol Cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AD. Reactivity of the Live Systems and the Energy Metabolism. Moscow: Nauka; 1979. Measurement of the succinatedehydrogenase activity. (in Russian). [Google Scholar]
- Wagner MC, Pfister KK, Bloom GS, Brady ST. Copurification of kinesin polypeptides with microtubule-stimulated Mg-ATPase activity and kinetic analysis of enzymatic properties. Cell Motil Cytoskeleton. 1989;12:195–215. doi: 10.1002/cm.970120403. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Knight AE, Kendrick-Jones J, Scholey JM. Sequences of sea urchin kinesin light chain isoforms. J Mol Biol. 1993;231:155–158. doi: 10.1006/jmbi.1993.1267. [DOI] [PubMed] [Google Scholar]
- Wright BD, Henson JH, Wedaman KP, Willy PJ, Morand JN, Scholey JM. Subcellular localization and sequence of sea urchin kinesin heavy chain: evidence for its association with membranes in the mitotic apparatus and interphase cytoplasm. J Cell Biol. 1991;113:817–833. doi: 10.1083/jcb.113.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analysis. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]