Abstract
RNA interference (RNAi) is a cellular process regulating gene expression and participating in innate defense in many organisms. RNAi has also been utilized as a tool to query gene function and is being developed as a therapeutic strategy for several diseases. Synthetic small interfering (siRNAs) or expressed stem–loop RNAs (short-hairpin RNAs [shRNAs] or artificial microRNAs [miRNAs]) have been delivered to cultured cells and organisms to inhibit expression of a variety of genes. A persistent question in the field, however, is which RNAi expression system is most suitable for distinct applications. To date, shRNA- and artificial miRNA-based strategies have been compared with conflicting results. In prior comparisons, sequences required for efficient RNAi processing and loading of the intended antisense strand into the RNAi-induced silencing complex (RISC) were not considered. We therefore revisited the shRNA–miRNA comparison question. Initially, we developed an improved artificial miRNA vector and confirmed the optimal shRNA configuration by altering structural features of these RNAi substrates. Subsequently, we engineered and compared shRNA- and miRNA-based RNAi expression vectors that would be processed to yield similar siRNAs that exhibit comparable strand biasing. Our results demonstrate that when comparison variables are minimized, the shRNAs tested were more potent than the artificial miRNAs in mediating gene silencing independent of target sequence and experimental setting (in vitro and in vivo). In addition, we show that shRNAs are expressed at considerably higher levels relative to artificial miRNAs, thus providing mechanistic insight to explain their increased potency.
Keywords: RNAi vectors, shRNA, miRNA, SCA1, HD
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved cellular process regulating gene expression and participating in innate defense against transposable elements and viral invasion (McManus and Sharp 2002). RNAi mediates sequence-specific gene silencing by double-stranded RNAs (dsRNAs), which may be processed by Dicer into functional small RNAs (small interfering RNAs [siRNAs] and microRNAs [miRNAs] among others) (Fire et al. 1998; Provost et al. 2002). Small RNAs associated with the RNA-induced silencing complex (RISC) or RISC-like complexes mediate post-transcriptional gene silencing by targeting transcripts for degradation or translational repression (Lee et al. 2004). Also, small RNAs in RISC-like complexes may direct heterochromatin formation to mediate transcriptional gene silencing (Verdel et al. 2004).
RNAi has been utilized as a tool to query gene function and is being developed as a modality for disease therapy. Exogenous RNAi has been expressed in cultured cells and organisms as short-hairpin RNAs (shRNAs) or artificial miRNAs (Fig. 1A; Elbashir et al. 2001; Xia et al. 2002; Zeng et al. 2002). The basic transcriptional unit of a shRNA is sense and antisense sequences connected by a loop of unpaired nucleotides. shRNAs are exported from the nucleus by Exportin-5, and once in the cytoplasm, are processed by Dicer to generate functional siRNAs (Provost et al. 2002; Yi et al. 2003; Lund et al. 2004). miRNA stem–loops, typically, are expressed as part of larger primary transcripts (pri-miRNAs) (Cai et al. 2004). The Drosha–DGCR8 complex excises the stem–loops generating intermediates known as pre-miRNAs, which are subsequently exported to the cytoplasm and diced into functional small RNAs (Lee et al. 2003; Gregory et al. 2004).
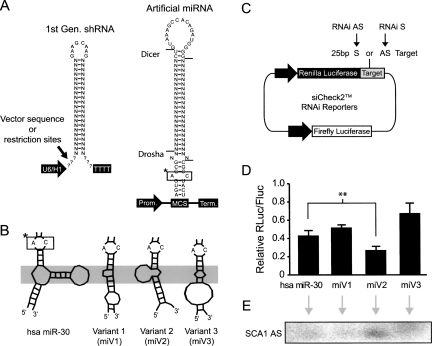
FIGURE 1.
Optimization of the human miR-30 shuttle. (A) General structures of shRNAs and artificial miRNAs (N's correspond to the siRNA-duplex region with sense and antisense being 5′ and 3′, respectively). Here, the antisense sequences are designed to target SCA1, HD, or GFP transcripts. Hash marks indicate the known major Drosha and Dicer cleavage sites of human miR-30 (Lee et al. 2003; Zeng and Cullen 2003; Silva et al. 2005). Processing sites of many shRNAs are unknown and vary based on the presence of short flanking sequences. (*) Boxed sequence is for orientation purposes in the panel. (B) Artificial miRNA variants were generated by altering the nearby flanking sequences, and portions of the predicted mFOLD (Zuker 2003) structures within the stem-base are shown. Instability (i.e., single-stranded nature) within the gray-shaded region may promote Drosha binding and cleavage (Zeng and Cullen 2005; Han et al. 2006). These variants contain identical RNAi sequences and structures in regions above the gray-shaded box. (C) Cartoon depicting RNAi reporters, generated by inserting target sequences into the 3′ UTR of Renilla luciferase. Reporter plasmids also contain a Firefly luciferase expression cassette for normalization. (D) Silencing activity of miR-30 variants. Artificial miSCA1 variants and RNAi reporter expression plasmids were cotransfected into HEK293 cells, and Dual-Glo Luciferase assays were performed after 24 h. Results of two independent experiments (each n = 3) are shown as mean±SEM relative to mock-treated (i.e., promoter only) controls and demonstrate that variant 2 (miV2) is the most effective artificial miRNA (even more effective than human miR-30, ** = P < 0.01). (E) Small transcript Northern blot was performed at 48 h post-transfection of miRNA variant expression plasmids into HEK293 cells. Results show that miV2 yields more mature antisense RNA (SCA1 AS) compared to the other artificial miRNA variants including the natural miR-30 shuttle.
Previous studies from several laboratories, including our own, demonstrate the utility and efficacy of shRNAs in vitro and in vivo (Elbashir et al. 2001; Song et al. 2003; Xia et al. 2004; Harper et al. 2005). However, the use of artificial miRNAs (pri-miRNA scaffolds shuttling siRNA sequences) as RNAi vectors has become an attractive alternative (Zeng et al. 2002; Chung et al. 2006). Artificial miRNAs more naturally resemble endogenous RNAi substrates and are more amenable to Pol-II transcription (allowing tissue-specific expression of RNAi) and polycistronic strategies (allowing delivery of multiple siRNA sequences) (Cai et al. 2004; Chung et al. 2006). A persistent question in the field, however, is which RNAi expression system is most optimal for achieving gene silencing in particular applications.
Optimization of RNAi strategies remains important due to the potential side effects caused by exogenously delivered RNAi. RNAi vectors may compete for endogenous RNAi machinery, stimulate cellular responses to dsRNA, or silence unintended target mRNAs due to partial complementarity (Bridge et al. 2003; Sledz et al. 2003; Fish and Kruithof 2004; Birmingham et al. 2006; Grimm et al. 2006; Castanotto et al. 2007). These side effects can produce severe cellular toxicity and even result in organismal death (Grimm et al. 2006). Improved RNAi-based gene silencing strategies may allow delivery of lower doses, thus reducing the likelihood of RNAi-related side effects.
To date, shRNA- and artificial miRNA-based strategies have been compared with conflicting results (Boden et al. 2004; Silva et al. 2005; Li et al. 2007). In certain comparisons, the shRNAs tested had suboptimal 5′ overhangs due to variable arrangements of transcription start and stop sequences, some caused inadvertently by the use of restriction enzyme sites during vector production (Boden et al. 2004; Silva et al. 2005). This raises concerns since recent reports have demonstrated that 2-nucleotide (nt) 3′ overhangs, often observed in natural pre-miRNAs, are optimal substrates for Exportin-5 and Dicer (Zeng and Cullen 2004; Vermeulen et al. 2005). Furthermore, none of the prior hairpin-based comparisons assessed the equivalency of strand biasing (Boden et al. 2004; Silva et al. 2005; Li et al. 2007). Strand biasing refers to which strand of the siRNA duplex enters the RISC and mediates gene silencing. In general, the strand with the weakest base-pairing near the 5′ end will be incorporated into the RISC (Khvorova et al. 2003; Schwarz et al. 2003). Hence, shRNA and artificial miRNA comparisons may be confounded if the vectors, following processing, release siRNAs that exhibit differential strand biasing (i.e., one loading the intended antisense strand and the other loading the unintended sense strand). Indeed, a single base-pair shift during RNAi substrate processing may alter which strand of the resulting siRNA preferentially enters the RISC, thus highlighting the importance of evaluating strand biasing in RNAi vector comparison studies.
In this work, we demonstrate how early generation shRNAs with poor strand biasing confound the comparison of hairpin-based RNAi approaches. Minimizing the variables for comparison reveals that, for three independent target sequences and in different settings (in vitro and in vivo), shRNAs are more potent than artificial miRNAs.
RESULTS
Development of an improved miR-30-based siRNA shuttle
In previous work, we demonstrated the therapeutic efficacy of shRNAs in mouse models of neurodegenerative diseases (Spinocerebellar ataxia type I [SCA1] and Huntington's disease [HD]) (Xia et al. 2004; Harper et al. 2005). To improve our RNAi strategy, we investigated the use of artificial miRNAs as siRNA shuttles. Multiple reports have shown that miRNA-based strategies are more potent than shRNAs (Boden et al. 2004; Silva et al. 2005). As in these previous studies, we modified the human miR-30 primary transcript by replacing the mature miR-30 region (∼20-base-pair [bp] duplex, which is excised via sequential Drosha and Dicer processing) with siRNA sequences (Fig. 1A). The structure of miR-30 has been well characterized, and the major Drosha and Dicer cleavage sites have been experimentally validated (Lee et al. 2003; Zeng and Cullen 2003; Silva et al. 2005). We generated artificial miRNAs containing the structural elements within the stem-base required for effective Drosha processing but otherwise devoid of natural miR-30 flanking sequences. With this strategy, the flanking sequences (∼35 nt upstream and ∼40 nt downstream) arise from the vector sequence within the multiple cloning site (MCS). Although the minimal sequence elements necessary for miR-30 processing are known, the effects of flanking sequences, and consequently the transcript structure within the nearby stem–loop base, on processing efficiency has not been evaluated. To test this, we generated miR-30 shuttle variants predicted to form unique RNA structures proximal to the base of an identical miRNA stem–loop (Fig. 1A,B). Luciferase reporters containing a siRNA target site (∼25 bp) within the 3′ UTR, herein referred to as RNAi luciferase reporters (Fig. 1C), were developed to assess silencing efficiencies in cotransfection studies. The variant flanking sequences influenced silencing efficacy relative to the same miRNA stem–loop flanked by natural human miR-30 sequences (Fig. 1D). Notably, one miRNA variant (miV2) yielded 20% more gene knockdown than the natural miR-30 structure (P < 0.01). Northern blot analysis demonstrated that miV2 generates more antisense RNA relative to the other variants (Fig. 1E); however, whether this results from improved Drosha processing or stability of the pri-miRNA transcript remains unknown. Interestingly, miV2 exhibits a region of single-stranded nature (Fig. 1B, shaded region) that may promote binding of the Drosha–DGCR8 complex, as supported by models for pri-miRNA processing (Zeng and Cullen 2005; Han et al. 2006).
Strand biasing confounds hairpin-based comparisons
We next developed miR-30 shuttles (miV1 and miV2) containing siRNA sequences based on HD2.1 (targets the transcript of the human HD gene) to compare to the short-hairpin shHD2.1 (a shRNA previously tested in therapeutic efficacy studies in a mouse model for HD) (Harper et al. 2005). The processing of CMV-driven miHD2.1 variants and U6-driven shHD2.1 was assessed by small transcript Northern blot analysis (Fig. 2A). Consistent with prior reports (Boden et al. 2004; Silva et al. 2005), the miRNA-based shuttles yielded more antisense RNA than the shRNA vector, which generated an abundance of unprocessed precursor stem–loop RNAs. Also, miV2 generated more antisense RNA than miV1 as previously observed in Figure 1E. Importantly, the improvement of the artificial miRNAs over the shRNA was not the result of promoter discrepancies since our U6-miRNA vectors are even more potent than our CMV-miRNA vectors (data not shown).
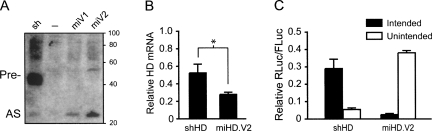
FIGURE 2.
Disparate strand biasing confounds shRNA and artificial miRNA comparisons. (A) Small transcript Northern blot performed at 48 h post-transfection of HD2.1 RNAi expression plasmids in HEK293 cells shows improved yield of processed antisense RNA (AS) from CMV-driven artificial miRNA variants 1 and 2 (miV1, miV2) relative to the U6-driven first-generation shHD2.1 and a mock-treated sample (–). Pre- designates the precursor stem–loop. (B) Q-PCR analysis for endogenous HD mRNA levels performed 48 h after transfection of HD2.1 RNAi expression plasmids into HEK293 cells. Results were normalized to GAPDH mRNA levels and are shown as mean±SEM relative to mock-treated samples (n =3, * = P < 0.05). (C) Strand biasing of U6-driven HD2.1 RNAi vectors. Strand biasing was assessed by measuring luciferase activity from reporters containing either sense (intended; binds RNAi antisense) or antisense (unintended; binds RNAi sense) target sequences in the 3′ UTR (Fig. 1C). RNAi reporter and RNAi expression plasmids were cotransfected into HEK293 cells, and Dual-Glo Luciferase assays were performed at 24 h. Results are shown as mean±SEM (n = 4) relative to mock-treated controls and demonstrate that shHD2.1 preferentially loads the unintended siRNA strand while miHD2.1 more often loads the intended strand.
The silencing efficiencies of CMV-miHD2.1(V2) and U6-shHD2.1 against endogenous HD transcripts were assessed by Q-PCR following transfection of RNAi expression vectors into human-derived HEK293 cells. Consistent with our Northern blot results, miHD2.1 demonstrated ∼50% improved silencing of endogenous HD mRNA relative to shHD2.1 (Fig. 2B, P < 0.05). These findings support prior data showing that miRNA-based strategies are more potent than first-generation shRNAs (Boden et al. 2004; Silva et al. 2005). However, because recent work has shown that strand biasing affects siRNA-mediated gene silencing (Khvorova et al. 2003; Schwarz et al. 2003), we tested whether shHD2.1 and miHD2.1 (both U6 driven) preferentially load the intended antisense strand or unintended sense strand. We designed RNAi luciferase reporters containing either sense or antisense target sites to evaluate the silencing activity of each strand. Cotransfection studies revealed that shHD2.1 preferentially silenced the unintended target strand, while miHD2.1 favored silencing of the intended strand (Fig. 2C). These data are important, as they demonstrate that disparate strand biasing can confound comparisons of shRNA- and miRNA-based approaches.
shRNA expression and potency is overhang dependent
To more fairly compare shRNA- and miRNA-based silencing strategies, we designed shRNAs using the predicted structure of Drosha-cleaved miR-30 stem–loops as a design guide. These shRNAs were designed to have minimized 3′ overhangs (2–4 U's resulting from Pol-III termination; Ng et al. 1979; Ohshima et al. 1981; Kunkel et al. 1986) to resemble the 2-nt 3′ overhangs that result from Drosha cleavage. Overhangs of this length provide optimal substrates for Exportin-5 and Dicer (Zeng and Cullen 2004; Vermeulen et al. 2005). In addition, target sequences were selected to account for the +1-G nucleotide of the mouse U6 promoter and to contain AU-rich 3′ ends, both of which promote loading of the antisense strand (Khvorova et al. 2003; Schwarz et al. 2003). While designing shRNAs in this manner is becoming common practice, the initial shRNA–miRNA comparison studies tested shRNAs with suboptimal overhangs. Hence, we revisited testing the effects of shRNA overhangs (5′ and 3′) on processing and silencing efficiency. We developed a series of vectors expressing a common shRNA but with altered overhang lengths, in certain cases, resembling those (i.e., 5′ variants) found in previous shRNA and artificial miRNA comparison studies (Fig. 3A; Boden et al. 2004; Silva et al. 2005). Northern blot and densitometry analyses showed that shRNAs with the minimal 3′-U2–4 overhangs yield three- and fourfold more pre-shRNA (unprocessed) and antisense RNA (processed), respectively, than shRNAs with 5′ overhangs (Fig. 3B). Unexpectedly, shRNA expression levels appear to decrease with lengthening 5′ or 3′ overhangs. We presume that suboptimal overhangs lower shRNA transcript stability, considering that all variants were expressed by the same mouse U6 promoter and only differ by up to 27 nt in length.
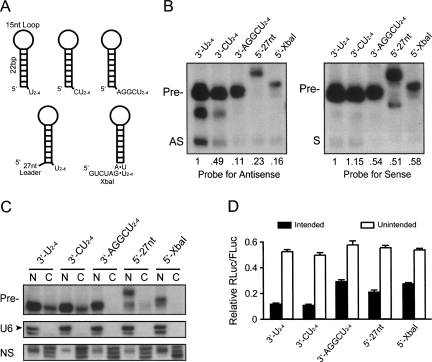
FIGURE 3.
shRNA processing and silencing efficiency is overhang dependent. (A) Diagrams depicting the various 5′ and 3′ overhangs tested on identical shRNA stem–loops. The 5′-27 nt (Silva et al. 2005) and 5′-XbaI (Boden et al. 2004) variants were used in prior shRNA and miRNA comparison studies. (B) Plasmids expressing the shRNA variants were transfected into HEK293 cells, and small transcript Northern blot (probing for antisense [AS] or sense [S] sequences) with densitometry analysis (values for AS and S bands are shown below blots) was performed 48 h later to assess shRNA processing efficiency (n = 3). Results show that 5′ overhang variants yield less precursor (Pre-) and antisense (AS) RNAs compared to the optimized shRNAs with U2–4 3′ overhangs (derived from Pol-III transcription termination (Ng et al. 1979; Ohshima et al. 1981; Kunkel et al. 1986). Appropriate strand loading was observed for each shRNA variant (i.e., AS:Pre->S:Pre-). (C) Plasmids expressing the shRNA variants were transfected into HEK293 cells, and nuclear/cytoplasmic fractionation and RNA isolation was performed 24 h later. Equal amounts of nuclear and cytoplasmic RNAs for each treatment group were analyzed by small transcript Northern blot (probing for antisense sequences) to evaluate where precursor (Pre-) shRNAs accumulate. Results demonstrate that the two most highly expressed shRNAs (3′-U2–4 or 3′-CU2–4) accumulate in both nuclear and cytoplasmic fractions, suggesting that saturation of both Exportin-5 and Dicer may occur. Conversely, buildup of the other shRNA variants was present primarily in the nucleus, suggesting lack of export due to suboptimal overhangs. Note: the presumed Drosha-cleavage product of the 5′-27 nt shRNA is exported and accumulates slightly in both compartments. The blot was stripped and reprobed for the nuclear U6 splicing RNA to control for fractionation and loading. Nonspecific bands (NS) present following the control probing are shown as additional fractionation and loading controls. (D) Silencing of intended (solid bars) or unintended (empty bars) target strands was assessed by cotransfection of shRNA-variant and RNAi luciferase reporter expression plasmids into HEK293 cells, and Dual Glo Luciferase assays were performed at 24 h. Results are shown as mean±SD (n = 3) relative to mock-treated controls and confirm that suboptimal overhangs decrease silencing efficiency. Notably, each shRNA preferentially silenced the intended target (transfected at 1:20 RNAi:target) relative to the unintended target (transfected at 3:1 RNAi:target).
In addition to this possibility, suboptimal overhangs may decrease export and processing efficiencies mediated by Exportin-5 and Dicer, respectively. To address this, we performed nuclear/cytoplasmic fractionation studies and evaluated if precursor shRNAs (Pre-) accumulate in those compartments. Our results demonstrate that the two most highly expressed shRNAs (3′-U2–4 or 3′-CU2–4) accumulate in both nuclear and cytoplasmic fractions, suggesting saturation of both Exportin-5 and Dicer (Fig. 3C). Conversely, buildup of the other shRNA variants occurs primarily in the nucleus, suggesting lack of export due to suboptimal overhangs.
In gene silencing studies targeting a cotransfected RNAi luciferase reporter, we found that shRNAs with minimal overhangs (3′-U2–4 or 3′-CU2–4) were most effective (P < 0.001), while efficiency is compromised upon increasing 5′ or 3′ overhangs (Fig. 3D). Northern blot analyses and gene silencing studies targeting sense or antisense RNAi luciferase reporters demonstrated that each shRNA variant favored loading of the intended antisense strand (Fig. 3B,D). While shRNAs with longer overhangs demonstrate slightly reduced intended:unintended silencing ratios, their decreased silencing efficiencies likely result from reduced transcript stability and processing as supported by our Northern blot data.
Minimizing the variables between artificial miRNA and shRNA vectors for comparison
Next, we sought to compare our improved artificial miRNAs (Fig. 1, miV2) and shRNAs (Fig. 4A, 3′-U2–4) for processing and silencing efficiency. To test for consistency of results independent of target and sequence, we designed shRNA- and miRNA-based vectors targeting SCA1, HD, and GFP mRNAs—the former two providing constructs to further test in therapeutic development. Importantly, and unique from other shRNA and miRNA comparisons (Boden et al. 2004; Silva et al. 2005; Li et al. 2007), our artificial miRNAs were adjusted to account for nucleotide restrictions associated with Pol-III transcription of shRNAs (Fig. 4). Hence, following Drosha cleavage, the pre-miRNAs would be comparable to the shRNAs, and subsequent processing of each stem–loop by Dicer would yield similar siRNA species. This is an important consideration, since a single base-pair shift may alter the strand biasing or potency of the resulting siRNA.
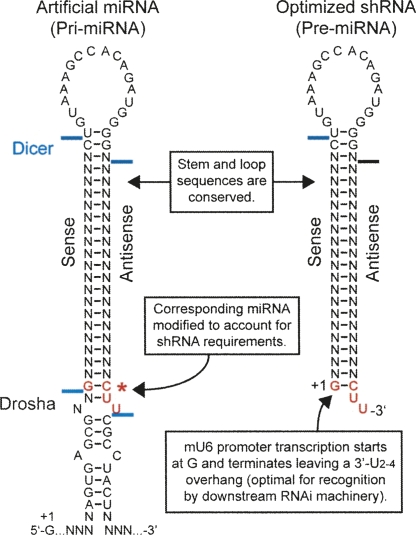
FIGURE 4.
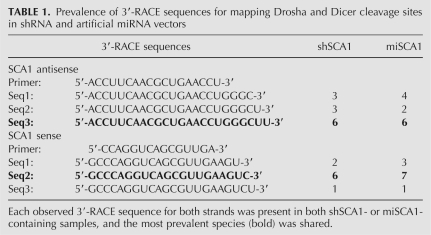
Design of comparable shRNA and artificial miRNA hairpins. Diagram depicting the design of hairpins for a fair comparison scheme. Relevant cleavage sites mapped by 3′-RACE are shown (Table 1). Vectors were designed to contain siRNAs targeting SCA1, HD, or GFP transcripts (Ns).
To evaluate the processing of our vectors, we performed 3′-RACE to identify the 3′ ends of the sense and antisense strands of siRNAs generated by corresponding artificial miRNA and shRNA expression vectors (Table 1). To date, none of the RNAi vector comparison studies have performed this important analysis to test the equivalency of processing between the compared vectors. Our 3′-RACE analyses revealed that both RNAi vectors generate at least four siRNA species that were common between the vectors. These findings are consistent with a previous report that demonstrated flexibility (i.e., base-pair shifting) in dsRNA cleavage by Dicer (Vermeulen et al. 2005). Notably, the most prevalent species generated by the RNAi vectors was shared, representing ∼50% of the 3′-RACE sequences analyzed (n = 10–12 per vector per strand). These results support the integrity of comparing artificial miRNAs and shRNAs designed with our strategy.
TABLE 1.
Prevalence of 3′-RACE sequences for mapping Drosha and Dicer cleavage sites in shRNA and artificial miRNA vectors
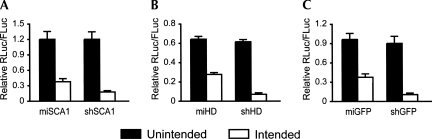
Next, we assessed the strand biasing of these RNAi vectors by targeting sense or antisense RNAi luciferase reporters, since inconsistent strand biasing confounded our original comparison (Fig. 2). In all three RNAi vector pairs (SCA1, HD, and GFP), a strong preference for silencing the intended strand was observed for both artificial miRNAs and shRNAs (Fig. 5). For the SCA1 and GFP vectors, neither of the RNAi expression strategies silenced the unintended targets, and only minimal activity of the unintended strand was observed with the HD RNAi vectors. These results demonstrate that the desired strand biasing is consistent for each RNAi vector pair, thus maintaining their suitability for further comparison studies.
FIGURE 5.
Comparable shRNA- and miRNA-based vectors exhibit appropriate strand biasing. (A–C) Strand biasing of SCA1, HD, and GFP RNAi vectors, respectively. Strand biasing was assessed using luciferase reporters containing either sense (intended) or antisense (unintended) target sequences. RNAi luciferase reporter and RNAi expression plasmids were cotransfected into HEK293 cells, and Dual-Glo Luciferase assays were performed at 24 h. Results of duplicate experiments (each n = 3) are shown as mean±SEM relative to mock-treated controls.
shRNAs show improved potency relative to artificial miRNAs
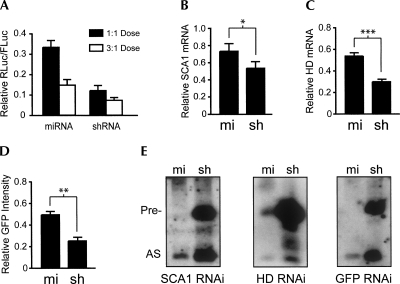
As an initial comparison of gene silencing efficacy, the RNAi vectors and their respective RNAi luciferase reporters were cotransfected into HEK293 cells. Luciferase assay data from several experiments (n = 4 SCA1, n = 2 HD, and n = 4 GFP) were compiled, revealing that optimized shRNAs are more potent than artificial miRNAs independent of the RNAi doses tested and target sequence (Fig. 6A, P < 0.001 and P < 0.05 for 1:1 and 3:1 [RNAi:target] doses, respectively). This improved efficacy of the shRNAs was also observed per vector pair in our previous strand biasing studies (Fig. 5). Notably, these results were not confounded by potential toxicity issues related to shRNA expression, as overall activities of nontargeted luciferase vectors were consistent independent of treatment (shRNA versus miRNA) for all vector pairs (P < 0.01) (data not shown).
FIGURE 6.
shRNAs are more potent than artificial miRNAs in vitro. (A) RNAi and RNAi luciferase reporter plasmids were cotransfected into HEK293 cells to assess gene silencing. Dual Glo Luciferase assays were performed at 24 h and results, shown as mean±SEM relative to mock-treated controls, were compiled from several experiments (4 GFP, 4 SCA1, and 2 HD; each n = 3). Dose is indicated as RNAi:target. P < 0.001 and P < 0.05 for 1:1 and 3:1 doses, respectively. (B,C) Plasmids expressing RNAi targeting SCA1 or HD were transfected into HEK293 cells, and Q-PCR analysis was performed at 48 h to measure reduction of endogenous transcripts. SCA1 and HD mRNA levels were normalized to GAPDH mRNA or 18S rRNA and are shown as mean±SEM (n ≥ 3, * = P < 0.05, *** = P < 0.001) relative to mock-treated controls. (D) GFP RNAi and eGFP expression plasmids were cotransfected into HEK293 cells, and fluorescence levels were evaluated 48 h later. Results are shown as mean±SEM (n = 4, ** = P < 0.01) relative to SCA1 RNAi-treated controls. (E) shRNA and artificial miRNA expression plasmids were transfected into HEK293 cells, and small transcript Northern blot was performed at 48 h to assess RNAi expression and processing. (Pre-) precursor; (AS) antisense RNA. Results show that shRNAs yield more than fourfold mature antisense RNA, relative to artificial miRNAs, independent of RNAi target sequence. These results were consistent among triplicate blots for each RNAi vector pair.
Though targeting RNAi luciferase reporters provides a quantitative and facile means for assessing gene silencing, we also wanted to evaluate the capacities for these RNAi vectors to silence their natural targets. In HEK293 cells transfected with RNAi expression plasmids, silencing trends of shRNA>miRNA were also observed when targeting endogenous SCA1 or HD transcripts (Fig. 6B,C, P < 0.05, P < 0.001, respectively). Furthermore, the shRNA targeting GFP demonstrated improved potency over the corresponding artificial miRNA following cotransfection of RNAi and GFP expression plasmids into HEK293 cells (Fig. 6D, P < 0.01).
Next, we compared the processing efficiencies of our artificial miRNA and shRNA vector pairs in vitro following transfection of RNAi expression plasmids. Small transcript Northern blots demonstrate that, unlike our first-generation shRNAs (Fig. 2A), the optimized shRNAs produce at least fourfold more antisense RNA as compared to artificial miRNAs independent of target (Fig. 6E). This improved yield likely results from higher expression levels, since optimized shRNAs still generate high levels of precursor, whereas artificial miRNAs show minimal buildup of pri- or pre-transcripts.
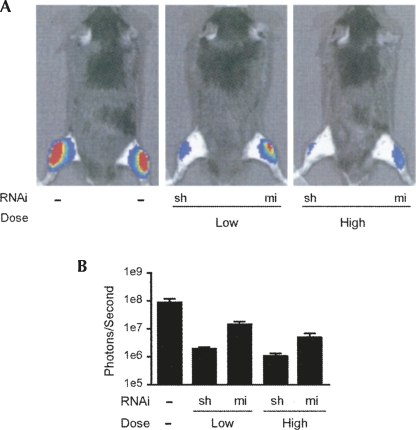
In vivo applications of RNAi are becoming widely used to study gene function or develop therapeutics. As such, we compared one of our vector pairs (shSCA1 and miSCA1) for gene silencing in mice. Since these constructs target human SCA1, and not mouse SCA1, we measured in vivo silencing of a co-delivered SCA1 RNAi luciferase reporter. RNAi and RNAi luciferase reporter plasmids were coelectroporated into the tibialis anterior muscles of mice, and bioluminescence imaging was performed to determine in vivo gene silencing at 4 and 8 d post-treatment. At 1:1 and 10:1 (RNAi:target) ratios, shRNAs show improved potency over artificial miRNAs (Fig. 7). These results, together with our in vitro data, demonstrate that shRNAs can be more potent than miRNAs independent of target and experimental setting (in vitro and in vivo).
FIGURE 7.
shRNAs are more potent than artificial miRNAs in vivo. (A) Gene silencing efficacy in vivo was compared by coelectroporating SCA1 RNAi and RNAi luciferase reporter plasmids into tibialis anterior muscles of 6–8-wk-old mice. Low and high doses are 1:1 and 10:1 (RNAi:target) ratios, respectively. Renilla luciferase activity was measured in vivo using bioluminescence imaging after 8 d. Representative “heat-map” images are shown along with quantitative analysis (B) represented as mean±SEM (n =4; P < 0.05 within each dose). Similar silencing trends were also observed at 4 d post-treatment (data not shown).
DISCUSSION
We have developed a novel miR-30-based siRNA shuttle that shows improved potency over the commonly used miR-30 shuttle, which contains natural miR-30 flanking sequences. Whether this improved potency is the result of increased Drosha processing or stability of the pri-miRNA transcript remains unknown. Further experiments employing in vitro Drosha cleavage assays or Drosha knock-out cell lines may help to elucidate the precise mechanism. As we only tested three miRNA variants, additional optimization of the flanking sequences may be warranted to improve the artificial miRNA strategy. miRNA-based approaches provide an attractive alternative to classic shRNAs; hence, we and others have compared the efficiencies of these systems in mediating gene silencing.
We present data demonstrating that shRNA and artificial miRNA comparison analyses must be carefully controlled, as differential strand biasing between the compared vectors may generate misleading results. This possibility, in addition to the use of shRNAs with suboptimal overhangs, may explain the discrepancies among previously published hairpin-based comparisons studies. Our analyses provide a fair comparison of shRNA- and miRNA-based vectors, taking into consideration the transcribed product, processing sites, and strand biasing. When variables between the two systems are minimized, shRNAs demonstrate improved silencing efficacy relative to their miRNA-based counterparts. While our results corroborate the Li et al. (2007) comparison, our experiments are unique in that we carefully controlled vector design (e.g., maintained stem length and loop structures between compared vectors), assessed the equivalency of siRNAs generated and strand biasing, and validated the improved potency of shRNAs in vivo. Furthermore, we demonstrate that shRNAs yield abundant levels of precursor and fully processed antisense RNA relative to artificial miRNAs, thus providing mechanistic insight to explain the increased potency. Whether shRNAs are transcribed more highly or show improved stability—the latter being more likely—is not clear at this time. The issue of transcript stability is quite interesting; particularly given our novel findings that the addition of a few nucleotides to shRNA overhangs can profoundly decrease expression levels (Fig. 3B). Identifying sequences or structural elements that stabilize RNAi transcripts may help to improve the potency of shRNA- and miRNA-based strategies.
General users of RNAi must carefully consider the balance of efficacy and toxicity when selecting the most suitable RNAi expression strategy. In our RNAi comparison studies, we evaluated gene silencing at 24–48 h post-transfection and found that the potential for RNAi-related toxicity did not confound our results. However, longer-term expression of RNAi vectors may induce cytotoxicity and alter the efficiency profile for a given RNAi strategy. For instance, the improved potency of optimized shRNAs may be offset by toxicity issues; high levels of RNAi substrates (processed and unprocessed) generated from shRNA expression vectors may saturate cellular RNAi machinery, thus interfering with endogenous miRNA biogenesis and function in cell culture and in vivo (Grimm et al. 2006; Castanotto et al. 2007). Recent work by our laboratory and Castanotto et al. (2007) reveals that miRNA-based strategies are less prone to interfering with these processes, suggesting that artificial miRNAs may be less toxic (McBride et al. 2008; R.L. Boudreau and B.L Davidson, unpubl.).
The robust expression of optimized shRNAs may be desirable for certain applications. For example, toxicity is likely minimized in low-copy applications such as the generation of stable cell lines via retroviral vectors, which typically transduce at a few to one copy per cell. Hence, shRNAs may be better suited than miRNA-based vectors where low-level expression would preclude sufficient silencing. Alternatively, shRNAs may be advantageous when silencing highly expressed targets that may outcompete low levels of antisense RNA; for example, therapeutic targeting of invasive viruses or cancers, where limiting toxicity may not be the highest priority.
For some applications, high vector doses must be delivered to achieve efficient transduction of a cell population. Hence, shRNA strategies may be undesirable if vector copies per cell cannot be readily limited, as often occurs in vivo, and minimizing toxicity is of high priority. In addition to saturation-based toxicity, high levels of siRNAs may promote silencing of unintended targets via partial complementarity (Birmingham et al. 2006). Hence, utilizing artificial miRNAs, which exhibit lower expression, may provide a unique opportunity to limit RNAi substrate levels when large vector doses must be administered to achieve efficient cell targeting throughout a given tissue. This concept is particularly relevant to the development of therapeutic RNAi, where transduction efficiency and vector safety are of the utmost importance.
Inevitably, the selection of which hairpin-based RNAi expression system is most suitable for a given application relies on several factors: project goals, delivery options, target expression levels, and desired silencing efficiency. Thus, empirical testing of these RNAi expression systems may be required for each target and tissue on a per study basis. The approach outlined here, where variables of strand biasing and processing are minimized, can help guide researchers in identifying suitable vectors for their RNAi-based applications.
MATERIAL AND METHODS
Vector design
An expression cassette (5′NheI–3′BglII) containing the CMV promoter, multiple cloning site (MCS), and SV40 polyadenylation (polyA) signal was cloned into TOPO-BluntII (XbaI–BamHI; Invitrogen) in reverse orientation. A Pol-III expression cassette containing mouse U6 promoter, MCS, and Pol-III-terminator (6T's) was cloned into TOPO-BluntII with the same strategy. Artificial miRNAs were generated by polymerase extension of overlapping DNA oligonucleotides (IDT). Polymerase-extended products were digested with XhoI–SpeI and cloned into the MCS of the expression vectors XhoI–SpeI (miV1) or XhoI–XbaI (miV2). miV3 resulted from an unexplained cloning error. shHD2.1 has been previously described (Harper et al. 2005). Other shRNAs were cloned using a tailed-PCR strategy. PCR products containing U6–shRNA–6T's were TOPO cloned into the BluntII vector, and positive clones were considered to be in reverse orientation to maintain consistency with the other expression vectors.
For in vivo studies, miRNA or shRNA expression cassettes driven by the mouse U6 promoter were cloned in the same orientation upstream of a CMV-hrGFP-SV40polyA expression cassette.
RNAi luciferase reporter vectors were constructed using psiCheck2 (Promega). Tailed-PCR was used to amplify a 3′ fragment of Renilla luciferase containing a single, perfect complementary RNAi target site (∼25 bp: 21-bp target+2-nt 5′ and 3′) downstream from the stop codon. This PCR product was digested with AatII–XhoI and cloned into the same sites within psiCheck2.
See Supplemental Table for vector construction information.
3′-RACE analyses
HEK293 cells grown in six-well plates were transfected (Lipofectamine 2000; Invitrogen) with RNAi expression plasmids (200 ng or 2 μg for shRNA and artificial miRNA plasmids, respectively). Total RNA was isolated at 48 h post-transfection using 1 mL TRIzol reagent (Invitrogen). Subsequently, 250 ng of total RNA were treated with A-Plus Poly(A) polymerase (Epicentre) and then subjected to first-strand cDNA synthesis using an anchored oligo-dT primer containing linker sequence (RLM-RACE, Ambion; TaqMan reverse transcription reagents, Applied Biosystems). PCR was performed using the following primers:
Linker primer: 5′-TTAATACGACTCACTATAGGT-3′;
SCA1 antisense RACE: 5′-ACTTCAACGCTGACCT-3′; and
SCA1 sense RACE: 5′-CCAGGTCAGCGTTGA-3′.
Products were TOPO cloned into the pCR2.1 vector (Invitrogen) and sequenced.
Northern blot analyses
HEK293 cells grown in six-well plates were transfected with 1.5 μg of RNAi expression plasmid. Total RNA was isolated at 48 h post-transfection using 1 mL of TRIzol reagent; alternatively, the small RNA fraction was harvested using mirVana Isolation Kit (Ambion). We resolved 15–20 μg total RNA or 1–2 μg small RNA fraction on a 15% acrylamide gel. Small transcript sizes were determined with the Decade Ladder (Ambion). Consistent loading and RNA integrity was assessed by ethidium bromide stain. RNA was transferred to Hybond-XL membrane (Amersham Pharmacia) at 200–400 mA for 1–2 h and UV-cross-linked with the auto-cross-link function on a Stratalinker 1800 (Stratagene). Blots were prehybridized using UltraHyb-Oligo (Ambion) at 35°C, probed with 32P-labeled DNA oligonucleotides (Ready-To-Go T4 polynucleotide kinase; Amersham) at 30°C–35°C overnight, washed three times (5 min each) in 2× SSC, 0.1% SDS at 30°C–35°C, and exposed to film. Alternatively, blots were probed with biotin-labeled DNA oligonucleotides and analyzed using the Bio-Detect Kit (Ambion). Densitometry analyses were performed using a bioimaging system (UVP) coupled with LabWorks software (UVP). Blot stripping was performed by washing with 0.5% SDS at 65°C three times (10 min each).
For fractionation studies, nuclear and cytoplasmic lysates were harvested using the PARIS kit (Ambion), and RNA was isolated from each fraction using 1 mL of TRIzol reagent. Approximately 12 μg of nuclear and cytoplasmic RNAs were subjected to the Northern blot protocol described above. After probing for antisense sequences to detect precursor shRNAs, blots were stripped and reprobed to detect both nuclear U6 and cytoplasmic Met-tRNA transcripts. Met-tRNA bands of appropriate size could not be discerned due to intense U6 signal; thus, nonspecific bands between ∼20 and 30 nt were used to control for cytoplasmic RNA integrity and loading.
Quantitative real-time PCR analyses
HEK293 cells grown in 24-well plates were transfected with 700 ng RNAi-expressing plasmids. At 48 h post-transfection, total RNA was isolated with 0.5 mL TRIzol reagent, and random-primed first-strand cDNA synthesis was performed using 1 μg total RNA (TaqMan reverse transcription reagents; Applied Biosystems) per the manufacturer's protocol. Assays were performed on a sequence detection system using primers-probe sets specific for human HD, SCA1, or GAPDH mRNAs, or 18S rRNA (Prism 7900HT and TaqMan 2X Universal Master Mix; Applied Biosystems). Relative gene expression was determined by using the relative standard curve method.
GFP silencing analyses
HEK293 cells grown in 12-well plates were transfected with 1.2 μg and 300 ng of RNAi (SCA1 or GFP) and eGFP expression plasmids, respectively. At 48 h post-transfection, fluorescent photomicrographs were captured at 4× magnification using an Olympus IX70 (microscope) and DP70 (camera) coupled with Olympus DP Controller software. Mean fluorescence in each image was determined using the histogram function in Image J software (NIH). Results for GFP RNAi-treated cells were normalized to control SCA1 RNAi-treated cells.
In vitro luciferase assays
HEK293 cells grown in black 96-well plates (Costar 3603; Corning Inc.) were cotransfected in triplicate with RNAi-expressing plasmids (1–60 ng) and RNAi luciferase target or nontargeted luciferase plasmids (10–20 ng). In dosing studies, empty-vector was supplemented to low doses to match total DNA load. Firefly and Renilla luciferase activities were assessed 24 h post-transfection using the Dual-Glo Luciferase Assay System (Promega) per the manufacturer's instructions, using 50 μL per substrate. Luminescent readings were acquired with a 96-well plate luminometer (Dynex). Results were calculated as the quotient of Renilla/Firefly luciferase activities.
In vivo luciferase assays
Animal studies were approved by the University of Iowa Animal Care and Use Committee. Eight-week-old male C57/BL6 mice (Jackson Laboratories) were anesthetized with ketamine-xylazine and injected with 30 μL of 0.4 U/μL hyaluronidase (Sigma) into the tibialis anterior (TA) muscle. Two hours later, plasmids in 30 μL saline were injected into the TA muscle of reanesthetized mice. All groups (n = 4 muscles) received 1 μg RNAi luciferase reporter plasmid along with 10 μg empty vector or RNAi plasmid (high dose) or 9 μg empty vector and 1 μg RNAi plasmid (low dose). Plasmids were prepared using the EndoFree Plasmid Maxi Kit (Qiagen). Plasmid-injected muscles were electroporated as previously described (McMahon et al. 2001) applying 175 V/cm in ten 20-ms pulses at 2 Hz (ECM 830 electroporator, BTX). At 4 and 8 d post-treatment, mice were sedated with isoflurane, and 30 μL of coelenterazine (0.3 mg/mL, Promega) were injected into the TA muscles. Bioluminescence imaging was performed immediately using an IVIS200 imaging system (Xenogen). Light emissions were analyzed using Living Image software (Xenogen) and Igor Pro image analysis software (WaveMetrics Inc.). Data collected at 4 and 8 d revealed similar silencing trends (4 d not shown).
Statistical analyses
Student's t-Test was used for all studies where P-values are provided. In all statistical analyses, P < 0.05 was considered significant.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This research was supported by funds from the NIH (NS-50210, HD-44093, DK-54759), the Hereditary Disease Foundation, the National Ataxia Foundation, and the Roy J. Carver Trust. R.L.B. is supported by the Lori C. Sasser Fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1062908.
REFERENCES
- Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Silbermann R., Lee F., Tucker L., Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., Sakurai K., Lingeman R., Li H., Shively L., Aagaard L., Soifer H., Gatignol A., Riggs A., Rossi J.J. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.H., Hart C.C., Al-Bassam S., Avery A., Taylor J., Patel P.D., Vojtek A.B., Turner D.L. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S.Q., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fish R.J., Kruithof E.K. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC Mol. Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kunkel G.R., Maser R.L., Calvet J.P., Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl. Acad. Sci. 1986;83:8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li L., Lin X., Khvorova A., Fesik S.W., Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: Implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J.M., Signori E., Wells K.E., Fazio V.M., Wells D.J. Optimization of electrotransfer of plasmid into skeletal muscle by pretreatment with hyaluronidase—Increased expression with reduced muscle damage. Gene Ther. 2001;8:1264–1270. doi: 10.1038/sj.gt.3301522. [DOI] [PubMed] [Google Scholar]
- McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Parker C.S., Roeder R.G. Transcription of cloned Xenopus 5S RNA genes by X. laevis RNA polymerase III in reconstituted systems. Proc. Natl. Acad. Sci. 1979;76:136–140. doi: 10.1073/pnas.76.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Okada N., Tani T., Itoh Y., Itoh M. Nucleotide sequences of mouse genomic loci including a gene or pseudogene for U6 (4.8S) nuclear RNA. Nucleic Acids Res. 1981;9:5145–5158. doi: 10.1093/nar/9.19.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Silva J.M., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A., Behlen L., Reynolds A., Wolfson A., Marshall W.S., Karpilow J., Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Mao Q., Paulson H.L., Davidson B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a mouse model of SCA1. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]