Abstract
Terminal oligopyrimidine (TOP) mRNAs (encoded by the TOP genes) are identified by a sequence of 6–12 pyrimidines at the 5′ end and by a growth-associated translational regulation. All vertebrate genes for the 80 ribosomal proteins and some other genes involved, directly or indirectly, in translation, are TOP genes. Among the numerous translation factors, only eEF1A and eEF2 are known to be encoded by TOP genes, most of the others having not been analyzed. Here, we report a systematic analysis of the human genes for translation factors. Our results show that: (1) all five elongation factors are encoded by TOP genes; and (2) among the initiation and termination factors analyzed, only eIF3e, eIF3f, and eIF3h exhibit the characteristics of TOP genes. Interestingly, these three polypeptides have been recently shown to constitute a specific subgroup among eIF3 subunits. In fact, eIF3e, eIF3f, and eIF3h are the part of the functional core of eIF3 that is not conserved in Saccharomyces cerevisiae. It has been hypothesized that they are regulatory subunits, and the fact that they are encoded by TOP genes may be relevant for their function.
Keywords: translational control, translation factor, polysomal analysis, protein synthesis
INTRODUCTION
An important fraction of the mRNAs of vertebrate cells (up to 20%) have a characteristic 5′ UTR, which is relatively short and starts with a sequence of 6–12 pyrimidines, called the 5′-terminal oligopyrimidine (5′ TOP) sequence (Meyuhas and Hornstein 2000). Consequently, these mRNAs are known as TOP mRNAs, and the genes which encode them, as TOP genes. The TOP sequence was first noted in vertebrate genes encoding ribosomal proteins (RPs), confirming the earlier indication of an abundant group of mRNAs starting with m7GpppPy (Schibler et al. 1977). TOP gene expression has been gaining increasing attention in the last few years. In fact, ribosome biogenesis, far from being a passive constitutive process, has been linked to cell-size control and tumorigenesis (Ruggero and Pandolfi 2003; Ruvinsky and Meyuhas 2006). However, the number and the regulation of TOP genes are not yet clearly defined. The TOP sequence is a cis-acting element necessary for a growth-associated translation regulation. In fact, TOP mRNAs are mostly localized on polysomes in actively growing cells but mostly sequestered in inactive mRNPs in quiescent or growth-arrested cells (Geyer et al. 1982; Loreni and Amaldi 1992). A number of studies have implicated the phosphoinositide 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) signaling pathway in the translational activation of TOP mRNAs (Fumagalli and Thomas 2000; Ruvinsky and Meyuhas 2006). However, the contribution of mTOR to the translational control of TOP mRNAs may be dependent on the cellular context; in some studies, inhibition of mTOR has little or no effect on TOP mRNA translation (Ruvinsky and Meyuhas 2006). More clear is the involvement of the PI3K which could activate TOP mRNA translation through mTOR and/or undefined additional pathways (Tang et al. 2001; Stolovich et al. 2002; Caldarola et al. 2004; Pende et al. 2004). However, the final effector(s) that directly affect TOP mRNAs have not been clearly identified. Some putative TOP mRNA translational regulators have been proposed: the autoantigen La, which binds the TOP sequence (Pellizzoni et al. 1996; Crosio et al. 2000; Zhu et al. 2001; Cardinali et al. 2003; Schwartz et al. 2004), and the cellular nucleic acid binding protein (CNBP), which binds a downstream region (Pellizzoni et al. 1997). The precise role of these proteins in TOP mRNA translational regulation as well as their relation with PI3K signaling pathways is still to be defined.
A canonical TOP mRNA is identified by two essential criteria: (1) the presence of a TOP sequence at the 5′ end and (2) a growth-associated translational regulation. According to this definition, all messengers coding for vertebrate RPs analyzed so far in various experimental systems are TOP mRNAs. Moreover, some additional structural features present in all 80 RP mRNA (as, for instance, a very short 3′ UTR) suggest that all of them are indeed coordinately regulated TOP mRNAs (Ledda et al. 2005). However, this assumption remains to be demonstrated, and a recent microarray analysis of translational regulation suggested the existence of subsets of RP mRNAs with different sensitivities to eukaryotic initiation factor (eIF) 4E levels (Mamane et al. 2007). Among the non-RP TOP mRNA, a common feature is the correlation of the encoded proteins with some aspects of translation. For instance, messengers for some eukaryotic elongation factors (eEFs) as well as poly(A)-binding protein and hnRNP A1 have been shown to exhibit “TOP” characteristics (Loreni et al. 1993; Terada et al. 1994; Camacho-Vanegas et al. 1997; Hornstein et al. 1999). This subset of TOP mRNA includes mRNAs with ambiguous TOP characteristics, such as, for instance, tissue-specific translational regulation (Avni et al. 1997). Therefore, the question of what kind of and how many TOP mRNAs are expressed in the cell is still open. A few cDNA array studies on translational regulation in different experimental systems have found several known TOP genes among other coding sequences (Mikulits et al. 2000; Lu et al. 2006; Provenzani et al. 2006; Spence et al. 2006). The identification of new TOP genes was not, however, the aim of these studies, and the identified messengers were not subjected to further investigation (such as the presence of a TOP sequence). To investigate the group of TOP genes, we decided to use a more direct and specific strategy by selecting genes on the basis of some rational criteria and then analyzing them by structural and functional assays. The first group of genes selected includes the translation factors as there is an evident correlation with translation and some components of this group of proteins are encoded by known TOP genes (e.g., eEF1A, eEF2). Since, as indicated above, the two main characteristics of TOP genes are the pyrimidine sequence at the 5′ end of the mRNA and the growth-associated translation regulation, we first selected possible TOP candidates on the basis of the presence of a pyrimidine sequence at their 5′ end. Candidate genes were then experimentally tested by the polysome association assay to verify their regulation at the translational level. We have found that all elongation factors are encoded by TOP genes, whereas nearly all the other translation factors are not. The few exceptions identified—eIF3e, eIF3f, and eIF3h—may suggest specific common functions for these factors.
RESULTS AND DISCUSSION
Structural analysis of candidate genes
The sequences coding for all translation factors were analyzed for the presence of a TOP sequence at the 5′ end of the transcript. To identify the transcription start site (tss) of each gene of interest, we proceeded as follows:
(1) We searched the NCBI “Homo sapiens (human) genome view” (http://www.ncbi.nlm.nih.gov/mapview) where, in particular, the UniGene map shows mRNA and EST sequences aligned to the assembled human genomic sequence, thus indicating a putative tss.
(2) In some cases, we considered it necessary to verify the alignment by a “blast” analysis of the human ESTs versus the genome sequence surrounding the promoter/tss region.
(3) The results were compared with the tss as presented in the Transcriptional Start Sites Database (DBTSS: http://dbtss.hgc.jp), which is based on the analysis of a full-length cDNA library constructed by the “oligo-capping” method (Ota et al. 2004).
(4) Finally, in a few cases (eEF1β, eEF1γ, eEF1δ, and eIF3e) we experimentally analyzed the tss by primer extension. The results, shown in Figure 1, basically confirmed the in silico analysis. Therefore, we did not extend this experimental approach to the other mRNAs.
FIGURE 1.
Primer extension. A 5′ end-labeled primer (specific for the indicated mRNAs) was annealed to 20 μg of total RNA from HeLa cells and then extended with reverse transcriptase. Extension products were separated on 6% polyacrylamide gel and exposed for autoradiography. The major extension band is indicated by the arrow. The nucleotide corresponding to the major extension band is indicated by an “*” in the sequence reported in the lower part of each panel which include the surrounding of the tss. M, 10-bp size marker; seq, unrelated sequence ladder; PE, primer extension products.
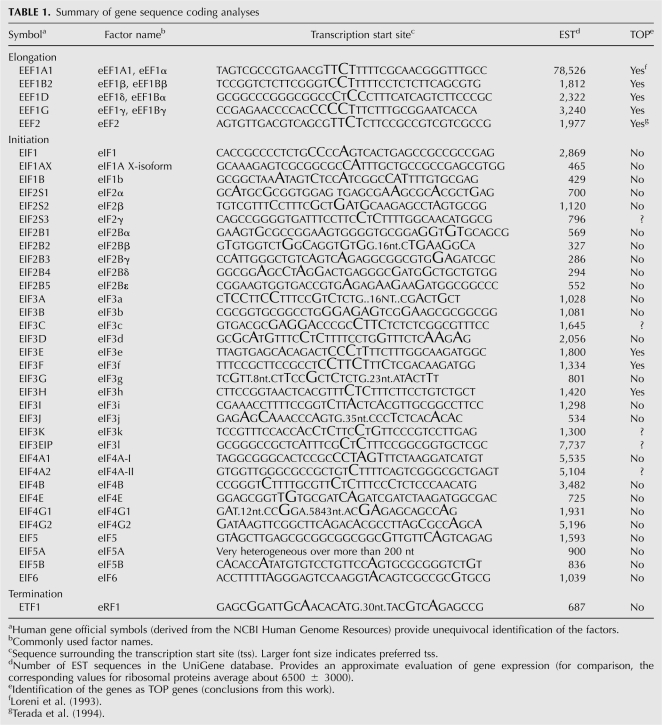
A summary of these analyses is presented in Table 1. The sequences surrounding the major tss of the genes for human translation factors are reported with the tss nucleotides indicated by a larger font size, approximately proportional to frequency. Most of the tss do not appear precisely localized on a single position, being often dispersed on a few nucleotides. The genes coding for the five elongation factors and a small number of those for the numerous initiation factors present a 5′-terminal oligopyrimidine sequence, thus representing good candidates for further analysis. On the other hand, the genes for most of the remaining initiation factors and for the termination factor eRF1 do not present a TOP sequence at the 5′ end. A few cases remain uncertain, mainly due to the above-mentioned tss heterogeneity, resulting in only a fraction of the mRNA, starting with the TOP sequence. We have also derived the gene expression profiles from NCBI UniGene database. The results, reported in column 4 of Table 1, indicate that the relative abundance of the mRNAs considered in this report is quite variable. The analysis of mRNA abundance, tss, and translational regulation (see below), suggests that there is no relationship between the expression level of a gene and its belonging to the TOP gene group.
TABLE 1.
Summary of gene sequence coding analyses
Analysis of growth-associated translation regulation
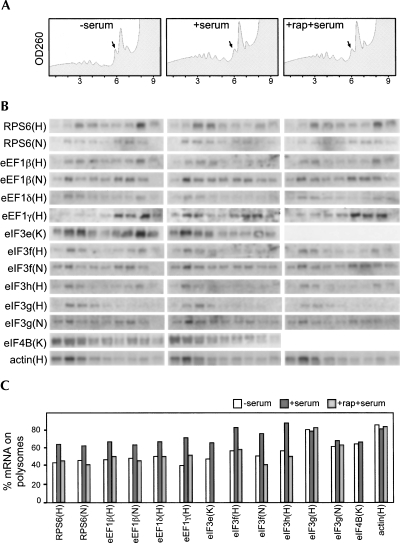
We have analyzed our candidate genes by the polysome association assay to verify their regulation at the translational level during nutritional shifts. For this purpose we used HeLa, HEK293, and NIH3T3 cells in the following growth conditions: (1) serum-starved for 2 h (HeLa and HEK293) or overnight (NIH3T3); (2) serum-stimulated (after starvation) for 1 or 2 h; and (3) serum-stimulated for 1 or 2 h in the presence of rapamycin (treatment started 30 min before stimulation), which is known to suppress specifically TOP mRNA translation (Fumagalli and Thomas 2000). Cytoplasmic extracts were prepared and fractionated through sucrose gradients (see Materials and Methods). Nine fractions were collected from each gradient while recording the absorbance profile. The RNA extracted from the gradient fractions was then analyzed by agarose gel electrophoresis and Northern blot hybridization with probes for: (1) the candidate TOP mRNAs (or as a control translation factor mRNAs without the TOP sequence); (2) a known TOP mRNA (RPS6 or RPS19); and (3) a non-TOP mRNA (β-actin). Quantification of the radioactive signals in the polysomal and nonpolysomal regions of the gradients was then used to evaluate the percentage of a given mRNA associated with the polysomes. The experiments were performed in HeLa, HEK293, and NIH3T3 cells. However, since the results with few of the probes were similar in all three cell lines, some of the candidate TOP genes were analyzed only in one or two of them. The most significant results are reported in Figure 2, which includes: (1) an example of absorbance profiles of the gradients (Fig. 2A); (2) the Northern analysis of gradient fractions (Fig. 2B); and (3) the quantitative analysis of the percentage of mRNA on polysomes for the different mRNAs (Fig. 2C). It can be observed that: (1) β-actin mRNA is prevalently associated with polysomes both in growing and in resting cells; (2) RPS6 mRNA is mainly associated with subpolysomal particles in resting cells but mostly associated with polysomes in growing cells, this translation activation being inhibited by rapamycin treatment; (3) eEF1β, eEF1δ, eEF1γ, eIF3e, eIF3f, and eIF3h mRNAs behave similarly to RPS6, although with minor quantitative differences; and (4) eIF3g, eIF4B (shown as an example), eIF3a, eIF3b, eIF3d, and eIF3j (not shown) mRNAs show a pattern similar to β-actin since no variation of polysomal association can be observed in the different growth conditions. All the other cases with a clear absence of pyrimidines around the tss were not analyzed. A few cases with some pyrimidines at the 5′ end produced no results, possibly due to the low abundance of mRNAs (eIF2γ, eIF3c, eIF3k, eIF3l, and eIF4AII). Table 1 reports the summary of our studies and considerations. In particular, column 5 shows the identification of a gene as TOP on the basis of our analyses. All five elongation factors and three initiation factors (eIF3e, eIF3f, eIF3h) are encoded by TOP genes. Most remaining initiation factors and the unique termination factor did not satisfy one or both criteria which identify TOP genes. It is interesting to note that a few pyrimidines around the tss are not sufficient to confer translational regulation to the mRNA (see eIF4B). A small number of cases remain undetermined, due to technical difficulty in assessing the growth-associated translational regulation.
FIGURE 2.
Polysomal mRNA profile in different growth conditions. HeLa (H), HEK293 (K), and NIH3T3 (N) cells were incubated in medium lacking serum (−serum) and then stimulated for 1 h with complete medium without (+serum) or with (+rap+serum) rapamycin. Cytoplasmic extracts were separated on sucrose gradients, and fractions were collected while absorbance was monitored at 260 nm. (A) Examples of absorbance profiles from HeLa cells are outlined with black arrows indicating the positions of the 80S ribosomes; the first fraction corresponds to the bottom of the gradient. (B) RNA extracted from each fraction was analyzed on Northern blots with the indicated probes. (C) Quantification of the signals is reported as a column plot of the percentage of mRNA on polysomes, obtained by adding up the values of fractions 1–5.
The turnover of some of the identified TOP mRNAs was also analyzed by actinomycin D treatment of cultured cells followed by Northern blot. The results (not shown) indicates that, in all cases, the half-life of new TOP mRNAs is comparable with the known TOP mRNAs (>24 h).
CONCLUSIONS
The discovery of a peculiar 5′ end linked to a growth-associated translational regulation allowed the classification of vertebrate RP mRNAs as TOP mRNAs (Meyuhas et al. 1996). After more than 20 years since the initial studies on the expression of RP genes, it is evident (although not formally proven in all cases) that all vertebrate RP mRNAs exhibit the two essential characteristics of TOP mRNAs: i) a 5′ TOP sequence and ii) growth-associated translational regulation. Moreover, the two characteristics are functionally linked, since the integrity of the TOP sequence is necessary for regulation (Meyuhas and Hornstein 2000). Experimental evidence indicates that translational control of TOP mRNAs is used by the cell to coordinate the syntheses of the different RPs and of RPs with other components of the translational apparatus. However, it is not clear how many other non-RP genes can be considered TOP. Here we report that the genes for all five translation elongation factors and those for three of the numerous initiation factors (eIF3e, eIF3f, eIF3h) are typical TOP genes. On the other hand, the genes for most initiation factors and for the single termination factor eRF1 are not TOP genes. One possible hypothesis to rationalize our findings is that, since elongation factors perform their function on the ribosome during protein synthesis process, they could be viewed as a peculiar subset of RP. Thus, these factors would need to share the specific structure and regulation with “regular” RPs. By contrast, initiation factors operate by assembling together mRNA, met-tRNAi, and ribosomal subunits to form the initiation complex. This is substantially independent of ribosome function during the process of protein synthesis, and therefore the requirements of such factor syntheses are different from those of the “ribosomal” components. As for the exceptions eIF3e, eIF3f, and eIF3h, which are encoded by TOP genes, an interesting perspective is suggested by a very recent study of human eIF3 (Masutani et al. 2007). In this work, the authors show that six of the 11 subunits make up the functional core of the factor (eIF3a, eIF3b, eIF3c, eIF3e, eIF3f, and eIF3h). The last three are present in Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana (Asano et al. 1997) but not in Saccharomyces cerevisiae. Masutani et al. (2007) propose that such less-conserved core subunits (eIF3e, eIF3f, and eIF3h), besides stabilizing the eIF3 complex, could also mediate general or specific eIF3-dependent translational regulation. Consistent with this hypothesis, several other reports link these subunits with regulative aspects of translation. For instance, eIF3e and/or eIF3f have been shown to bind S6K1 (Holz et al. 2005), eIF4G (LeFebvre et al. 2006), and mTOR (Harris et al. 2006). eIF3f has been suggested as a negative regulator of translation (Shi et al. 2006), whereas eIF3h has been implicated in the efficient translation of mRNAs with upstream open reading frames (Kim et al. 2007). A recent study on the overexpression of eIF3 subunits showed an opposite effect of eIF3h compared to eIF3e and eIF3f. In fact, ectopic expression of eIF3h stimulates cell growth and induces neoplastic transformation possibly by enhancing translation of mRNAs involved in cell proliferation. On the contrary, overexpression of eIF3e and eIF3f moderately inhibits cell growth and proliferation (Zhang et al. 2007). We have found that the three eIF3 subunits (e, f, and h) are encoded by TOP genes. Although at present we cannot reach a specific hypothesis, we do think that our observation supports the idea that belonging to the TOP gene class is, for a gene, a functionally relevant feature.
MATERIALS AND METHODS
Cell cultures
Human HeLa, HEK293, and mouse NIH3T3 cells were grown at 37°C in Dulbecco's Modified Eagle Medium supplemented with 10% fetal calf serum, 2 mM glutamine, 50 units/mL penicillin, and 50 mg/mL streptomycin. To induce serum starvation (resting cells), cells were washed twice in PBS, trypsinized, transferred into a new dish, and grown for 2 h (HeLa, HEK293) or 16 h (NIH3T3) in serum-free medium. To stimulate the cells, serum was added to the medium and the incubation continued for 2 h. To analyze mRNA half-life, cell cultures were treated with actinomycin D at the concentration of 2 μg/mL. At time intervals, RNA was extracted and analyzed by Northern blot hybridization.
Primer extension
A synthetic 20-mer complementary to a defined positions with respect to the initiation AUG codon of target mRNA was 5′-end-labeled with 25 μCi of [γ32P]ATP and T4 polynucleotide kinase, according to standard protocols. Labeled primer was annealed to 20 μg of total RNA from HeLa cells and extended with MMLV reverse transcriptase. Extension products were separated on a 6% polyacrylamide gel and exposed for autoradiography or to a PhosphorImager screen.
Polysome analysis
Cell lysis, sucrose gradient sedimentation, and analysis of the polysome/mRNP distribution of mRNAs were performed essentially as previously described (Caldarola et al. 2004). In particular, cells [(1−2) × 106] that had been washed once with phosphate-buffered saline buffer (150 mM NaCl, 2.7 mM KCl, 8 mM NaH2PO4, and 1.4 mM K2PO4) were lysed directly on the plate with 300 μL of lysis buffer (10 mM NaCl, 10 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 36 U/mL RNase inhibitor [Pharmacia], 1 mM dithiothreitol) and transferred into a microcentrifuge tube. After 5 min of incubation on ice with occasional vortexing, the lysate was centrifuged for 8 min at 10,000 rpm at 4°C. The supernatant was frozen in liquid nitrogen and stored at −70°C to be analyzed later or immediately layered in a 15%–50% (w/v) sucrose gradient containing 30 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgCl2, and centrifuged in a Beckman SW41 rotor for 110 min at 37,000 rpm. Fractions were collected while monitoring the optical density at 254 nm, and total RNA was extracted from each fraction by the proteinase K method (Sambrook et al. 1989). For Northern analysis, RNA was fractionated on formaldehyde–agarose gels and transferred to GeneScreen Plus membrane (PerkinElmer Life Sciences). Northern blotting was performed essentially as recommended by the manufacturer. Radioactive probes were prepared by the random primer technique (Sambrook et al. 1989) using DNA fragments isolated from plasmids containing PCR-amplified cDNA sequences. Primers for amplification were designed according to sequences present in the GenBank/EBI Data Bank. Quantitation of Northern blots was performed using a PhosphorImager and the Image-Quant software (Amersham Biosciences).
ACKNOWLEDGMENTS
We thank Marcello Giorgi for expert technical assistance. This work was supported by MIUR-FIRB and MIUR-PRIN grants to F.A. and F.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1037108.
REFERENCES
- Asano K., Vornlocher H.P., Richter-Cook N.J., Merrick W.C., Hinnebusch A.G., Hershey J.W. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J. Biol. Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- Avni D., Biberman Y., Meyuhas O. The 5′-terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldarola S., Amaldi F., Proud C.G., Loreni F. Translational regulation of terminal oligopyrimidine mRNAs induced by serum and amino acids involves distinct signaling events. J. Biol. Chem. 2004;279:13522–13531. doi: 10.1074/jbc.M310574200. [DOI] [PubMed] [Google Scholar]
- Camacho-Vanegas O., Weighardt F., Ghigna C., Amaldi F., Riva S., Biamonti G. Growth-dependent and growth-independent translation of messengers for heterogeneous nuclear ribonucleoproteins. Nucleic Acids Res. 1997;25:3950–3954. doi: 10.1093/nar/25.19.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B., Carissimi C., Gravina P., Pierandrei-Amaldi P. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J. Biol. Chem. 2003;278:35145–35151. doi: 10.1074/jbc.M300722200. [DOI] [PubMed] [Google Scholar]
- Crosio C., Boyl P.P., Loreni F., Pierandrei-Amaldi P., Amaldi F. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S., Thomas G. S6 phosphorylation and signal transduction. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 695–717. [Google Scholar]
- Geyer P.K., Meyuhas O., Perry R.P., Johnson L.F. Regulation of ribosomal protein mRNA content and translation in growth-stimulated mouse fibroblasts. Mol. Cell. Biol. 1982;2:685–693. doi: 10.1128/mcb.2.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.E., Chi A., Shabanowitz J., Hunt D.F., Rhoads R.E., Lawrence J.C., Jr mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J. 2006;25:1659–1668. doi: 10.1038/sj.emboj.7601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz M.K., Ballif B.A., Gygi S.P., Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Git A., Braunstein I., Avni D., Meyuhas O. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J. Biol. Chem. 1999;274:1708–1714. doi: 10.1074/jbc.274.3.1708. [DOI] [PubMed] [Google Scholar]
- Kim B.H., Cai X., Vaughn J.N., von Arnim A.G. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8:R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda M., Di Croce M., Bedini B., Wannenes F., Corvaro M., Boyl P.P., Caldarola S., Loreni F., Amaldi F. Effect of 3′UTR length on the translational regulation of 5′-terminal oligopyrimidine mRNAs. Gene. 2005;344:213–220. doi: 10.1016/j.gene.2004.09.023. [DOI] [PubMed] [Google Scholar]
- LeFebvre A.K., Korneeva N.L., Trutschl M., Cvek U., Duzan R.D., Bradley C.A., Hershey J.W., Rhoads R.E. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreni F., Amaldi F. Translational regulation of ribosomal protein synthesis in Xenopus cultured cells: mRNA relocation between polysomes and RNP during nutritional shifts. Eur. J. Biochem. 1992;205:1027–1032. doi: 10.1111/j.1432-1033.1992.tb16870.x. [DOI] [PubMed] [Google Scholar]
- Loreni F., Francesconi A., Amaldi F. Coordinate translational regulation in the syntheses of elongation factor 1 alpha and ribosomal proteins in Xenopus laevis . Nucleic Acids Res. 1993;21:4721–4725. doi: 10.1093/nar/21.20.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., de la Pena L., Barker C., Camphausen K., Tofilon P.J. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–1061. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M., Sonenberg N., Yokoyama S., Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Hornstein E. Translational control of TOP mRNAs. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 671–693. [Google Scholar]
- Meyuhas O., Avni D., Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Mathews M.B., et al., editors. Translational control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 363–388. [Google Scholar]
- Mikulits W., Pradet-Balade B., Habermann B., Beug H., Garcia-Sanz J.A., Mullner E.W. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Cardinali B., Lin-Marq N., Mercanti D., Pierandrei-Amaldi P. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′UTR of ribosomal protein mRNAs in vitro: Implication of a protein factor in complex formation. J. Mol. Biol. 1996;259:904–915. doi: 10.1006/jmbi.1996.0368. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Lotti F., Maras B., Pierandrei-Amaldi P. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J. Mol. Biol. 1997;267:264–275. doi: 10.1006/jmbi.1996.0888. [DOI] [PubMed] [Google Scholar]
- Pende M., Um S.H., Mieulet V., Sticker M., Goss V.L., Mestan J., Mueller M., Fumagalli S., Kozma S.C., Thomas G. S6K1−/−/S6K2−/−mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzani A., Fronza R., Loreni F., Pascale A., Amadio M., Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27:1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Meyuhas O. Ribosomal protein S6 phosphorylation: From protein synthesis to cell size. Trends Biochem. Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schibler U., Kelley D.E., Perry R.P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Schwartz E.I., Intine R.V., Maraia R.J. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5′-terminal oligopyrimidine mRNA metabolism. Mol. Cell. Biol. 2004;24:9580–9591. doi: 10.1128/MCB.24.21.9580-9591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Kahle A., Hershey J.W., Honchak B.M., Warneke J.A., Leong S.P., Nelson M.A. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- Spence J., Duggan B.M., Eckhardt C., McClelland M., Mercola D. Messenger RNAs under differential translational control in Ki-ras-transformed cells. Mol. Cancer Res. 2006;4:47–60. doi: 10.1158/1541-7786.MCR-04-0187. [DOI] [PubMed] [Google Scholar]
- Stolovich M., Tang H., Hornstein E., Levy G., Cohen R., Bae S.S., Birnbaum M.J., Meyuhas O. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 2002;22:8101–8113. doi: 10.1128/MCB.22.23.8101-8113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hornstein E., Stolovich M., Levy G., Livingstone M., Templeton D., Avruch J., Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N., Patel H.R., Takase K., Kohno K., Nairn A.C., Gelfand E.W. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Pan X., Hershey J.W. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J. Biol. Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- Zhu J., Hayakawa A., Kakegawa T., Kaspar R.L. Binding of the La autoantigen to the 5′-untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim. Biophys. Acta. 2001;1521:19–29. doi: 10.1016/s0167-4781(01)00277-9. [DOI] [PubMed] [Google Scholar]