Abstract
It is well known that nearly all eukaryotic mRNAs contain a 3′ poly(A) tail. A polyadenylation signal (AAUAAA) nearby the 3′ end of pre-mRNA is required for poly(A) synthesis. The protein complex involved in the pre-mRNA polyadenylation is coupled with RNA polymerase II during the transcription of a gene. According to the commonly accepted view, only RNAs synthesized by RNA polymerase II can be polyadenylated in an AAUAAA-dependent manner. Here we report the polyadenylation of short interspersed elements (SINEs) B2 and VES transcripts generated by RNA polymerase III. HeLa cells were transfected with SINE constructs with or without polyadenylation signals. The analyses of the SINE transcripts showed that only the RNAs with the AAUAAA-signal contained poly(A) tails. Polyadenylated B2 RNA was found to be much more stable in cells than B2 RNA without a poly(A) tail.
Keywords: RNA polymerase III, polyadenylation signal, transcriptional terminator, poly(A), SINEs of class T+, B2 RNA
INTRODUCTION
Poly(A) at the 3′ end of eukaryotic mRNAs plays an important role in regulation of their stability, export from the nucleus to cytoplasm, and translation initiation (Jacobson and Peltz 1996; Hall 2002). Poly(A) is synthesized post-transcriptionaly in the nucleus, and its length can reach 250 nt. mRNA processing includes the cleavage of pre-mRNA and synthesis of poly(A) at the 3′ end. This processing requires that the pre-mRNA contains: (1) a polyadenylation signal, AAUAAA, 10 to 30 nucleotides (nt) upstream of the cleavage site, and (2) a GU-rich element 20 to 40 nt downstream of the cleavage site (Manley 1995; Zarudnaya et al. 2003). A complex consisting of at least five protein factors including poly(A) polymerase is involved in this process. These factors interact with the nascent RNA and are associated with the elongating polymerase complex through the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (pol II) (Hirose and Manley 2000; Bentley 2005).
Until recently, only transcripts synthesized by pol II were thought to be polyadenylated. However, defective molecules of tRNA and 5S rRNA, transcription products of RNA polymerase III (pol III), are subject to polyadenylation followed by their degradation by the exosome (Anderson 2005; Reinisch and Wolin 2007). Hence, polyadenylation can participate in surveillance and turnover of noncoding RNAs including pol III-synthesized ones. However, such polyadenylation essentially differs from polyadenylation of mRNA. Poly(A)-containing tRNA and 5S rRNA have a very short lifetime in normal cells, and they are detected only in cells with defective exosomes. The particular poly(A) polymerase and other protein factors are involved in polyadenylation of noncoding RNAs. Finally, this process requires no AAUAAA hexamer in RNA. Thus, it is generally accepted that only pol II transcripts are subject to AAUAAA-dependent polyadenylation.
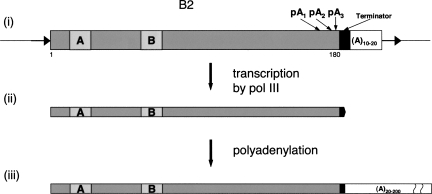
Previously, we demonstrated a short poly(A)-containing RNA generated by pol III transcription of multiple genomic copies of the B2 short interspersed element (SINE) in mouse cells (Kramerov et al. 1982; Kramerov et al. 1985a; Kramerov et al. 1990). Poly(A) length varied from 20 to 200 nt, and this contributed to the heterogeneity of the B2 RNA length (Kramerov et al. 1985b). We assumed that this poly(A) is synthesized post-transcriptionally on the 3′ end of this RNA (Fig. 1). The B2 SINE structure supports this hypothesis: (1) many copies of B2 SINE have a region containing two to four polyadenylation signals (AATAAA) and a transcriptional terminator (TCTTT) near their 3′ end (Haynes and Jelinek 1981; Krayev et al. 1982); (2) there is an oligo(dA)10–20 at the 3′ end of the B2 SINE; at the same time, a much longer poly(A) tail (up to 200 nt) is found in B2 RNA, i.e., this tail must be generated post-transcriptionally. Thus, we proposed (Kramerov et al. 1985b; Kramerov et al. 1990) that B2 SINE transcription by pol III terminates at TCTTT, and a long poly(A) tail is synthesized at the 3′ end of the resulting 180-nt RNA (Fig. 1). In mammals, at least 10 SINE families whose tails are similar to that of B2 were found. Such SINEs were assigned to the T+ class, whereas the class of SINEs without a polyadenylation signal (AATAAA) and transcriptional terminator (TCTTT or TTTT) was referred to as T– (Borodulina and Kramerov 2001). Pol III transcripts of all T+ class SINE families were proposed to be polyadenylated in an AAUAAA-dependent manner.
FIGURE 1.
A probable way of generation of poly(A)-containing B2 RNA, a pol III transcript of B2 SINE. (i) SINE is depicted as a rectangle; the following functional elements are shown: boxes A and B of a pol III promoter, potential polyadenylation signals (pA1, pA2, and pA3), a pol III transcriptional terminator (filled box), and an oligo(A) tail (open box). Thin lines indicate DNA sequences flanking SINE including TSD (target site duplication) or short direct repeats (horizontal arrows). (ii) A 180-nt B2 RNA, a product of pol III transcription of B2 SINE. (iii) Heterogeneous in length B2 RNA, a product of polyadenylation of the 180-nt transcript. A poly(A) tail is shown as an open rectangle.
SINE copies (Alu, B1, and ID) that recently integrated in the genome proved to have long (up to 50 nt) poly(dA) tails (Roy-Engel et al. 2002; Odom et al. 2004). In addition, human Alu SINE could be effectively amplified by the retroposition mechanism given it has a long poly(A) tail (Dewannieux and Heidmann 2005). Roy-Engel et al. (2005) examined whether polyadenylation of pol III transcripts of SINEs may generate long poly(A) tails. Since human Alu have neither polyadenilation nor transcriptional terminator signals (T− class), these investigators introduced AATAAA and TTTT sequences into the Alu tail and transfected mouse cells with this construct. Nevertheless, such Alu RNA with a polyadenylation signal was not polyadenylated. Roy-Engel et al. (2005) concluded that pol III-generated transcripts cannot be polyadenylated in tissue culture. This prompted us to carry out similar experiments with the B2 SINE. In this paper we unambiguously demonstrate pol III transcripts with a 3′-terminal AAUAAA signal that can be effectively polyadenylated in mammalian cells. Moreover, polyadenylation dramatically increased the lifetime of such RNAs.
RESULTS
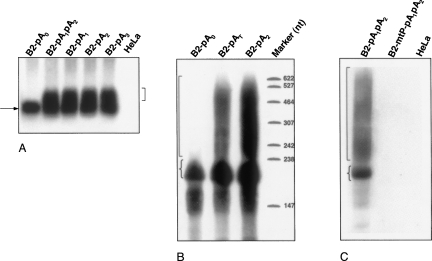
To study the capability of pol III transcripts to be polyadenylated, we used a B2 SINE copy from clone Mm 14 (Krayev et al. 1982) located in the third intron of mouse Cxcl16 gene. This B2 copy contains normal A and B boxes of pol III promoter, two potential polyadenylation signals (AATAAA), and a pol III terminator (TCTTTT) located in its A-rich tail (Fig. 2A). By means of PCR and DNA cloning, five plasmids were constructed that contained the mouse 5′ flanking sequence (84 bp) and B2 with an A-rich tail harboring AATAAA-signals in different positions and numbers (Fig. 2B). Plasmids carrying these constructs were transiently transfected in HeLa cells, RNA was isolated 20 h after transfection, and B2 SINE transcripts were detected by Northern hybridization. Change of a T with a C in both AATAAA hexamers (B2-pA0 construct) resulted in a quite narrow band of B2 RNA, whereas in the case of a native construct (B2-pA1pA2) longer heterogeneous RNAs were also observed (Fig. 3A). We interpreted the longer RNAs as polyadenylated B2 transcripts. The same hybridization pattern was observed in the case of B2-pA1 and B2-pA2 constructs with the only polyadenylation signal (Fig. 3A). The polyadenylation of B2 RNA also took place when cells were transfected with construct B2-pA3 lacking space between an AATAAA hexamer and a pol III terminator (Fig. 3A).
FIGURE 2.
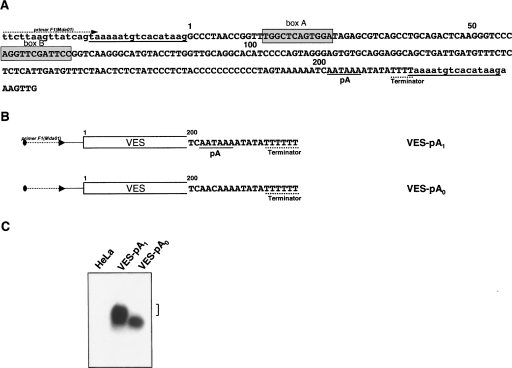
(A) Nucleotide sequence of the mouse B2 SINE copy used for preparation of constructs. The SINE and its flanking sequences are shown in upper and lower cases, respectively. TSD flanking SINE is underlined. A pol III promoter (box A and box B), potential polyadenylation signals pA1 and pA2 (underlined), and a pol III terminator (underlined with dotted line) are indicated in the B2 sequence. (B) The structure of six constructs used in the study of the polyadenylation capability of B2 SINE pol III transcripts. The first 150 bp of B2 are depicted as a rectangle, whereas a terminal region of the B2 constructs is represented as a nucleotide sequence. Potential polyadenylation signals are underlined; a terminator is underlined with dotted line. Note that an additional T residue was introduced in the terminator, whereas an oligo(A) tail was removed from all the constructs.
FIGURE 3.
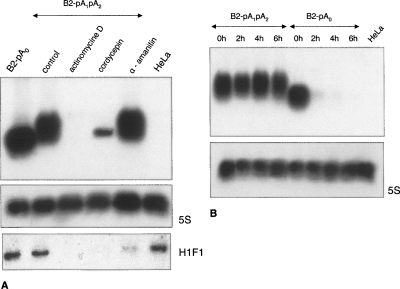
Northern blot analysis of B2 SINE transcripts isolated from HeLa cells that were transfected with B2-containing constructs with or without polyadenylation signals (see Fig 2B) as well as the construct with mutant pol III promoter (B2-mtP-pA1pA2). The blot analysis was performed by separating total cellular RNA by electrophoresis in an agarose (A) or polyacrylamide (B, C) gel. A180-nt B2 RNA is indicated by an arrow or brace. Longer forms of B2 RNA are marked with square brackets.
In order to estimate B2 RNA length, Northern hybridization of RNA from transfected cells fractionated by electrophoresis in PAAG was performed. The construct without a polyadenylation signal (B2-pA0) generated a 180-nt RNA, whereas a construct with AATAAA produced heterogeneous RNAs from 200 nt to 500 nt, besides the 180-nt band (Fig. 3B). This result suggests that the poly(A) length in the B2 RNA is variable, and can be up to 300 nt.
In the same experiment we examined whether a noncanonical polyadenylation signal (ATTAAA) that occurs in 12%–15% of mRNAs (Zarudnaya et al. 2003) directs polyadenylation of B2 RNA. As shown in Figure 3B, this hexanucleotide does indeed direct polyadenylation of the B2 RNA (construct B2-pAT), but probably less effectively.
A similar experiment was carried out with the B2mtP-pA1pA2 construct containing a trinucleotide substitution (TTC → CCT) in box B of pol III promoter. In the RNA isolated from cells transfected with this construct, neither the 180-nt transcript nor long heterogeneous RNAs were detected (Fig. 3C). This result demonstrates the synthesis of both RNA species by pol III.
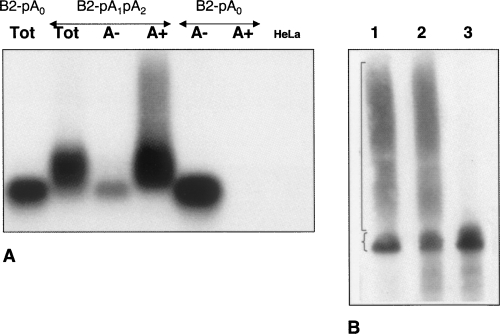
To obtain additional direct evidence for polyadenylation of B2 transcript, the following experiments were performed. RNA from transfected cells was fractionated on oligo(dT) cellulose columns to separate poly(A)+ and poly(A)– fractions (Fig. 4A). All B2 RNA molecules from cells transfected with the B2-pA0 construct (containing no AATAAA signal) were detected in the poly(A)– fraction. At the same time, the major part (90%) of the B2 RNAs from cells transfected with the B2-pA1pA2 construct were found in the poly(A)+ fraction. Noteworthy, the B2 RNA from the poly(A)+ fraction was much longer than the RNA from the poly(A)– fraction. These results directly argue for the effective polyadenylation of transcripts of the B2 SINE containing an AATAAA signal.
FIGURE 4.
(A) Blot analysis of RNA fractionated on oligo(dT) cellulose from HeLa cells transfected with B2 SINE constructs with (B2-pA1pA2) or without (B2-pA0) polyadenylation signals. By means of chromatography on oligo(dT)-cellulose, total cellular RNA preparations were separated into poly(A)+ and poly(A)– RNA fractions (A+ and A–, respectively). These RNA fractions and the total cellular RNA (Tot) including that from nontransfected cells (HeLa) were separated by electophoresis in an agarose gel followed by blot hybridization with B2 probe. (B) Removal of poly(A) tracts from B2 RNA. RNA from cells transfected with the B2-pA1pA2 construct (lane 1) was digested with RNAse H following incubation with (lane 3) or without (lane 2) oligo(dT)12–18. A B2 RNA was detected by Northern hybridization following PAGE. A 180-nt and long B2 RNA are indicated by brace and square brackets, respectively.
We used one more approach to confirm the presence of poly(A) in the long hetrorogeneous B2 RNA. The RNA from cells transfected with the B2-pA1pA2 construct was hybridized to oligo(dT)12–18 and digested with RNAse H to remove the sequences that hybridized to oligo(dT). Such treatment eliminated the long B2 RNA and increased the 180-nt B2 RNA band intensity (Fig. 4B). This result confirms the presence of poly(A) in the long B2 RNA.
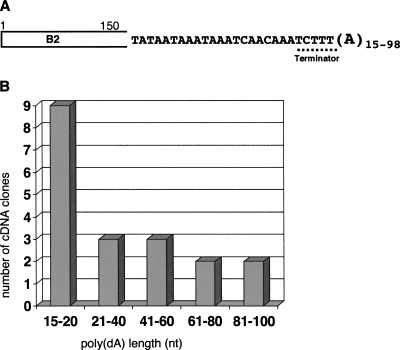
The most compelling evidence of B2 RNA polyadenylation can be obtained by means of cDNA cloning. RNA from HeLa cells transfected with plasmid B2-pA1pA2 was reverse transcribed using an oligo(dT)-containing primer. cDNA was amplified by PCR with a pair of primers, one of them being specific for the 5′ end of B2. This system of primers allowed us to minimize poly(A) shortening during PCR (see Materials and Methods). Cloned B2-containing cDNAs were sequenced. Examples of such sequencing are shown in Supplemental Figure 1. In all cDNA clones, B2 sequences contained poly(dA) fragments from 15 nt to 98 nt in length (Fig. 5). These values are a lower than expected poly(A) segment length in B2 RNA molecules because oligo(dT) primes cDNA synthesis by matching with poly(A) in random sites. In any case, the result of this experiment unambiguously shows that B2 RNA contains a poly(A) tail, which could form only as a result of post-transcriptional polyadenylation. (Note that the B2 construct used for cell transfection did not contain oligo[A]14,which was located at the end of the original B2 copy; Fig. 2).
FIGURE 5.
(A) Consensus of nucleotide sequences of 20 cloned cDNAs corresponding to the 3′ end of the B2 RNA (a rectangle depicts the rest of cDNA sequence). RNA was isolated from HeLa cells transfected with a B2-pA1pA2 construct, and B2 RNA-derived cDNAs were synthesized, cloned, and sequenced. Noteworthy are terminator sequence shortening (TCTTT) and the emergence of poly(A). (B) Diagram showed distribution of the numbers of B2 cDNA clones analyzed against length of poly(A) segments in them. The number of residues in the poly(A) segments from each clone was counted and numbers of the clones with poly(A) segment length fallen into each size range were summarized.
The aim of the following experiments with inhibitors was to confirm that B2 RNA is not synthesized by pol II, and that it is subject to polyadenylation. In vitro (Haynes and Jelinek 1981; Sakamoto et al. 1984; Singh et al. 1985; Carey et al. 1986) and in vivo (Kramerov et al. 1985a; Kramerov et al. 1990) experiments demonstrated the generation of a small RNA in process of transcription B2 SINE by pol III. Additionally, we (Kramerov et al. 1990) and other investigators (Kravchenko et al. 2005) had previously shown that the treatment of cells with α-amanitin at a concentration of 20 μg/mL inhibits pol II (but not other RNA polymerases). One hour following transfection, we added actinomycin D (inhibitor of transcription), α-amanitin (pol II inhibitor), or cordycepin (inhibitor of poly(A) polymerase) to cells and isolated the RNA in 7 h. Northern hybridization revealed that actinomycin D completely prevented B2 RNA synthesis, α-amanitin did not change its formation, and cordycepin, while slightly decreasing the level of the primary B2 transcript, completely inhibited the generation of longer (polyadenylated) B2 RNA molecules (Fig. 6A). Thus, as one might expect, pol II is not involved in B2 RNA synthesis in transfected cells, and poly(A) polymerase is responsible for the generation of longer B2 RNA molecules.
FIGURE 6.
(A) Blot analysis of the RNA from HeLa cells transfected with construct B2-pA1pA2 and treated with actimomycin D, cordycepin, α-amanitin, or neither of the inhibitors (control). An RNA from untransfected cells (HeLa) or cells transfected with construct B2-pA0 was also analyzed for comparison. Hybridization with the 5S rRNA probe was used as a sample-loading control. The short-lived histone H1F1 mRNA was detected by hybridization in order to verify the inhibition of pol II by α-amanitin. Normalized data on H1F1 mRNA hybridization indicate an 80% inhibition of pol II. (B) Study of B2 RNA stability in HeLa cells transfected with construct B2-pA1pA2 or B2-pA0. The RNA was isolated in indicated time periods following addition of actinomycin D to transfected cells.
To verify whether pol III transcripts of other SINEs can be polyadenylated, an experiment with VES SINE from the bat, Myotis daubentoni, was carried out. Nucleotide sequence of this SINE demonstrates no similarity with that of B2 except for the presence of a pol III promoter (A and B boxes), a hexamer AATAAA, and a pol III terminator, TTTT (Borodulina and Kramerov 1999). A VES element (copy Mda01) with AATAAA (construct Ves–pA1) or AACAAA (construct Ves–pAo) in its A-rich tail was obtained (Fig. 7A). Transfection of HeLa cells and the subsequent detection of VES RNA showed that the construct VES–pA1 generated longer RNA molecules than the construct Ves–pAo did (Fig. 7B). Thus, transcripts of this SINE can also be polyadenylated in the AAUAAA-dependent manner.
FIGURE 7.
(A) A nucleotide sequence of the bat VES SINE copy used for preparation of constructs. The SINE and its flanking sequences are shown in upper and lower cases, respectively. TSD flanking SINE is underlined. In the VES sequence, a pol III promoter (box A and box B), a potential polyadenylation signal pA (underlined), and a pol III terminator (underlined with dotted line) are indicated. (B) The structure of two constructs used to study the polyadenylation ability of VES SINE pol III transcripts. The first 200 bp of VES are depicted as a rectangle, whereas a terminal region of the SINE constructs is represented as a nucleotide sequence. A potential polyadenylation signal is underlined; a terminator is underlined with a dotted line. Note that two additional T residues were introduced in the terminator of the both constructs. (C) Blot analysis of VES SINE transcripts isolated from HeLa cells transfected with VES-containing constructs with (VES-pA1) or without (VES-pA0) a polyadenylation signal. Long forms of VES RNA were indicated with a square bracket.
The presence of poly(A) at the 3′ end of mRNAs is known to increase their lifetime in cells (Bernstein and Ross 1989). Perhaps polyadenylation also stabilizes B2 RNA in cells. To test this hypothesis, actinomycin D was added to HeLa cells transfected with the constructs B2-pA1pA2 or B2-pA0, RNA was isolated after every 2 h, and B2 RNA was detected by Northern hybridization. B2 RNA from cells transfected with B2 carrying polyadenylation signals was stable, while the polyadenylation signal mutation resulted in a dramatic decrease in the lifetime of the B2 RNA (Fig. 6B). In a special experiment, the half-life of the poly(A)– B2 RNA was estimated as 40 min (Supplemental Fig. 2). Thus, polyadenylation of pol III transcript of B2 SINE plays an important role in the stabilization of this RNA in cells.
DISCUSSION
Polyadenylation of pol III transcripts
In this study, we showed that transcripts synthesized by pol III, similarly to mRNA, can be polyadenylated in mammalian cells, and this post-transcriptional modification depends on the presence of an AAUAAA-signal at the 3′ end of the RNA. Although some evidence for polyadenylation of B2 SINE pol III transcripts in mouse tumor cells have already been reported (Kramerov et al. 1985a,b; Bladon et al. 1990; Kramerov et al. 1990), those data were based on a study of the mixture of multiple B2 transcripts. The presence of the B2 copies with mutated transcriptional terminators (Borodulina and Kramerov 2001) or with rather long (40–50 nt) A-tails (Odom et al. 2004), as well as the existence of the mRNAs containing B2 sequences in their 3′-nontranslated regions, could potentially lead to misinterpretation of those results. Here we used human cells transfected with B2 and VES SINE constructs with or without AATAAA-signals, to obtain more direct evidence of the AATAAA-dependent polyadenylation of pol III transcripts. It is usually assumed that the presence of a polyadenylation signal upstream of a transcriptional terminator is the requirement for poly(A) synthesis at the 3′ end of such transcripts. Our data suggest that effective polyadenylation occurs even if sequences AATAAA and TCTTT (or TTTT) locate immediately adjacent to each other (Figs. 2, 7). This fact demonstrates a difference in polyadenylation between pol III and pol II transcripts because cleavage and polyadenylation of pre-mRNA occur only if the polyadenylation signal and the cleavage site are spaced with at least 20 nt (Zarudnaya et al. 2003; Bentley 2005).
Previously, three groups reported the results of cell transfection experiments using chimeric constructs with a pol III-transcribed gene fused to a protein-coding fragment (Lewis and Manley 1986; Sisodia et al. 1987; Gunnery and Mathews 1995). Such constructs were transcribed by pol III due to the presence of an internal promoter for pol III in their 5′ region. Lewis and Manley (1986) found that the chimeric transcripts can be accurately cleaved and polyadenylated. Sisodia et al. (1987) worked with the same constructs and drew the opposite conclusion. They proposed that Lewis and Manley observed polyadenylation of RNA synthesized by pol II (rather than pol III) from fortuitous transcription initiation sites. Gunnery and Mathews (1995) also concluded that artificial pol III-synthesized mRNA-like transcripts were not polyadenylated. Thus, the results of these studies are quite contradictory. Our results can easily explain this inability of the chimeric pol III transcript to be polyadenylated: the chimeric gene had no polyadenylation signal in one construct (Gunnery and Mathews 1995), while the other construct had such signal but had no pol III terminator nearby (Sisodia et al. 1987).
A more intriguing fact is a difference between our results and those of Roy-Engel et al. (2005) on the inability to be polyadenylated of Alu transcripts artificially provided with a polyadenylation signal and pol III terminator. Alu, unlike tRNA-derived SINE B2, originates from 7SL RNA, a component of ribonucleoprotein signal-recognition particle (SRP) involved in synthesis of membrane and secreted polypeptides (Weiner 1980; Ullu and Tschudi 1984). Pol III transcripts of Alu (but not B2 RNA) bind some protein components of SRP (SRP9 and SRP14) (Hsu et al. 1995), which could in some way prevent polyadenylation of Alu RNA. In any case, the following questions arise: (1) Being supplied with a polyadenylation signal, pol III transcripts of which genes can be subjected to this post-transcriptional modification? (2) What structural features (besides the presence of AAUAAA upstream from the 3′ end) of pol III transcripts are required for their polyadenylation?
The discovery of AAUAAA-dependent polyadenylation of RNA generated by pol III can expand our knowledge about mechanisms of formation of poly(A) at the RNA 3′ ends. Polyadenylation of pre-mRNA is known to be tightly linked to pol II: a protein complex involved in the 3′ end formation of pre-mRNA by cleavage and polyadenylation binds to the CTD of the large subunit of pol II at the transcription initiation stage (Hirose and Manley 2000). This complex attaches to pol II during gene transcription and becomes active when polymerase reaches a polyadenylation site (Bentley 2005). It is not clear whether pol III is able to bind to polyadenylation protein factors; however, a pol II CTD homologs domain is known to be absent from pol III (Bentley 2005). Most likely the mechanism of AAUAAA-dependent polyadenylation of pol III transcripts essentially differs from mRNA polyadenylation mechanisms. Perhaps the polyadenylation of RNA generated by pol III is not directly linked to transcriptional process.
The following hypothesis can be proposed. It was recently found that the B2 RNA can directly bind pol II and thus repress transcription (Espinoza et al. 2004). Pol II-bound B2 RNA can come in contact with poly(A) polymerase associated with pol II and specific protein factors. Thus, the B2 RNA can be polyadenylated by the poly(A) polymerase enzyme that synthesizes poly(A) on the 3′ end of the mRNA.
Polyadenylation as a source for long A-tails in T+ class SINEs
Long poly(A) is known to be necessary for the retroposition of LINE-1-mobilized SINEs (Dewannieux and Heidmann 2005). The generation of long poly(A) tails in SINEs is being discussed in the literature. According to one of the hypotheses, the A-tail growth involves template switching from the A-tail region of LINE-1 to the RNA SINE during reverse transcription (Roy-Engel et al. 2005). As we showed here, polyadenylation of transcripts can be the mechanism of the generation of long poly(A) in SINEs of the class T+ (i.e., SINEs with an AATAAA-signal and terminator in their tails). Such SINEs have been found in mammals, and they comprise about a third of all known SINE families from this vertebrate order (Borodulina and Kramerov 2001; Kramerov and Vassetzky 2005). Probably the different SINE families independently acquired tails specific for the T+ class SINEs during evolution. (It can be considered as an example of convergence on the molecular level.) Studies on evolution of armadillo DAS SINE (Churakov et al. 2005) showed that only relatively young, complex, and effective DAS subfamilies had a T+ class-specific tail.
It should be pointed out that the pol III transcription of a canonical T+ class SINE results in shortening of the terminator sequence in the SINE transcript. As a consequence of two to three rounds of transcription–retroposition the shortening of a terminator (up to TCTT) can lead to a loss of its function. Many copies of T+ class SINEs with such mutated terminators do occur in genomes. However, there may be a mechanism of the terminator recovery associated with an uridylation activity in cells (Shen and Goodman 2004; Kwak and Wickens 2007), which could increase the number of U residues at the 3′ end of a SINE transcript (e.g., UCUU could be converted to UCUUU). Due to such a process novel potential master genes of SINEs would arise.
Here we have shown that not only an AAUAAA-signal but a noncanonical variant of this signal (AUUAAA) can also mediate polyadenylation of a pol III transcript. Hence, transcripts of SINEs with an essentially mutated tail containing a hexamer AUUAAA instead of AAUAAA (such SINE copies often occur in the genome) should also be able to be polyadenylated. Finally, some copies of the T– class SINEs can contain a single hexamer AATAAA or ATTAAA at their A-tail as well as a pol III terminator (e.g., TTTT) in the nearby flanking sequence. Pol III transcripts of such copies seem also to be subject to polyadenylation. In other words, SINEs of class T– could rarely acquire long poly(A) tails due to polyadenylation of some of their pol III transcripts.
MATERIALS AND METHODS
Plasmid constructs
Schematics of the constructs are shown in Figures 2 and 7. Mouse B2 SINE from clone Mm14 (Krayev et al. 1982) and bat VES SINE from clone Mda01 (Acc. No Y19051) (Borodulina and Kramerov 1999) were used for preparation of the constructs. To remove the flanking sequences and to introduce nucleotides substitutions, PCRs with specific primers and Mm14 and Mda01 plasmid DNA were carried out. (Nucleotide sequences of the primers are shown in Figures 2 and 7; the “reverse” primers were complementary to those regions of constructs that are represented by nucleotide sequences in Figures 2B and 7B.) The 5′-GAAGGTCCTGAGCCTAATTCCCGGC-3′ primer was used to generate B2 SINE with a mutation in box B of the pol III promoter (the trinucleotide substitution is underlined). PCR products were purified by electrophoresis in 3% agarose gel and cloned into plasmid pGEM-T (Promega) following the manufacturer's protocol. Plasmids were isolated using QIAGEN Plasmid Midi Kit. All constructs were subjected to DNA sequence analysis in order to exclude plasmids with random nucleotide substitutions induced by PCR.
Cell transfections and use of inhibitors
Transfections were carried out in the HeLa cell line. Monolayer was grown to 80% confluency in Petri dishes (60-mm diameter). Cells were transiently transfected with 5 μg of plasmid DNA using ExGen 500 in vitro Transfection Reagent (Fermentas) following the manufacturer's protocol. RNA was isolated 20 h post-transfection using the guanidinium thiocyanate method.
Actinomycin D, α-amanitin, or cordycepin were added to the medium to final concentrations of 5 μg/mL, 20 μg/mL, and 40 μg/mL, respectively. RNA was isolated in the time indicated in text.
RNA analysis
RNA was treated with 100 μg/mL RNase-free DNase I (Fermentas) at 37°C for 30 min. Following inactivation of DNase, RNA was resolved by denaturing electrophoresis in either 1.5% agarose or 5% polyacrylamide gel (PAAG). RNA was transferred from the agarose gel onto a Hybond-XL membrane by capillary blotting, whereas RNA transfer from PAAG onto the membrane was performed by semidry electroblotting at 5 V for 2 h.
A hybridization probe corresponding to the first 94 nt of B2 SINE (clone Mm14) was prepared using PCR with primers 1 and 2 as it was described previously (Serdobova and Kramerov 1998). The probe was labeled with α[32P] dATP by PCR using only primer 2; such a probe detects only B2 RNA but not possible transcripts of the opposite strand of B2 SINE. A VES-specific probe was obtained by PCR using clone Mda01 DNA as a template and 5′-GGTTGAGCGTCGGCCT-3′ and 5′-GATCAGCTGCCTCCTGCA-3′ as primers (the latter was used in the probe labeling reaction). The blots were hybridized in 50% formamide, 5× Denhardt solution, 4× SSC, 1% SDS, and 0.1 mg/mL salmon sperm DNA at 42°C. Washes were performed in 0.1% SSC and 0.1% SDS at 52°C.
Hybridization of RNA to oligo(dT)12–18 and RNase H (Ambion) digestion was carried out as described elsewhere (Bladon et al. 1990).
cDNA cloning
Reverse transcription reaction was carried out at 42°C for 60 min in the mixture that contained 10 μg RNA from transfected cells, primer XbaI-T14 (5′-GGCGCGCGTCGAGTCTAGATTTTTTTTTTTTTT-3′), 320 μM dNTPs, 100 U MMLV reverse transcriptase, and reaction buffer; 1/10 of the cDNA was added to the PCR mixture containing primer XmaI-B2 (5′-TACCCGGGCTGGAGAGATGGC-3′) and primer XbaI-T2 (5′-GGCGCGCGTCGAGTCTAGATT-3′), dNTP, Taq polymerase, and reaction buffer. The PCR products were separated by electrophoresis in 1.5% agarose gel, 250–600 bp DNA was isolated, digested with XmaI and XbaI, cloned in pGEM-7z vector (Promega), and sequenced.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Andrej Kramerov for editorial assistance. This research was supported by the Russian Foundation for Basic Research (Grant 08-04-00350) and the Molecular and Cellular Biology Program of the Russian Academy of Sciences.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1006608.
REFERENCES
- Anderson J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005;15:R635–R638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci. 1989;14:373–377. doi: 10.1016/0968-0004(89)90011-x. [DOI] [PubMed] [Google Scholar]
- Bladon T.S., Fregeau C.J., McBurney M.W. Synthesis and processing of small B2 transcripts in mouse embryonal carcinoma cells. Mol. Cell. Biol. 1990;10:4058–4067. doi: 10.1128/mcb.10.8.4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodulina O.R., Kramerov D.A. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 1999;457:409–413. doi: 10.1016/s0014-5793(99)01059-5. [DOI] [PubMed] [Google Scholar]
- Borodulina O.R., Kramerov D.A. Short interspersed elements (SINEs) from insectivores. Two classes of mammalian SINEs distinguished by A-rich tail structure. Mamm. Genome. 2001;12:779–786. doi: 10.1007/s003350020029. [DOI] [PubMed] [Google Scholar]
- Carey M.F., Singh K., Botchan M., Cozzarelli N.R. Induction of specific transcription by RNA polymerase III in transformed cells. Mol. Cell. Biol. 1986;6:3068–3076. doi: 10.1128/mcb.6.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G., Smit A.F., Brosius J., Schmitz J. A novel abundant family of retroposed elements (DAS-SINEs) in the nine-banded armadillo (Dasypus novemcinctus) Mol. Biol. Evol. 2005;22:886–893. doi: 10.1093/molbev/msi071. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Espinoza C.A., Allen T.A., Hieb A.R., Kugel J.F., Goodrich J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Gunnery S., Mathews M.B. Functional mRNA can be generated by RNA polymerase III. Mol. Cell. Biol. 1995;15:3597–3607. doi: 10.1128/mcb.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.M. Poly(A) tail synthesis and regulation: recent structural insights. Curr. Opin. Struct. Biol. 2002;12:82–88. doi: 10.1016/s0959-440x(02)00293-2. [DOI] [PubMed] [Google Scholar]
- Haynes S.R., Jelinek W.R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc. Natl. Acad. Sci. 1981;78:6130–6134. doi: 10.1073/pnas.78.10.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hsu K., Chang D.Y., Maraia R.J. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J. Biol. Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- Jacobson A., Peltz S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Kramerov D.A., Vassetzky N.S. Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- Kramerov D.A., Lekakh I.V., Samarina O.P., Ryskov A.P. The sequences homologous to major interspersed repeats B1 and B2 of mouse genome are present in mRNA and small cytoplasmic poly(A) + RNA. Nucleic Acids Res. 1982;10:7477–7491. doi: 10.1093/nar/10.23.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D.A., Tillib S.V., Lekakh I.V., Ryskov A.P., Georgiev G.P. Biosynthesis and cytoplasmic distribution of small poly(A)-containing B2 RNA. Biochim. Biophys. Acta. 1985a;824:85–98. doi: 10.1016/0167-4781(85)90084-3. [DOI] [PubMed] [Google Scholar]
- Kramerov D.A., Tillib S.V., Ryskov A.P., Georgiev G.P. Nucleotide sequence of small polyadenylated B2 RNA. Nucleic Acids Res. 1985b;13:6423–6437. doi: 10.1093/nar/13.18.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D.A., Tillib S.V., Shumyatsky G.P., Georgiev G.P. The most abundant nascent poly(A) + RNAs are transcribed by RNA polymerase III in murine tumor cells. Nucleic Acids Res. 1990;18:4499–4506. doi: 10.1093/nar/18.15.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko J.E., Rogozin I.B., Koonin E.V., Chumakov P.M. Transcription of mammalian messenger RNAs by a nuclear RNA polymerase of mitochondrial origin. Nature. 2005;436:735–739. doi: 10.1038/nature03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A.S., Markusheva T.V., Kramerov D.A., Ryskov A.P., Skryabin K.G., Bayev A.A., Georgiev G.P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982;10:7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.D., Manley J.L. Polyadenylylation of an mRNA precursor occurs independently of transcription by RNA polymerase II in vivo. Proc. Natl. Acad. Sci. 1986;83:8555–8559. doi: 10.1073/pnas.83.22.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. A complex protein assembly catalyzes polyadenylation of mRNA precursors. Curr. Opin. Genet. Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Odom G.L., Robichaux J.L., Deininger P.L. Predicting mammalian SINE subfamily activity from A-tail length. Mol. Biol. Evol. 2004;21:2140–2148. doi: 10.1093/molbev/msh225. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Okada N. SINEs and LINEs: Symbionts of eukaryotic genomes with a common tail. Cytogenet. Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- Reinisch K.M., Wolin S.L. Emerging themes in non-coding RNA quality control. Curr. Opin. Struct. Biol. 2007;17:209–214. doi: 10.1016/j.sbi.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Roy-Engel A.M., Salem A.H., Oyeniran O.O., Deininger L., Hedges D.J., Kilroy G.E., Batzer M.A., Deininger P.L. Active Alu element “A-tails”: Size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel A.M., El-Sawy M., Farooq L., Odom G.L., Perepelitsa-Belancio V., Bruch H., Oyeniran O.O., Deininger P.L. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet. Genome Res. 2005;110:365–371. doi: 10.1159/000084968. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Kominami R., Mishima Y., Okada N. The 6S RNA transcribed from rodent total DNA in vitro is the transcript of the type 2 Alu family. Mol. Gen. Genet. 1984;194:1–6. doi: 10.1007/BF00383489. [DOI] [PubMed] [Google Scholar]
- Serdobova I.M., Kramerov D.A. Short retroposons of the B2 superfamily: Evolution and application for the study of rodent phylogeny. J. Mol. Evol. 1998;46:202–214. doi: 10.1007/pl00006295. [DOI] [PubMed] [Google Scholar]
- Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Singh K., Carey M., Saragosti S., Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985;314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- Sisodia S.S., Sollner-Webb B., Cleveland D.W. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol. Cell. Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980;22:209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- Zarudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: Primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]