Abstract
Yeast and human Clp1 proteins are homologous components of the mRNA 3′-cleavage-polyadenylation machinery. Recent studies highlighting an association of human Clp1 (hClp1) with tRNA splicing endonuclease and an intrinsic RNA-specific 5′-OH polynucleotide kinase activity of hClp1 have prompted speculation that Clp1 might play a catalytic role in tRNA splicing in animal cells. Here, we show that expression of hClp1 in budding yeast can complement conditional and lethal mutations in the essential 5′-OH RNA kinase module of yeast or plant tRNA ligases. The tRNA splicing activity of hClp1 in yeast is abolished by mutations in the kinase active site. In contrast, overexpression of yeast Clp1 (yClp1) cannot rescue kinase-defective tRNA ligase mutants, and, unlike hClp1, the purified recombinant yClp1 protein has no detectable RNA kinase activity in vitro. Mutations of the yClp1 ATP-binding site do not affect yeast viability. These findings, and the fact that hClp1 cannot complement growth of a yeast clp1Δ strain, indicate that yeast and human Clp1 proteins are not functional orthologs, despite their structural similarity. Although hClp1 can perform the 5′-end-healing step of a yeast-type tRNA splicing pathway in vivo, it is uncertain whether its kinase activity is necessary for tRNA splicing in human cells, given that other mammalian counterparts of yeast-type tRNA repair enzymes are nonessential in vivo.
Keywords: RNA end healing, mRNA 3′ processing, polynucleotide kinase activity
INTRODUCTION
Intron-containing tRNAs are found in all eukaryal and archaeal species. The intron is usually located in the anticodon loop of the pre-tRNA and must be removed for the tRNA to function in protein synthesis. A tRNA splicing endonuclease recognizes the fold of the pre-tRNA and makes incisions at the exon–intron borders to yield 2′,3′ cyclic phosphate and 5′-OH termini at both break sites (Peebles et al. 1983). The human tRNA splicing endonuclease is a component of a stable nuclear protein complex that includes human Clp1 (hClp1), a conserved eukaryal polypeptide required for 3′ processing of pre-mRNAs (Paushkin et al. 2004). In yeast, the splicing endonuclease resides on the outer mitochondrial surface; consequently, yeast tRNA splicing is a cytoplasmic event (Yoshihisa et al. 2003, 2007). Notwithstanding these differences in localization, the structures and mechanisms of tRNA splicing endonucleases are conserved among lower and higher eukaryal species and archaea (Trotta et al. 1997, 2006; Li et al. 1998; Paushkin et al. 2004; Xue et al. 2006).
In contrast, the tRNA sealing reactions of the splicing pathway diverge in archaea and eukarya. The ends of cleaved archaeal tRNAs are rejoined in a single step by attack of the 5′-OH on the 2′,3′ cyclic phosphodiester to form a 3′,5′ phosphodiester at the splice junction (Zofallova et al. 2000; Salgia et al. 2003). The responsible archaeal enzyme is not known. Yeast tRNA splicing takes a totally different biochemical route. A single tRNA ligase enzyme, Trl1, heals and seals the broken tRNA ends. Trl1 performs three reactions, each catalyzed by a discrete protein domain: (1) the 2′,3′ cyclic phosphate of the proximal tRNA half-molecule is hydrolyzed to a 3′-OH, 2′-PO4 by a cyclic phosphodiesterase (CPD); (2) the 5′-OH of the distal half-molecule is phosphorylated by an NTP-dependent polynucleotide kinase; and (3) the 3′-OH, 2′-PO4, and 5′-PO4 ends are sealed by an ATP-dependent RNA ligase to form a spliced tRNA containing an unconventional 2′-PO4, 3′-5′ phosphodiester at the splice junction (Greer et al. 1983). The 2′-PO4 at the splice junction is removed by the 2′-phosphotransferase Tpt1 (Spinelli et al. 1998).
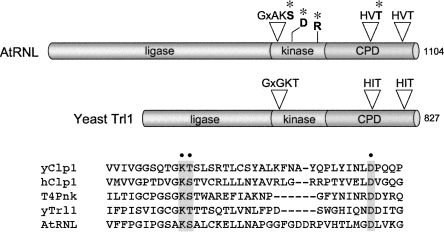
Yeast Trl1 consists of an N-terminal ligase domain and a C-terminal end-healing domain (Fig. 1). The ligase domain is an ATP-dependent strand-sealing enzyme that belongs to the ligase family exemplified by bacteriophage T4 RNA ligase 1 (Wang and Shuman 2005). The end-healing domain is composed of distinct kinase and CPD modules (Fig. 1). The kinase module resembles T4 polynucleotide kinase (Pnk), which belongs to the P-loop phosphotransferase superfamily (Fig. 2; Wang et al. 2002, 2006). The CPD component of yeast tRNA ligase belongs to the 2H phosphotransferase superfamily (Mazumder et al. 2002; Wang et al. 2006), which is defined by two HxT motifs (Fig. 1), and includes the mammalian 2′,3′ cyclic nucleotide phosphodiesterase, CNP (Braun et al. 2004). An Arabidopsis tRNA ligase ortholog (AtRNL) is a 1104-amino acid polypeptide with a similar tripartite domain structure (Fig. 1; Englert and Beier 2005; Wang et al. 2006). Plant AtRNL is capable of performing all three essential splicing reactions in vivo in yeast cells that lack Trl1 (Wang et al. 2006).
FIGURE 1.
Domain organization of yeast and plant tRNA ligases and conservation of polynucleotide kinase motifs in tRNA repair kinases and yeast and human Clp1. (A) The tRNA ligases of yeast (Trl1) and plant (AtRNL) are composed of three discrete catalytic domains: an N-terminal ligase module; a central 5′-OH polynucleotide kinase module; and a C-terminal RNA 2′,3′ cyclic phosphodiesterase (CPD) module. The positions of the P-loop motif and essential aspartate at the kinase active site and the two HxT motifs that comprise the CPD active site are depicted above the AtRNL polypeptide. (*) Lethal and conditional AtRNL alleles used in the present study contained substitutions for these residues. (B) Alignment of the amino acid sequences in the vicinity of the P-loop of yeast and human Clp1, yeast and plant tRNA ligases, and T4 Pnk. (Gray boxes, dots) The conserved lysine, serine/threonine, and aspartate residues subjected to alanine mutagenesis.
FIGURE 2.
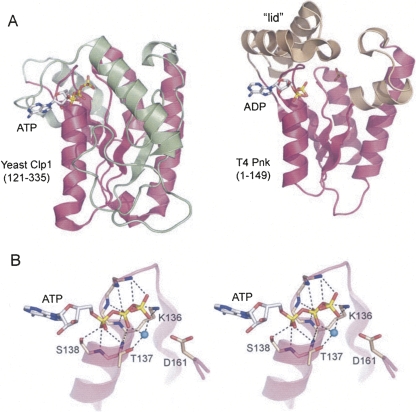
Structural homology between Clp1 and T4 Pnk. (A) The structures of the yClp1 phosphoesterase domain (PDB ID 2NPI) and the T4 Pnk kinase domain (PDB ID 1LTQ and 1LY1) were superimposed and then offset horizontally. The folds are shown as ribbon traces with shared structural elements (magenta). (Green) Distinctive structural components of yClp1, (beige) distinctive structural components of T4 Pnk. The ATP and ADP ligands are depicted as stick models. In the T4 Pnk structure, a sulfate ion occupies the position of the terminal phosphodiester of the 5′-OH polynucleotide substrate. (B) Stereo view of the ATP-binding site of yClp1 highlighting the oxyanion hole formed by main-chain amides and the side chains of the P-loop motif QTGKTS138. (Cyan sphere) Magnesium ion, (dashed lines) atomic contacts of yClp1 with ATP and magnesium.
The mechanism of tRNA exon ligation in animal cells is poorly understood. Initial studies demonstrated a direct tRNA end-joining reaction in human cell extracts in which the 2′,3′ cyclic phosphate is retained as a 3′–5′ phosphodiester at the splice junction of the mature tRNA (Filipowicz and Shatkin 1983; Filipowicz et al. 1983; Laski et al. 1983). The identity of the responsible enzyme remains a mystery. Later work suggested that human cells also have a yeast-like pathway of tRNA splicing (Zillman et al. 1991) in which the junction 3′–5′ phosphate in mature tRNA derives from ATP and the starting 2′,3′ cyclic phosphate is converted to a 2′-PO4 that is ultimately removed. The existence of a yeast-like tRNA repair system in metazoa is consistent with the presence of Tpt1-type phosphotransferase, 2′,3′ cyclic phosphodiesterase, and RNA-specific 5′-OH polynucleotide kinase activities in human cells (Shuman and Hurwitz 1979; Spinelli et al. 1998; Braun et al. 2004).
Solving the “division of labor” problem for the two distinct human tRNA exon-joining pathways hinges on identifying the relevant enzymes and probing their function genetically. Recent studies in this vein have cast doubt on the importance of the yeast-type ligation pathway for splicing of animal tRNAs. For example, mammalian CNP and Tpt1 have been shown to complement the essential functions of their yeast homologs, thereby attesting to their capacity to catalyze 3′-end-healing and junction dephosphorylation reactions of tRNA splicing in vivo (Spinelli et al. 1998; Schwer et al. 2008). However, neither CNP nor Tpt1 is essential in mice (Lappe-Siefke et al. 2003; Harding et al. 2008). The relatively benign effects of completely deleting CNP in mice seem to exclude an essential role for the yeast-type splicing pathway, if one assumes that CNP, which is the source of all measurable 2′,3′-cyclic phosphodiesterase activity in mouse brain (Lappe-Siefke et al. 2003), is the only available source of 3′-end-healing for the yeast-like splicing pathway. Similarly, Tpt1 is the sole source of measurable tRNA 2′-phosphotransferase activity in mouse cells, yet homozygous tpt1 −/− null mice develop normally and are healthy and fertile (Harding et al. 2008). These results appear to relegate the yeast-type pathway in mammals to either no role, or a functionally redundant role, in tRNA splicing. Further consolidation of this inference will require genetic assessments of the ligase and kinase components of the yeast-type pathway in mammalian cells.
A recent advance on this front was the assignment of RNA 5′-OH kinase activity to the human Clp1 protein (Weitzer and Martinez 2007). An RNA-specific 5′-kinase was isolated from HeLa cell nuclei and characterized biochemically nearly 30 years ago (Shuman and Hurwitz 1979). It was speculated then that the human RNA kinase might provide sealable 5′-monophosphate ends for an RNA ligase in tRNA splicing (Shuman and Hurwitz 1979), notwithstanding that the 5′-OH terminus of the cleaved tRNA ends and requirements for a polynucleotide kinase in yeast tRNA splicing, plant RNA ligation, and bacteriophage T4 tRNA restriction/repair had not yet been established (Knapp et al. 1979; Konarska et al. 1981, 1982; Greer et al. 1983; Amitsur et al. 1987). Interest in metazoan RNA kinase was revived by the realization that exogenous siRNAs acquire a 5′-monophosphate end in order to be effective agents of RNAi. Weitzer and Martinez (2007) purified an siRNA kinase from HeLa cells and identified hClp1 (425 amino acids) as the active principle. A recombinant GST-hClp1 fusion protein sufficed for siRNA kinase activity. Remarkably, the kinase purified from human cells consisted of a complex of hClp1 with all of the subunits of the human tRNA splicing endonuclease, hSEN (Weitzer and Martinez 2007). The hClp1-hSEN complex could phosphorylate the 5′-OH end of cleaved pre-tRNA. Based on siRNA knockdown experiments, Clp1 was suggested to be an agent of mammalian tRNA splicing (Weitzer and Martinez 2007). The model for hClp1 tRNA splicing function remains to be tested by generating a viable mammalian cell line or organism in which the tRNA kinase activity of Clp1 is eliminated without affecting other aspects of Clp1 function. This is a potentially complicated task, given the essential role of Clp1 in mRNA 3′ processing (de Vries et al. 2000; Gross and Moore 2001).
Yeast Clp1 (yClp1; 445 amino acids) is essential for the viability of Saccharomyces cerevisiae. yClp1 is a subunit of pre-mRNA 3′ cleavage factor 1 (CF1); the other subunits are Rna14, Rna14, Hrp1, and Pcf11 (Amrani et al. 1997; Minvielle-Sebastia et al. 1997; Gross and Moore 2001). The crystal structure of yeast Clp1 bound to a peptide derived from Pcf11 revealed a central P-loop phosphotransferase module to which ATP and magnesium were bound (Fig. 2; Noble et al. 2007). Efforts to demonstrate ATP hydrolysis by yClp1 were unsuccessful (Noble et al. 2007), which makes sense in light of the subsequent findings that hClp1 catalyzes transfer of the ATP γ-phosphate to a 5′-OH RNA terminus, not to water (Weitzer and Martinez 2007). The fold of the central yClp1 segment resembles that of bacteriophage T4 polynucleotide kinase (Wang et al. 2002; Galburt et al. 2002). T4 Pnk and yClp1 share a central parallel four-strand β sheet flanked by three homologous α helices (colored magenta in Fig. 2A). The loop between the first β strand and α helix includes the signature GxGKTS motif that coordinates the ATP phosphates and the divalent cation (Fig. 2B). The two structures diverge considerably outside the shared core. Yeast Clp1 has no counterpart of the essential T4 Pnk “lid” module that covers the NTP phosphoryl donor (Fig. 2A). Indeed, yClp1 has a larger central β sheet (seven strands) than does T4 Pnk, and it includes several α helices and loops that have no equivalent in the phage kinase (Fig. 2A).
To our knowledge, it is not established if the yClp1 ATP-binding site, or an imputed phosphoryl transferase activity of yClp1, is required for yeast viability. A corollary question is whether yClp1, like its human homolog, is an RNA-specific kinase and whether that activity is relevant to RNA metabolism in S. cerevisiae. Available evidence would argue against yClp1 being the catalyst of the 5′-end-healing step of yeast tRNA splicing, insofar as Trl1 itself has the requisite kinase function and kinase-inactivating mutations in Trl1 are lethal in vivo (Sawaya et al. 2003). However, a different scenario might pertain to human cells.
The findings that tRNA repair enzymes from heterologous sources can complement the catalytic function of one or all of the domains of yeast Trl1 (Schwer et al. 2004, 2008; Wang et al. 2006) provides a way to interrogate whether candidate proteins can perform tRNA splicing reactions in vivo. Here, we apply this approach to show that hClp1 can indeed execute the 5′-end-healing step of tRNA splicing in yeast. Major functional differences between the yClp1 and hClp1 are revealed by the following findings: (1) Complementation of kinase-defective tRNA ligase alleles by hClp1 is abolished by mutations in the hClp1 kinase active site; (2) equivalent mutations of the yClp1 ATP-binding site do not affect yeast viability. (3) overexpression of yClp1 cannot rescue kinase-defective tRNA ligase mutants; (4) yClp1 has no detectable RNA kinase activity in vitro; and (5) hClp1 cannot complement growth of a yeast clp1Δ strain.
RESULTS AND DISCUSSION
Human Clp1 complements conditional kinase-defective tRNA ligase mutations in yeast
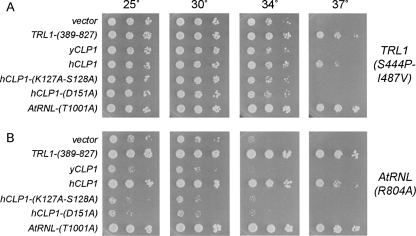
To query whether hClp1 can act as a tRNA splicing enzyme, we expressed the full-length hClp1 protein in S. cerevisiae under the control of a constitutive promoter on a 2μ plasmid and asked whether hClp1 could complement the temperature-sensitive growth defect caused by the S444P-I487V double mutation in the kinase module of yeast Trl1 (Schwer et al. 2008). S444P-I487V cells failed to form colonies at 37°C. Growth of S444P-I487V cells at restrictive temperature was revived by a plasmid-borne copy of TRL1-(389-827) (the yeast kinase-CPD domain), but not by an empty plasmid vector (Fig. 3A). Growth was also rescued by the T1001A allele of plant tRNA ligase AtRNL, which has an inactivating mutation in the CPD active site (Wang et al. 2006; Fig. 3A). Thus, the ability to complement the S444P-I487V strain is a specific readout of 5′-kinase function in tRNA splicing. The instructive finding was that hCLP1 expression restored growth of S444P-I487V cells at the restrictive temperature, signifying that the mammalian enzyme could act in lieu of the Trl1 kinase (Fig. 3A).
FIGURE 3.
Complementation by hClp1 of kinase-ts mutations in yeast and plant tRNA ligases. Functional complementation was tested by spotting on SD-His− agar plates serial 10-fold dilutions of trl1Δ pTRL1-(S444P-I487A) (TRP1 CEN) yeast cells (A) or trl1Δ AtRNL1-R804A (TRP1 CEN) yeast cells (B) that had been transformed with a CEN HIS3 TRL1-(389-827) plasmid or a CEN HIS3 AtRNL-T1001A plasmid (positive controls) or with 2μ HIS3 plasmids bearing genes encoding either wild-type yCLP1, wild-type hCLP1, or hCLP1 kinase active site mutants K127A-S128A or D151A. Cells transformed with the empty 2μ HIS3 vector served as a negative control. The plates were photographed after incubation for 3 d at 25°C, 30°C, 34°C, or 37°C.
Plant tRNA ligase AtRNL can provide all of the essential tRNA healing and sealing functions in a yeast trl1Δ background. The AtRNL-R804A allele has a single mutation in the kinase module that causes a slow-growth phenotype in yeast at 25°C and 30°C and abolishes colony formation at 37°C (Fig. 3B). This growth defect was reversed by introduction of a 2μ plasmid expressing the yeast kinase-CPD domain or the CPD-defective plant tRNA ligase, but not by the empty vector control (Fig. 3B). These results signify that AtRNL-R804A is defective purely in its kinase activity. The key finding was that expression of hCLP1 restored growth of AtRNL-R804A cells to wild-type levels at 25°C, 30°C, and 34°C (gauged by colony size) and rescued colony formation at 37°C (Fig. 3B).
hClp1 activity in yeast is abolished by mutations of the kinase active site
The central kinase domain of hClp1 includes conserved counterparts of the ATP-binding P-loop motif (GxxxxGKS128 in hClp1) and the downstream aspartate (Asp151) that are essential for the kinase activity of T4 Pnk and yeast and plant tRNA ligases (Fig. 1; Wang and Shuman 2001, 2002; Sawaya et al. 2003; Wang et al. 2006). The P-loop motif forms an oxyanion hole for the binding of the NTP phosphates via a network of hydrogen bonds to the phosphate oxygens from the main chain amides and the signature lysine side chain (Fig. 2B). The P-loop serine/threonine side chain coordinates the divalent cation cofactor for phosphoryl transfer (Fig. 2B). The downstream aspartate is located in the active site near the 5′-OH terminus of the polynucleotide (Wang et al. 2002; Eastberg et al. 2004), where it is proposed to activate the 5′-OH for its attack on the ATP γ phosphorus (Wang et al. 2002). Weitzer and Martinez (2007) have shown that hClp1 kinase activity in vitro is abolished by P-loop mutations K127A and S128A. Here, we found that a K127A-S128A double mutation abolished the capacity of hClp1 to complement conditional kinase-defective mutants of either yeast Trl1 or plant AtRNL (Fig. 3). Moreover, we find that a single alanine substitution of the hClp1 active site Asp151 also abolished hClp1 tRNA splicing activity in yeast (Fig. 3). We surmise that hClp1 kinase activity is critical for its execution of the 5′ healing step of yeast tRNA splicing.
Human Clp1 complements lethal kinase-defective tRNA ligase mutations in yeast
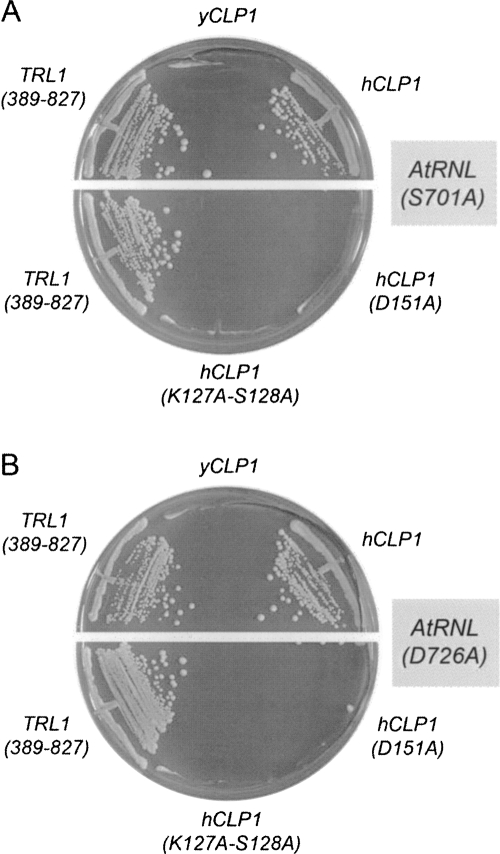
To consolidate the inference from these initial experiments that the 5′ kinase activity of hClp1 can replace the kinase activity of yeast or plant tRNA ligases, we tested the ability of hClp1 to substitute for lethal mutations in the kinase active site of AtRNL (Wang et al. 2006). Yeast trl1Δ strains carrying TRL1 on a CEN URA3 plasmid were transformed with CEN TRP1 plasmids bearing lethal S701A and D726A alleles of AtRNL. We introduced into these strains HIS3 plasmids expressing wild-type yeast kinase-CPD domain, wild-type hClp1, or the hClp1 active site mutants K127A-S128A and D151A. AtRNL-701A and AtRNL-D726A were unable to sustain growth on medium containing the drug FOA (5-fluoroorotic acid), which selects against the URA3 TRL1 plasmid. Growth on FOA was restored by yeast kinase-CPD, as expected. The salient findings were that hClp1 complemented the lethal kinase mutations, whereas the hClp1 active site mutants did not (Fig. 4).
FIGURE 4.
hClp1 complements lethal mutations in plant tRNA ligase. Yeast trl1Δ p360-TRL1 (URA3 CEN) cells containing CEN TRP1 plasmids bearing lethal AtRNL alleles S701A (A) or D726A (B) were transformed with 2μ HIS3 plasmids bearing genes encoding either wild-type yCLP1, wild-type hCLP1, or hCLP1 kinase active site mutants K127A-S128A and D151A. Control cells were transformed with the empty 2μ HIS3 vector (negative control) or a CEN HIS3 plasmid expressing the wild-type kinase-CPD domain TRL1-(389-827) (positive control). Trp+ His+ transformants were selected at 30°C and then streaked on agar medium containing 0.75 mg/mL FOA. The plates were photographed after 4 d at 30°C.
Increased gene dosage of yeast Clp1 fails to rescue kinase-defective tRNA ligases
The finding that hClp1 can catalyze the 5′ healing step of tRNA splicing in yeast raises the question: Why does endogenous yeast Clp1 not perform this function when the Trl1 kinase domain is disabled? We considered the prospect that the basal levels of yClp1 protein might be sequestered in the pre-mRNA 3′-end forming CF1 complex and therefore unavailable for tRNA splicing. We endeavored to increase yClp1 expression by placing the CLP1 gene on a multicopy plasmid under the control of the strong yeast TPI1 promoter. Increased gene dosage of yCLP1 failed to complement any of the conditional or lethal kinase-defective alleles of yeast or plant tRNA ligases that were readily rescued by expression of hClp1 (Figs. 3, 4). Thus, yeast and human Clp1 proteins have distinct activity profiles with respect to tRNA repair in vivo.
Yeast Clp1 has no detectable polynucleotide kinase activity
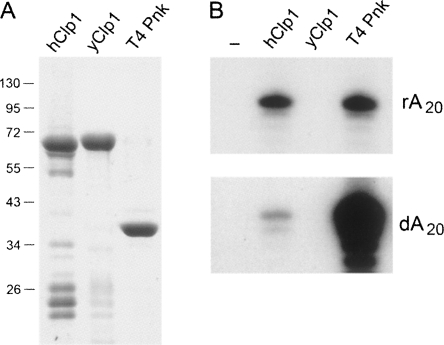
One simple explanation for the inability of yClp1 to serve as a tRNA repair enzyme could be that it does not have an RNA kinase activity, notwithstanding its membership in the P-loop superfamily and its primary structure similarity to hClp1. To address this issue, we produced yClp1 in bacteria as a His10-Smt3 fusion protein and purified the recombinant yClp from a soluble bacterial extract by Ni-agarose chromatography. The preparation was enriched with respect to the His10-Smt3-yClp1 polypeptide (Fig. 5), the identity of which was verified by its susceptibility to cleavage by the Smt3-specific protease Ulp1 (not shown). In parallel, we purified His10-Smt3-tagged hClp1 and His10-tagged T4 Pnk (Fig. 5). The proteins were tested for RNA and DNA kinase activities by label transfer from [γ32P]ATP to oligo(rA)20 and oligo(dA)20 substrates. As expected, T4 Pnk was adept at phosphorylating RNA and DNA strands, while hClp1 displayed a strong preference for RNA as the phosphoryl acceptor (Fig. 5). A protein titration experiment indicated that the kinase specific activity of hClp1 with oligo(rA)20 was ∼80-fold higher than with oligo(dA)20 (not shown). We did not detect any RNA or DNA kinase activity associated with the yClp1 preparation (Fig. 5). The assay sensitivity is such that we can estimate that yClp1 has <0.03% of the RNA kinase activity of hClp1 under the assay conditions used (not shown). A mixing experiment verified that the yClp1 preparation did not contain a trans-acting inhibitor of the RNA kinase activities of T4 Pnk or hClp1 (not shown). We surmise that yClp1 either lacks the ATP phosphotransferase activity imputed to it based on the crystal structure, or that its activity is targeted to non-nucleic acid substrates as phosphate acceptors.
FIGURE 5.
Assays of RNA kinase activity of yeast and human Clp1 and T4 Pnk. (A) Aliquots of recombinant hClp1, yClp1, and T4 Pnk proteins (adjusted to contain 5 μg of full-length polypeptide) were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are shown on the left. (B) Polynucleotide kinase reaction mixtures (10 μL) containing 70 mM Tris-HCl (pH 7.8), 12 mM imidazole, 15 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 μM [γ-32P]ATP, 50 pmol 5′-OH oligoribonucleotide rA20, or a 5′-OH oligodeoxyribonucleotide dA20, as specified, and 1 μg of hClp1, yClp, or T4 Pnk (full-length polypeptide) were incubated for 30 min at 37°C. Enzyme was omitted from control reactions (lane −). The reactions were quenched with 15 μL of 95% formamide, and the products were analyzed by electrophoresis through a 20-cm 18% polyacrylamide gel (19:1) containing 7 M urea in TBE (80 mM Tris-borate, 1 mM EDTA). The radiolabeled oligonucleotide products were visualized by autoradiography of the gel. (Top panel) 20-min exposure, (bottom panel) 2.5-h exposure. The extent of label transfer to the oligonucleotide was quantified by scanning the gel with a Typhoon phosphorimager (GE Life Sciences). The rA20 substrate was phosphorylated quantitatively by T4 Pnk and hClp1. The dA20 substrate was phosphorylated quantitatively by T4 Pnk; hClp1 phosphorylated ∼2% of the DNA acceptor. No kinase activity was detected for yClp1.
Human Clp1 cannot perform the essential functions of yeast Clp1 in vivo
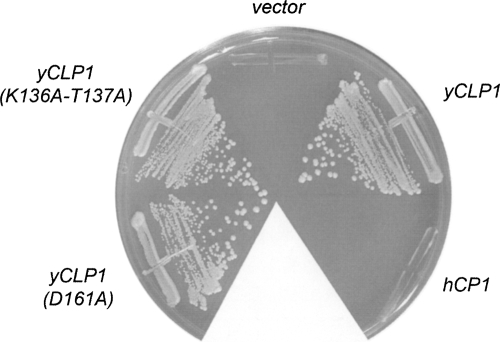
The findings that hClp1 and yClp1 are not orthologous with respect to the RNA end-repair capacity raises the issue of whether hClp1 can perform the essential functions of yClp1 in vivo. To answer the question, we established a plasmid shuffle assay for complementation of a lethal clp1Δ mutation. A haploid S. cerevisiae strain in which the chromosomal CLP1 gene is disrupted depends for growth on a wild-type yCLP1 gene carried on a URA3 plasmid. This clp1Δ strain is unable to grow on agar medium containing FOA, which selects against retention of the URA3 yCLP1 plasmid. Transformation of clp1Δ cells with a 2μ HIS3-marked plasmid containing another copy of yCLP1 allows them to form FOA-resistant colonies, whereas transformation with an empty 2μ vector does not (Fig. 6). The 2μ yCLP1 plasmid that complemented clp1Δ in this experiment is the same vector that failed to rescue kinase-defective mutations in tRNA ligase. By contrast, a 2μ HIS3 hCLP1 plasmid failed to confer on clp1Δ cells the ability to form FOA-resistant colonies (Fig. 6), signifying that human Clp1 cannot perform the essential functions of yClp1 in S. cerevisiae. Note that the 2μ hCLP1 plasmid that failed to complement clp1Δ is the same vector that did successfully complement kinase-defective tRNA ligases. These results suggest that human and yeast Clp1 proteins are not true orthologs.
FIGURE 6.
Test of clp1Δ complementation by hClp1 and yClp1 and mutants thereof. Yeast clp1Δ pYX212-yCLP1 cells were transformed with 2μ HIS3 plasmids bearing wild-type hCLP1, wild-type yCLP1, or yCLP1 mutants K136A-T137A or D161A. His+ transformants were selected at 30°C and then streaked on agar medium containing 0.75 mg/mL FOA. The plate was photographed after 3 d at 30°C.
Yeast Clp1's essential function in vivo is unaffected by mutations in the “active site”
If it is the case that phosphoryl transfer by yClp1 to an as yet unknown substrate is essential in vivo, we would expect that mutations in the yClp1 “active site” should elicit an obvious phenotype in S. cerevisiae. Accordingly, we introduced a double alanine mutation K136A-T137A in the yClp1 P-loop and a single alanine mutation D161A at the putative catalytic aspartate. The Asp161 side chain of yClp1 occupies the same spatial position in the crystal structure as Asp35 of T4 Pnk (not shown). The equivalent alanine mutations in the P-loop and distal aspartate of T4 Pnk abolish its kinase activity (Wang and Shuman 2001, 2002). Here, we found that the K136A-T137A and D161A alleles of yCLP1 were active in vivo in supporting the growth of clp1Δ cells on medium containing FOA (Fig. 6). Moreover, the viable yCLP1-(K136A-T137A) and yCLP1-D161A strains grew as well as wild-type CLP1 cells on YPD agar medium at 25°C, 30°C, 34°C, and 37°C (not shown). We surmise that a putative phosphotransferase activity of yClp1, if it does exist, is not an essential part of yClp1's functional repertoire. We do not exclude the prospect, discussed by Noble et al. (2007), that ATP binding might be important for yClp1 function, because it is quite possible that the mere loss of the atomic contacts between Lys136 and ATP in the P-loop mutant would not seriously compromise ATP binding, given that there are nine other hydrogen bonds between the protein and the ATP ligand (Fig. 2B).
Implications for Clp1 function in tRNA splicing
Here, we provided genetic evidence that human Clp1 is an RNA repair enzyme in vivo and is capable of performing the 5′-end-healing step of yeast tRNA splicing. hClp1 has structural homology with the kinase domain of bacteriophage, yeast, and plant tRNA repair systems, and the outcomes of their kinase reactions are the same: a 5′-monophosphate end that could be a substrate for a “conventional” RNA ligase. The RNA end-healing function of hClp1 might be relevant to either of two cases of programmed RNA breakage and repair in mammals: (1) tRNA splicing and (2) mRNA splicing during the endoplasmic reticulum unfolded protein response (UPR) (Ron and Walter 2007). The existence of two different RNA repair pathways in mammalian cells (yeast-type and archaeal-type) raises the issue of whether they are functionally redundant or dedicated to particular physiological substrates. The key features of the yeast-type pathway are the need for end-healing prior to sealing and removal of the 2′-PO4 at the splice junction. The fact that at least two mammalian homologs of yeast-type tRNA splicing enzymes (CNP and Tpt1) are nonessential in mice hints strongly that the yeast-type pathway is not required for mammalian tRNA splicing or mRNA splicing in the UPR (Lappe-Siefke et al. 2003; Harding et al. 2008).
Although the physical association of hClp1 with human tRNA splicing endonuclease is tantalizing (Paushkin et al. 2004; Weitzer and Martinez 2007), it is difficult to envision that the Clp1 kinase activity is essential for tRNA splicing in animal cells, when other enzymatic components of the yeast-type pathway are not, and because the archaeal-type pathway in animal cells specifically requires an unhealed 5′-OH terminus. To the extent that the RNA kinase activity of hClp1 might be critical for Clp1 function in mammalian cells, we suspect that it is directed to RNA substrates other than cleaved intron-containing pre-tRNAs. Possible candidates include: (1) processed siRNAs or miRNAs that have undergone dephosphorylation at their 5′ ends and would required re-phosphorylation by hClp1 to restore their activity in gene silencing; or (2) decapped or cleaved mRNAs that have undergone dephosphorylation at their 5′ ends and that require re-phosphosphorylation by hClp1 to restores their ability to be degraded by Xrn1-type 5′ exoribonucleases.
A key step in establishing a physiological role for hClp1 kinase activity in RNA sealing will be to identify the ligase component of the yeast-like pathway in mammalian cells. To our knowledge, no Rnl1-type RNA sealing enzyme has been identified in a mammalian organism, and there is no apparent homolog of Rnl1 or the Trl1 ligase domain in any mammalian proteome. The surrogate genetic system we describe here might afford a means to do so, by screening mammalian cDNA libraries for functional complementation of a yeast strain with a conditional mutation in the RNA-sealing domain of yeast or plant tRNA ligases.
Major functional distinctions between yeast and human Clp1
Yeast and human Clp1 are closely related in primary structure, with 107 positions of chain identity plus 74 positions of side chain similarity over the 425-amino-acid hClp1 polypeptide. This similarity persists throughout the three discretely folded structural domains of yCl1p: an N-terminal β sandwich module, a central P-loop-type phosphotransferase domain, and a C-terminal globular α/β module (Noble et al. 2007). yClp1 is essential in S. cerevisiae, where it comprises part of the mRNA 3′-end-forming machinery. Many protein–protein interactions have been documented for yClp1 by affinity capture, two-hybrid, and biochemical reconstitution methods (see Saccharomyces Genome Database at www.yeastgenome.org). Prominent among these is the interaction between yClp1 and Pcf11, another essential component of the CF1 complex (Amrani et al. 1997; Gross and Moore 2001). The binding site for Pcf11 is located on a surface groove of yClp1 at the interface between the central and C-terminal domains; this site is quite remote from the ATP-binding site of yClp1 (Noble et al. 2007). To our knowledge, there is little published information concerning structure–activity relations for yClp1. Here, we show that the conserved amino acid constituents of the putative phosphotransferase active site are not essential for yeast viability. This result contrasts sharply with findings that the same residues in hClp1 are required to sustain its ability to function in yeast tRNA splicing. Although the RNA processing repertoire of hClp1 appears to be broader than that of yClp1 (which lacks RNA kinase activity), it was somewhat surprising to us that hClp1 does not function in lieu of yClp1 in vivo. It is conceivable that the full network of protein–protein interactions involving yClp1 cannot be fulfilled by hClp1.
It is noteworthy that the original purification of human pre-mRNA cleavage factor IIm yielded a complex that contained stoichiometric amounts of hClp1 and hPcf11 (de Vries et al. 2000). Subsequent independent purifications of human tRNA splicing endonuclease and human RNA kinase yielded complexes containing stoichiometric amounts of hClp1 and hSEN subunits, but no reported presence of hPcf11 (Paushkin et al. 2004; Weitzer and Martinez 2007). It is conceivable that hClp1 exits as a subunit of distinct macromolecular complexes that play different roles in the maturation of mammalian tRNAs and mRNAs. The importance of the hClp1 RNA kinase activity in the function of such complexes is not fully understood. The present study shows that RNA kinase is not a conserved or essential property of the yeast Clp1 homolog. Perhaps there is no selection pressure to maintain an RNA kinase activity of Clp1 in budding yeast, a species that lacks an siRNA pathway and clearly has another essential RNA kinase enzyme (in Trl1) that is dedicated to tRNA splicing. It will be of interest in future studies to survey Clp1 homologs from diverse eukarya for their ability to phosphorylate RNA 5′OH ends.
MATERIALS AND METHODS
Yeast vectors for expression of hClp1 and yClp1
A 1.3-kb cDNA fragment spanning the hCLP1 ORF was amplified from total HeLa cell RNA by RT-PCR using the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen). The hCLP1 gene-specific primers were designed to introduce an EcoRI site immediately upstream of the ATG codon and an XhoI site downstream of the stop codon. A DNA fragment spanning the yCLP1 ORF was amplified from a S. cerevisiae genomic DNA plasmid library by PCR with primers designed to introduce an EcoRI site immediately upstream of the ATG codon and a HindIII site downstream of the stop codon. The DNA fragments were cleaved at their terminal restriction sites and then inserted into pYX212 (2μ URA3), a shuttle vector in which expression of hCLP1 and yCLP1 is under the transcriptional control of the constitutive TPI1 promoter. The TPI1-hCLP1 and TPI1-yCLP1 expression cassettes, contained within 3.5-kb PvuII fragments, were excised from the pYX212 plasmids and inserted into PvuII-cut yeast shuttle vector pRS423 (2μ HIS3). Alanine mutations were introduced into the hCLP1 and yCLP1 genes by two-stage PCR with mutagenic primers; mutated genes were inserted into pRS423. The inserts of all clones were sequenced to exclude the presence of unwanted mutations.
Recombinant yClp1, hClp1, and T4 Pnkp proteins
DNA fragments encoding hClp1 and yClp1 were amplified by PCR using pYX212-hCLP1 and pYX212-yCLP1 as templates and primers designed to introduce SacI restriction sites at the ATG codons and an XhoI or HindIII site, respectively, adjacent to the stop codon. The PCR products were digested and inserted into pET28-His10Smt3 so that the CLP1 genes are fused in-frame to a leader sequence encoding an N-terminal His10Smt3 polypeptide. The inserts were sequenced to exclude the presence of unwanted mutations and confirm that the ORFs were in-frame with the leader. The pET28-His10Smt3-hClp1 and pET28-His10Smt3-ScClp1 plasmids were transformed into Escherichia coli BL21 PLUS (DE3) (Invitrogen). Single kanamycin-resistant colonies were inoculated into LB medium containing 0.05 mg/mL kanamycin and incubated at 37°C until A 600 reached 0.6–0.8. The pET28-His10Smt3-hClp1 culture (1000 mL) was placed for 30 min on ice and then incubated for 18 h at 17°C with constant shaking. The pET28-His10Smt3-yClp1 culture (500 mL) was adjusted to 0.4 mM IPTG and 2% (v/v) ethanol prior to incubation for 18 h at 17°C with constant shaking. Cells were harvested by centrifugation, and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Cell pellets were thawed on ice and suspended in buffer A (50 mM Tris-HCl at pH 7.4, 250 mM NaCl, 10% sucrose) to attain 5% of the original culture volume. Lysozyme was added to 0.2 mg/mL final concentration. After 45 min, the lysates were adjusted to 0.1% Triton X-100 and then sonicated to reduce viscosity. Insoluble material was removed by centrifugation for 20 min at 15,000 rpm in a Sorvall SS34 rotor. The supernatants were withdrawn and mixed with 1 mL of Ni-NTA-agarose resin (Qiagen) that had been equilibrated with buffer A. The slurries were poured into columns, which were then washed with 20 mL of buffer B (50 mM Tris-HCl at pH 7.4, 250 mM NaCl, 10% glycerol) containing 25 mM imidazole. Bound polypeptides were eluted in a stepwise fashion with buffer B containing 100, 200, 300, 400, and 500 mM imidazole. The protein compositions of the eluate fractions were monitored by SDS-PAGE. The 200 mM and 300 imidazole fractions containing hClp and yClp1, respectively, were stored at −80°C. Protein concentrations were determined using the Bio-Rad dye-binding assay with bovine serum albumin as a standard. His10-tagged T4 Pnk was produced in E. coli (Wang and Shuman 2001) and purified from a soluble lysate by Ni-agarose chromatography, in parallel with yClp and hClp1, as described above.
Deletion of CLP1 in S. cerevisiae
The entire CLP1 open reading frame was deleted in the S. cerevisiae and replaced with a bacterial kanamycin-resistance gene as follows. Diploid strain W303 was transformed with a linearized pUC19-clp1∷kanR fragment composed of a 1.44-kb kanR cassette (a BglII–SacI fragment excised from pUG6 [Güldener et al. 1996]) flanked by 505 base pairs (bp) of yeast genomic DNA 5′ of the CLP1 translation start codon and 390 bp of DNA 3′ of the CLP1 translation stop codon. Correct gene targeting was confirmed by Southern blotting of G418-resistant transformants. The CLP1 clp1∷kanR diploid was transformed with pYX212-yClp1 (2μ URA3 yCLP1). The resulting Ura+ diploid was sporulated and tetrads were dissected. We thereby recovered viable clp1∷kanR haploids that were resistant to G418 and unable to grow on medium containing 0.75 mg/mL FOA, a drug that selects against the URA3 yCLP1 plasmid. The clp1Δ strain yAR1 (Mat a ura3 ade2 trp1 his3 leu2 can1 clp1∷kanR, pYX212-yClp1 [2μ URA3]) was used to test plasmid-borne alleles of yCLP1 and hCLP1 for clp1Δ complementation by plasmid shuffle.
ACKNOWLEDGMENTS
This work was funded by NIH grant GM42498. S.S. is an American Cancer Society Research Professor. B.S. is supported by funds from the William Randolph Hearst Foundation to the Department of Microbiology and Immunology. A.R. is an NIH predoctoral trainee supported by grant T32-GM008539.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1142908.
REFERENCES
- Amitsur M., Levitz R., Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet M., Wyers F., Dufour M., Aggerbeck L., Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P.E., Lee J., Gravel M. 2′,3′-Cyclic nucleotide 3′-phoshodiesterase: Structure, biology and function. Myelin Biol. Disord. 2004;1:499–521. [Google Scholar]
- de Vries H., Rüegsegger U., Hübner W., Friedlein A., Langen H., Keller W. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;21:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastberg J.H., Pelletier J., Stoddard B.L. Recognition of DNA substrates by bacteriophage T4 polynucleotide kinase. Nucleic Acids Res. 2004;32:653–660. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M., Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Shatkin A.J. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Konarska M., Gross H.J., Shatkin A.J. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt E.A., Pelletier J., Wilson G., Stoddard B.L. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- Greer C.L., Peebles C.L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Gross S., Moore C.L. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Lackey J.G., Hsu H.C., Zhang Y., Deng J., Xu R.M., Dhama M.J., Ron D. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M., Filipowicz W., Domdey H., Gross H.J. Formation of a 2′-phosphomonoester, 3′,5′-phosphodiester linkage by a novel RNA ligase in wheat germ. Nature. 1981;293:112–116. doi: 10.1038/293112a0. [DOI] [PubMed] [Google Scholar]
- Konarska M., Filipowicz W., Gross H.J. RNA ligation via 2′-phosphomonoester, 3′,5′-phosphodiester linkage: Requirement of 2′,3′-cyclic phosphate termini and involvement of a 5′-hydroxyl polynucleotide kinase. Proc. Natl. Acad. Sci. 1982;79:1474–1478. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P.E., Griffiths I.R., Nave K.A. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Laski F.A., Fire A.Z., RajBhandary U.L., Sharp P.A. Characterization of tRNA precursor splicing in mammalian extracts. J. Biol. Chem. 1983;258:11974–11980. [PubMed] [Google Scholar]
- Li H., Trotta C.R., Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- Mazumder R., Iyer L., Vasudevan S., Aravind L. Detection of novel members, structure–function analysis and evolutionary classification of the 2H phosphoesterase family. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker P.J., Wiederkehr T., Strahm Y., Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in pre-messenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C.G., Beuth B., Taylor I.A. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007;35:87–89. doi: 10.1093/nar/gkl1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Patel M., Furia B.S., Peltz S.W., Trotta C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′-end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- Peebles C.L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Salgia S.R., Singh S.J., Gurha P., Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: Joining of exons and circularization of introns. RNA. 2003;9:319–330. doi: 10.1261/rna.2118203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R., Schwer B., Shuman S. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003;278:43298–43398. doi: 10.1074/jbc.M307839200. [DOI] [PubMed] [Google Scholar]
- Schwer B., Sawaya R., Ho C.K., Shuman S. Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Aronova A., Ramirez A., Braun P., Shuman S. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:204–210. doi: 10.1261/rna.858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S.L., Malik H.S., Consaul S.A., Phizicky E.M. A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli . Proc. Natl. Acad. Sci. 1998;95:14136–14141. doi: 10.1073/pnas.95.24.14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta C.R., Miao F., Arn E.A., Stevens S.W., Ho C.K., Rauhut R., Abelson J.N. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- Trotta C.R., Paushkin S.V., Patel M., Li H., Peltz S.W. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature. 2006;441:375–377. doi: 10.1038/nature04741. [DOI] [PubMed] [Google Scholar]
- Wang L.K., Shuman S. Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. 2001;276:26868–26874. doi: 10.1074/jbc.M103663200. [DOI] [PubMed] [Google Scholar]
- Wang L.K., Shuman S. Mutational analysis defines the 5′ kinase and 3′ phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.K., Shuman S. Structure–function analysis of yeast tRNA ligase. RNA. 2005;11:966–975. doi: 10.1261/rna.2170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.K., Lima C.D., Shuman S. Structure and mechanism of T4 polynucleotide kinase—An RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.K., Schwer B., Englert M., Beier H., Shuman S. Structure–function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006;34:517–527. doi: 10.1093/nar/gkj441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S., Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:2222–2226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Xue S., Calvin K., Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T., Yunoki-Esaki K., Ohshima C., Tanaka N., Endo T. Possibility of cytoplasmic pre-tRNA splicing: The yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T., Ohshima C., Yunoki-Esaki K., Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae . Genes Cells. 2007;12:285–297. doi: 10.1111/j.1365-2443.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- Zillman M., Gorovsky M.A., Phizicky E.M. Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell. Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofallova L., Guo Y., Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G., Ogden R.C., Peebles C.L., Abelson J. Splicing of yeast tRNA precursors: Structure of the reaction intermediates. Cell. 1979;18:37–45. doi: 10.1016/0092-8674(79)90351-9. [DOI] [PubMed] [Google Scholar]
- Shuman S., Hurwitz J. 5′-Hydroxyl polyribonucleotide kinase from HeLa cell nuclei. Purification and properties. J. Biol. Chem. 1979;254:10396–10404. [PubMed] [Google Scholar]