Abstract
RNA polymerase III (Pol III) expression systems for short hairpin RNAs (U6 shRNAs or chimeric VA1 shRNAs) or individually expressed sense/antisense small interfering RNA (siRNA) strands have been used to trigger RNA interference (RNAi) in mammalian cells. Here we show that individually expressed siRNA expression constructs produce 21-nucleotide siRNAs that strongly accumulate as duplex siRNAs in the nucleus of human cells, exerting sequence-specific silencing activity similar to cytoplasmic siRNAs derived from U6 or VA1-expressed hairpin precursors. In contrast, 29-mer siRNAs separately expressed as sense/antisense strands fail to elicit RNAi activity, despite accumulation of these RNAs in the nucleus. Our findings delineate different intracellular accumulation patterns for the three expression strategies and suggest the possibility of a nuclear RNAi pathway that requires 21-mer duplexes.
Keywords: siRNA, tandem expression vectors, PTGS, nucleus, RNAi
INTRODUCTION
RNAi in mammalian cells is a phenomenon by which short double-stranded RNAs (dsRNAs) direct enzyme-dependent cleavage of specific target RNAs. In mammalian systems, sequence-specific RNAi effects have been observed either by direct introduction of synthetic siRNAs or by in vivo expression of siRNAs and shRNAs using both Pol II and Pol III promoters (McManus and Sharp 2002; Dykxhoorn et al. 2003). Two different approaches for Pol III expression trigger RNAi. One utilizes two tandem U6 promoters to produce separate sense and antisense single-stranded 21–23 mers, which can anneal and form siRNAs (pU6 siRNAs) (Lee et al. 2002; Miyagishi and Taira 2002). The other is modeled after naturally occurring microRNAs (miRNAs) and results in the production of duplex precursors with 19–29 base stems and connecting loops that are further processed into siRNAs (pU6 shRNAs), presumably by Dicer (Dykxhoorn et al. 2003). Exportin5 is believed to export nuclear-expressed shRNAs into the cytoplasm (Yi et al. 2003), but it is not known whether siRNAs produced by separate sense and antisense transcription are exported via this pathway.
Since the major protein components involved in the RNAi machinery, the RNA-induced silencing complex (RISC) and Dicer, are thought to assemble and function in the cytoplasm (Carmell et al. 2002; Provost et al. 2002; Tabara et al. 2002; Zeng and Cullen 2002), it is widely believed that RNAi-mediated cleavage of targeted mRNAs occurs only in the cytoplasm. However, recent studies have demonstrated that siRNAs can induce sequence-specific transcriptional gene silencing (TGS) (Morris et al. 2004) and post-transcriptional gene silencing (PTGS) (Langlois et al. 2005; Robb et al. 2005; Berezhna et al. 2006) in the nucleus of human cells.
Here we show that tandem siRNA expression constructs (pU6 siRNAs) produce independently transcribed complementary 21-nucleotide (nt) sense and antisense RNA duplexes that accumulate to large extents in the nucleus of human cells, exerting silencing activity similar to cytoplasmic siRNAs derived from U6 or VA1-expressed hairpin precursors. However, 29-mer siRNAs expressed from tandem constructs (pU6 siRNA–29 mer) fail to elicit RNAi activity, despite the accumulation of these RNAs in the nucleus. These results suggest the possibility of a nuclear PTGS pathway that requires 21-mer duplexes to mediate RNAi.
RESULTS
We have previously demonstrated that pU6 siRNAs can elicit RNAi against both a reporter target eGFP fused to HIV-1 rev or HIV-1 rev itself (Lee et al. 2002). To better understand the differences between pU6 siRNAs and pU6 shRNAs, we carried out cotransfection experiments with different siRNA expression constructs and a Ponasterone A-inducible target (rev–eGFP). This approach facilitates studies of inhibition using both fluorescence microscopy and FACS analysis to monitor RNAi function via knockdown of eGFP expression. The independently transcribed complementary 21-mer RNA sequences targeted to HIV-1 rev (site II) were transcribed using two U6 promoters in tandem (pU6 si II) (Lee et al. 2002), and the short hairpin RNAs (sh II) contained a 21-mer RNA duplex and an 8-nt loop (Paddison et al. 2002) (pU6 sh II; Fig. 1A). Additionally, the sh II sequence was inserted within the stem–loop IV region of the VA1 RNA, which also functions as a Pol III promoter (Cagnon and Rossi 2000) (pVA1 sh II; Fig. 1A). We have previously used this system to express a functional hammerhead ribozyme that localizes in the cytoplasm (Cagnon and Rossi 2000). We reasoned that fusing the VA1 RNA with an shRNA might facilitate cytoplasmic transport of the shRNAs via Exportin5 (Gwizdek et al. 2003) and thus colocalize the hairpin with Dicer and the cytoplasmic RNAi machinery.
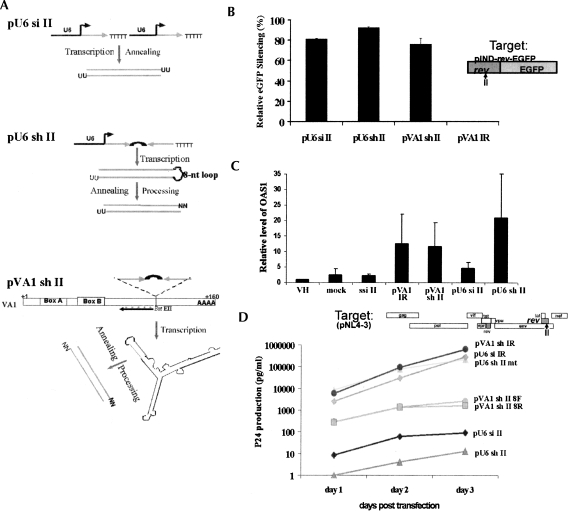
FIGURE 1.
Three approaches used in this study for intracellular expression of siRNAs from DNA vectors using Pol III systems in mammalian cells. (A) Schematic representation of three approaches. siRNAs targeted to HIV-1 rev (site II) (Lee et al. 2002) are produced by transcription of sense and antisense strands in trans using tandem U6 promoters (pU6 si II) (Lee et al. 2002), or in the form of a short hairpin RNA (shRNA) with an 8-nt loop (pU6 sh II) (Paddison et al. 2002). In addition, the sh II sequences are inserted in the BstEII site of VA1 (pVA1 sh II) (Cagnon and Rossi 2000). The VA1 Pol III promoter is represented by A and B boxes. The FISH probe site of the 50-mer antisense VA1 DNA-modified oligomers conjugated with Cy3 (red asterisks) is depicted as ←. (B) Silencing of eGFP expression (HIV-1 rev fused to eGFP) (Lee et al. 2002) by siRNAs from different vectors. 293/EcR cells were cotransfected with the target pIND rev–eGFP and different siRNA expression constructs and analyzed for eGFP expression by FACS (Lee et al. 2002). pVA1 sh IR (irrelevant siRNA sequence) was used as a control for sequence-specific inhibition. Other controls (pU6 si IR and pU6 sh II mt) were also used (data not shown). Standard errors are based on the average and standard error from five independent experiments. (C) qRT-PCR measured expression of 2′–5′ OAS mRNA in cells cotransfected with target and siRNA constructs. Standard errors are based on average measurements from four independent experiments. One experiment had high values of OAS1 mRNA for all the shRNA constructs, biasing the standard error. (D) siRNA-mediated silencing of HIV-1 expression. PNL4-3 proviral DNA was cotransfected with different siRNA expression constructs. At 1 d post-transfection, and at the indicated times, supernatant aliquots were withdrawn for HIV-1 p24 antigen assays. The various constructs used are indicated. mt, mutant containing four mismatches in the middle of the antisense strand; 8F/8R, 8-nt loop with forward and reverse sequences of shRNA, respectively.
Expression cassettes containing the si II and sh II genes were cotransfected into 293/EcR cells (Lee et al. 2002) with the inducible rev–eGFP target construct, and knockdown of eGFP expression was monitored via FACS analysis in replicate experiments (Fig. 1B). pU6 sh II showed the greatest knockdown (91 ± 5%), followed by the pU6 si II (85 ± 4%) and pVA1 sh II (75 ± 7%) constructs (Fig. 1B). Control constructs included irrelevant (pU6 si IR and pVA1 sh IR) or mutant siRNA sequences (pU6 shII mt) that did not alter the expression levels of rev–eGFP (Fig. 2B; data not shown), thus establishing sequence specificity for the decrease in eGFP expression.
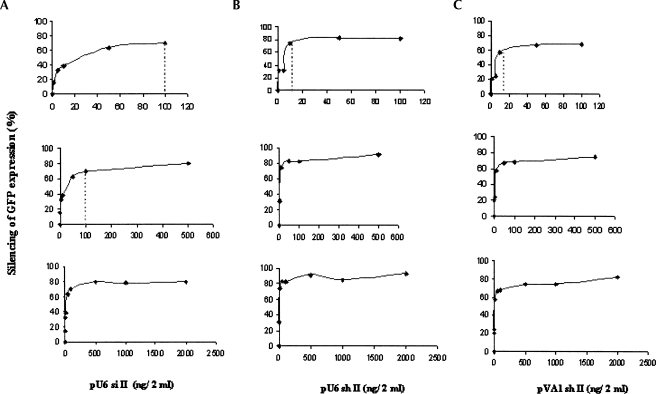
FIGURE 2.
Plasmid titration. Silencing of eGFP expression was examined at different concentrations of each construct: pU6 si II (A), pU6 sh II (B), and pVA1 sh II (C). 293/EcR cells grown in six-well plates were cotransfected with the target construct (2 μg/2 mL) and different concentrations of each construct (5 ng–2 μg/2 mL) for a total of 4 μg DNA/2 mL. FACS data were normalized with the transfection efficiency, which was determined using a LacZ expression vector in cotransfections.
To ensure that the constructs were not eliciting target knockdown via induction of interferon-related responses, we measured the up-regulation of 2′–5′ oligoadenylate synthetase 1 mRNA (OAS1), interferon β (IFNβ) mRNA induction, PKR phosphorylation, and MxA mRNA levels following transfection (Samuel 2001). A twofold increase in OAS1 mRNA expression over vector backbone-transfected cells was observed in cells expressing shRNAs from the VA1 promoter (pVA1 sh II and pVA1 IR), and a fivefold increase was observed from the pU6 sh II (Fig. 1C). Little or no change in OAS1 mRNA expression was observed from the pU6 si II (Fig. 1C). Levels of OAS1 mRNA in cells transfected with a long dsRNA (either a 700-bp GFP dsRNA or polyIC) were ∼1500-fold greater than mock-transfected cells (data not shown). No differences were observed in PKR phosphorylation, MxA, or IFNβ mRNA expression between control cells and those expressing siRNA or shRNA at 48-h post-transfection (data not shown). The small increase in 2′–5′ OAS mRNA expression was the only interferon-related response observed, and this was only observed with the shRNAs. Even with this minor interferon response, RNAi-mediated silencing was still specific for the targeted transcript in these cells (Fig. 1B,D).
We next tested the potential anti-HIV-1 activities of the various constructs in cotransfection assays with HIV-1 pNL4-3 proviral DNA in 293 cells (Fig. 1D). As with the rev–eGFP construct, pU6 sh II gave the most potent inhibition of p24 production, providing approximately four logs of inhibition relative to a four-base mismatched mutant (mt) control. pU6 si II and pVA1 sh II exerted ∼3.5 and ∼2.8 logs of inhibition of p24 production, respectively. These data demonstrate that both the pU6 si II and pVA1 sh II pathways are less effective than the pU6 sh II pathway.
To further examine the relative differences in target knockdown by the various constructs, we carried out plasmid titration assays (Fig. 2). The silencing activities of shRNAs mediated by both U6 and VA1 systems reached a plateau at ∼10 ng per 2 mL of cultured cells (Fig. 2B,C), whereas silencing from the tandem siRNA expression systems (pU6 si II) required a 10-fold higher concentration (∼100 ng/2 mL) to reach the same plateau (Fig. 2A). These data suggest that the pU6 si II pathway is less efficient, possibly due to a lack of stability and/or ability to form duplexes. Alternatively, the U6-expressed siRNAs may function through a pathway different from shRNAs.
To examine whether siRNAs from the pU6 si II construct were less stable than those from the shRNA constructs, we performed Northern gel blotting assays on total RNA from transfected cells (Fig. 3). Northern results revealed that expression of siRNAs from the pU6 si II construct was robust, with significantly more detectable 21-mer RNAs than observed from shRNA constructs (Fig. 3A,B). Paradoxically, despite the higher amounts of siRNAs, reduction of target mRNAs via the pU6 si II constructs was less effective than with the pU6 sh II (89 ± 5% relative to 96 ± 4%) (Fig. 3D). The RNA knockdown level determined by Northern analysis was thus consistent with fluorescent microscope imaging (85 ± 4%) and FACS data (91 ± 5%) (Fig. 1B). These data suggest that the extent of siRNA-directed silencing is not limited by the expression level or stability of the siRNAs. Interestingly, the levels of siRNA-sized products from the pVA1 sh II were comparable to that of the siRNAs from the pU6 sh II (Fig. 3A). Nevertheless, the silencing activity from the pVA1 sh II was less effective than from the pU6 sh II (75 ± 7% relative to 85 ± 4%) (Fig. 1B). The VA1-generated siRNAs were considerably less abundant (∼1/100) than the siRNAs expressed from pU6 si II (Fig. 3A), perhaps due to inefficient processing, but their biological silencing potencies were roughly equivalent (75 ± 7% relative to 85 ± 4%) (Fig. 1B).
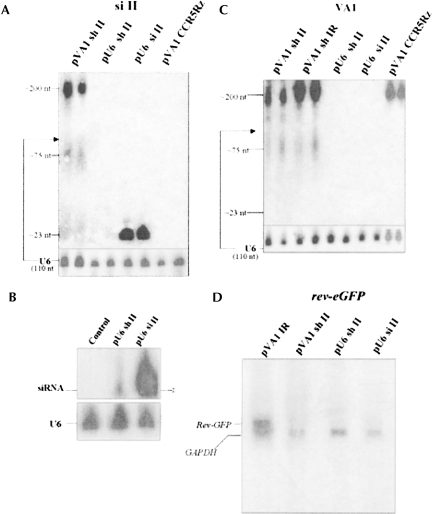
FIGURE 3.
Detection of siRNAs and target mRNAs by Northern assay. RNA samples were prepared from 293/EcR cells transiently cotransfected with the target construct and different siRNA constructs and Northern analyses were performed. Expression of siRNAs from various constructs is presented using a sense probe for site II (A,B) and a probe complementary to the VA1 transcript (C). In A and C, two independent RNA samples for each construct were loaded in PAGE gels. Endogenous U6 snRNA was used as an internal control and a size marker (∼110 nt). pVA1 CCR5Rz contained the hammerhead ribozyme targeted to CCR5, inserted into VA1 gene, and was used as a negative control substrate for Drosha processing of the VA1 chimeric transcripts. (D) Inhibition of target rev–eGFP mRNA by siRNAs expressed from the various constructs was examined by Northern analyses using a probe complementary to rev–eGFP. GAPDH mRNAs were used as an internal loading control. Quantification of rev–eGFP mRNAs was normalized to GAPDH using QuantamOne of α phosphor images. Standard error % is based on two independent experiments.
An interesting observation regarding the Northern analysis of the pVA1 sh II construct was the presence of an intermediate-sized product of ∼75 nt, as well as the 21–23-nt RNAs (Fig. 3A). This 75-nt product was reminiscent of pre-miRNA processing intermediates (Lee et al. 2003). Such an intermediate-sized product was not detected when a hammerhead ribozyme targeted to CCR5 was inserted into VA1 (pVA1 CCR5Rz) (Fig. 3C; Cagnon and Rossi 2000). We attributed this product to an intermediate processing event involving the shRNA inserted within domain IV of VA1. The intermediate product was present in cells transfected with two different shRNA sequences, one deriving from sh II and the other from an irrelevant shRNA targeting lacZ mRNA. It is important to note that there was a large quantity of unprocessed VA1 sh II RNA in addition to the processed products. Such unprocessed shRNAs were not detected in cells transfected with the pU6 sh II vector. We propose that the shRNA structure inserted into the VA1 RNA might be a substrate for Drosha, which cleaves pri-miRNAs to pre-miRNAs (Lee et al. 2003). Processing of the VA1 shRNA by Drosha would have to proceed prior to cytoplasmic export, and the large amount of unprocessed transcript may lead to a competition between processing and cytoplasmic export.
To investigate the subcellular localization of the siRNAs and shRNAs, we performed fluorescent in situ hybridization (FISH) in 293/EcR and HeLa cells using sense sequences (complementary to the antisense or guide sequences) labeled with Cy3 as a probe (Fig. 4). The siRNAs produced from pU6 si II accumulated to high levels in the nucleus and appeared in discrete foci (Fig. 4A,B). No siRNAs from the pU6 si II were detected in the cytoplasm. Since 21-mer duplexes mimic the products of Dicer cleavage, we asked whether a 29-mer duplex transcribed from pU6 si II–29 mer constructs could function in RNAi. We constructed 29-mer sense and antisense U6 transcription units that contained five extra 5′ nt and three extra 3′ nt complementary to the HIV-1 rev site II target (pU6 si II–29 mer) (Fig. 4C). The pU6 si II–29 mer constructs were cotransfected into 293/EcR cells with the rev–eGFP target construct, and their silencing activities were compared with those of the 21-mer siRNAs expressed from pU6 si II (Fig. 4D). No inhibition was observed when the pU6 si II–29 mer constructs were cotransfected into 293/EcR cells with the rev–eGFP construct (Fig. 4D). To ascertain whether the 29 mers were processed into 21 mers, we performed Northern analysis. The 29-mer RNAs were readily detected, but no processed 21-mer products were observed (Fig. 4E). Thus, the 29-mer RNAs expressed from the pU6 si II–29 mer constructs were not processed into functional 21-mer siRNAs. Since the 29 mers might not be functional Dicer substrates, we chemically synthesized the same 29 mers and transfected them into 293/EcR cells, along with the inducible rev–eGFP target construct. The transfected 29 mers elicited potent inhibition of eGFP expression (Fig. 4D), demonstrating that they could in fact be processed into functional 21-mer siRNAs in the cytoplasm. We conclude from these studies that U6-expressed 29-mer siRNAs are not transported to the cytoplasm for processing into 21 mers but are instead retained in the nucleus.
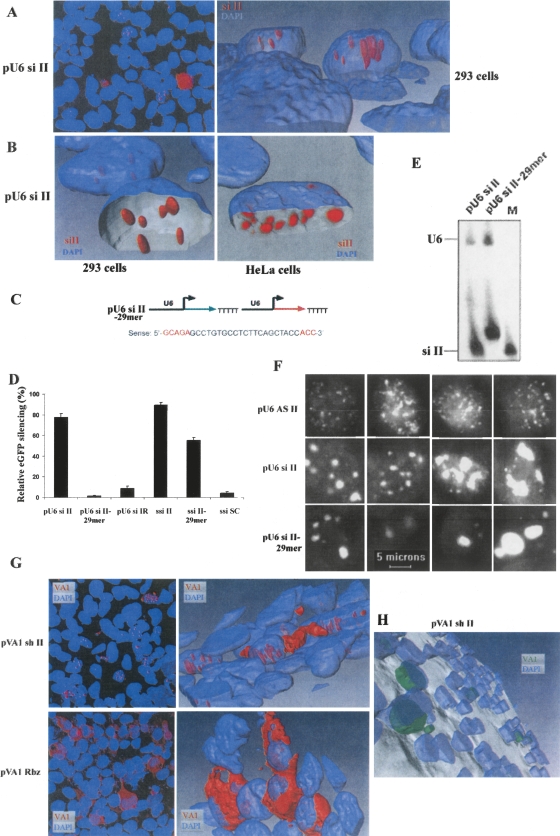
FIGURE 4.
FISH analysis of the subcellular localization of siRNAs and VA1-containing shRNAs. (A) A maximum intensity projection (left) of fluorescence in 293 cells using a cy3-labeled si II RNA probe (complementary to the antisense strand) (red) and two-photon imaging of DAPI (blue). A 3D surface model of the confocal data (right) shows the si II signal contained within the nucleus. (B) Removal of part of the DAPI surface shows the shape and orientation of the siII RNA within the nucleus. The right panel shows similar structures present in the nucleus of similarly transfected HeLa cells. For the HeLa cell image, the si II signal is presented as Voxel reconstruction of the cy3 data. (C) Sense sequences of 29 mers of pU6 si II–29 mer. (D,E) Function (D) and expression (E) of siRNAs from pU6 si II and pU6 si II–29 mer in cells. (D) FACS analyses of the inhibition of eGFP expression by siRNAs were examined in 293/EcR cells cotransfected with the target construct and different siRNA expression constructs, or synthetic siRNAs (ssiRNAs). PU6 si IR and ssi SC (scrambled) were used as controls. (E) Small RNAs from two different expression plasmids detected using Northern gel analyses. M is a radiolabeled sense 21-mer oligo as a size marker. U6 was used as an internal control. (F) Subcellular localization (cy3-labeled sense probes) of small RNAs expressed from different expression plasmids in 293 cells by FISH. Data are presented as maximum intensity projections of cy3 fluorescence (white) in a single nucleus of four different cells cotransfected with target and the different expression constructs. The outline of the nucleus (DAPI staining and two-photon imaging) can be seen in black. Individual images for each stack of 30 were captured at 0.5 μm spacing using a 60× oil immersion objective (NA=1.4). pU6 AS II containing only the antisense sequence was used as a control. (G) Subcellular localization of VA1 sh II (upper) or hammerhead ribozyme RNA (lower) in 293 cells by FISH using a cy3-labeled VA1 RNA probe (complementary to the antisense strand) (red) and two-photon imaging of DAPI (blue). The left panels show maximum intensity projections of confocal image stacks, the right panels show 3D surface reconstructions. (H) 3D image of subcellular localization of VA1 sh II using Amira.
To further examine the nature of the discrete, nuclear siRNA foci, FISH analysis was performed using the sense strand as a probe in cells transfected with only antisense RNA expression constructs (pU6 AS II). When the pU6 AS II constructs were transfected into 293/EcR cells, the discrete foci of hybridization were not detected in the nucleus. Instead, the nucleus was filled with smaller speckles (Fig. 4F, upper). In contrast, small RNAs produced from dual sense/antisense transcription of the pU6 si II and pU6 si II–29 mer constructs formed discrete, large foci in the nucleus (Fig. 4D, middle and bottom panels, respectively). The signals obtained when both sense and antisense 21-mer or 29-mer RNAs were expressed simultaneously (pU6 si II or pU6 si II–29 mer) were not due to cross-hybridization with DNA, since no hybridization signals were detected in cells transfected with only the sense RNA expression constructs (pU6 S II) (data not shown). These data suggest that the foci are comprised of dsRNAs, perhaps bound to nuclear proteins that form large foci with the bound siRNAs.
We next investigated the localization of transcripts from pVA1 sh II by performing FISH analysis using a probe complementary to the VA1 RNA portion of the constructs (upstream of the Bst EII site that is the shRNA insertion site; Fig. 1C). The VA1 chimeric transcripts containing sh II RNAs localized in the nucleus and cytoplasm (Fig. 4G, upper panel), whereas the VA1 chimeric transcripts containing a hammerhead ribozyme localized mainly in the cytoplasm, with almost no signal detected in the nucleus (Fig. 4G, bottom panel). The difference between an shRNA (∼50 mers) and a ribozyme (∼43 mers) is the presence of a dsRNA-containing stem–loop structure. VA1 RNAs containing ribozymes may be rapidly exported by Exportin5 (cytoplasmic localization, Fig. 4G), but a subpopulation of the VA1 shRNAs may be retained in the nucleus as a consequence of interacting with the miRNA processing machinery/Drosha to produce sh II (Fig. 4G). These shRNAs could then be exported by Exportin5, while the remaining VA1 RNA might be trapped in the nucleus. Since VA1 and shRNAs both use the same export system, the reduced efficacy of the VA1 shRNA relative to the U6 shRNA may be attributed to poor processing of the shRNA from the VA1 precursor or competition for Exportin5. Some VA1 chimeric transcripts containing sh II RNAs appeared to be protruding from the nuclear compartment toward the cytoplasm (Fig. 4H). Nuclear VA1 shRNAs formed thin tubes (Fig. 4G) or channels, which was especially noticeable in HeLa cells (Supplemental Fig. 1). These RNAs did not form nuclear foci like those observed with the U6-expressed si II RNAs in 293/EcR cells (Fig. 4A). The reduced efficacy of the VA1 shRNA could also be the consequence of a slightly different siRNA product being generated that might have reduced efficacy.
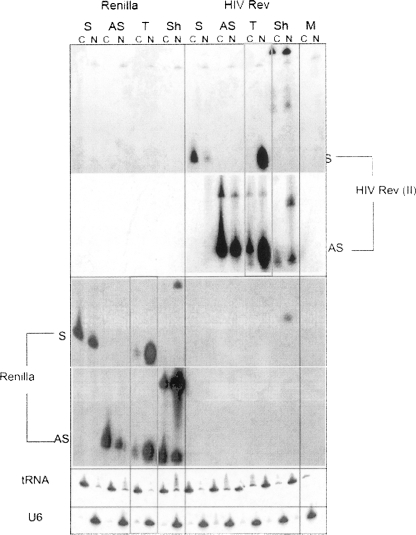
To exclude the possibility of nuclear localization being specific to the site II sequences of HIV-1 rev, we also tested another tandem U6 siRNA expression construct that targeted an unrelated Renilla luciferase gene (pU6 si Renilla). Both sense and antisense RNAs expressed from these tandem siRNA expression constructs (pU6 si II and pU6 si Renilla) were abundant in the nucleus (Fig. 5, red boxes), although some antisense strands were detected in the cytoplasm (Fig. 5), most likely from leakage during the nuclear/cytoplasmic fraction procedures, as we also observed some leakage of nuclear U6 snRNAs in the cytoplasm. Interestingly, only the antisense strands from pU6 sh II or pU6 sh Renilla were localized in both the nucleus and cytoplasm, whereas the sense RNAs from both constructs were not detectable (Fig. 6). The nuclear presence of processed antisense strands from these shRNA constructs could be due to nuclear import of activated RISC containing the antisense guide strand, although this could also be due to leakage from the cytoplasmic compartment into the nucleus during the fractionation procedure. Failure to detect the sense strand could also be due to differences in the hybridization efficiencies of probes specific for the AS and S strands. This result was different from the abundant levels of both S and AS RNAs (Fig. 5, red boxes) observed when expressed from tandem siRNA expression constructs (pU6 si II and pU6 si Renilla) (Fig. 6).
FIGURE 5.
Northern hybridization of nuclear (N) and cytoplasmic (C) extraction of 293/EcR cells cotransfected with siRNA expression vectors and their targets. Expression vectors are pU6 siRNA (Tandem, T), pU6 shRNA (Sh), pU6 Sense (S), and pU6 Antisense (AS) to the HIV rev site II and Renilla luciferase transcripts. tRNALys3 and U6 snRNAs are used as cytolasmic and nuclear marker controls.
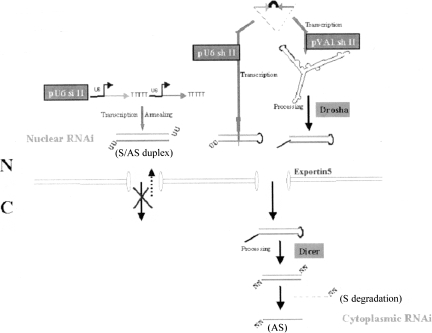
FIGURE 6.
Model of nuclear RNAi from a tandem siRNA expression construct (pU6 si II). Our tandem siRNA expression construct (pU6 si II) produces separate sense and antisense (red) single-stranded 21–23 mers from two tandem U6 promoters and forms duplexes (S/AS duplex) containing U overhangs at each 3′ end. The siRNAs accumulate in the nucleus of human cells and exert silencing activity (nuclear RNAi) similar to that obtained from siRNAs derived from the U6 or VA1 expressed hairpins.
We next compared the stability of both sense and antisense strands expressed in tandem with sense or antisense strands expressed alone. The sense and antisense strands expressed in tandem (T, pU6 si II, and pU6 si Renilla) were more stable than the singly expressed sense or antisense strands (S or AS, pU6 S II, and pU6 S Renilla, or pU6 AS II and pU6 AS Renilla) (Fig. 5). These results suggest that siRNAs are being efficiently formed as duplexes. Previous work from our lab, which used RNAse treatment of RNAs produced from the sense/antisense coexpression system for si II, confirmed the efficient formation of duplexes within this coexpression system (Lee et al. 2002). Our results are consistent with the work of Berezhna et al. (2006), in which siRNAs targeting the nuclear 7SK and U6 RNAs were localized in the nucleus as duplexes. Taken together, our data strongly support a double-stranded siRNA complex when sense and antisense strands are coexpressed in cells.
A possible mechanism by which nuclear-localized, U6-expressed si II RNAs mediate gene silencing is via TGS and heterochromatin formation. To investigate this possibility in 293/EcR cells cotransfected with pU6 si II and rev–eGFP constructs, we carried out chromatin immunoprecipitation (ChIP) assays of the rev target site of pU6 si II using antibodies against Argonaute-1 (Ago1) and trimethyl histone H3 lysine 27 (H3K27me3), as described previously (Kim et al. 2006). Both Ago1 and H3K27me3 were not enriched at the rev target site (Supplemental Fig. 2), suggesting that the inhibition was post-transcriptional as opposed to transcriptional in nature. Additional supporting experiments in 293/EcR cells using trichostatin A, a histone deacetylase inhibitor, and 5-aza-2′-deoxycytidine, a DNA methyltransferase inhibitor, showed no effect on the gene silencing function of pU6 si II (data not shown), indicating that pU6 si II was acting through a nuclear PTGS pathway.
DISCUSSION
RNAi is a multistep process involving the generation of siRNAs from long dsRNAs through the action of Dicer (Carmell and Hannon 2004). The resulting 21- to 23-nt siRNAs direct RISC toward degradation of their cognate mRNAs. We have utilized Pol III promoters to express small RNAs that can elicit RNAi. Constructs that transcribe RNAs that can be processed into 21–23-mer duplexes or that directly produce such duplexes are effective triggers of RNAi. Silencing activity mediated by siRNAs produced from pU6 sh II was the most potent of the constructs tested, perhaps because these shRNAs could be rapidly exported to the cytoplasm and subsequently processed by Dicer for entry into RISC. We also observed that shRNAs expressed as VA1 shRNA chimeras were processed in a two-step reaction into siRNAs, similar to miRNA precursors.
In human cells, mono- and poly-cistronic primary precursors for miRNAs (pri-miRNAs) undergo processing by nuclear-localized Drosha. This enzyme generates ∼70-nt pre-miRNAs (Murchison and Hannon 2004). These pre-miRNAs are then exported from the nucleus by Exportin5. In addition to export of pre-miRNAs, Exportin5 is also a transport carrier for shRNAs and other mini-helix-containing RNAs, including the VA1 RNA. Despite the high transcript levels of the VA1 shRNA constructs, their poor efficacy relative to U6 promoter expression of the same shRNA may be a consequence of competition between Exportin5-binding, export of the chimeric VA1 sh II transcripts, and/or the added step of Drosha processing. Additionally, even if the unprocessed VA1 shRNA chimeric transcripts enter the cytoplasm, they may be poor substrates for processing by Dicer, since they lack a terminal 3′ single-stranded region which is bound by Dicer on pre-miRNAs.
Most protein components of the RNAi machinery, including Dicer and RISC, are believed to assemble and function in the cytoplasm (Carmell et al. 2002; Provost et al. 2002; Tabara et al. 2002; Zeng and Cullen 2002). Recently, Hock and colleagues reported interactions of Ago1 and Ago2 with proteins that have nuclear-localization properties (Hock et al. 2007), and several lines of evidence have indicated that RNAi may occur in the nucleus of other organisms and mammals. In Caenorhabditis elegans, nuclear proteins are reportedly required for RNAi (Dudley et al. 2002). It has also been suggested that 21- to 22-nt siRNAs are produced in the nucleus of plants by a nuclear Dicer-like protein (Papp et al. 2003), and that PTGS can occur in the nucleus of plants (Dong et al. 2003; Papp et al. 2003). Additionally, siRNA-mediated degradation of the nuclear-localized 7SK RNA (Robb et al. 2005) and DMPK mRNA (Langlois et al. 2005) has been shown to occur in human cells. Furthermore, siRNAs targeting promoter regions are involved in TGS in the nucleus of human cells (Morris et al. 2004).
In this study, we have shown that one of our in vivo siRNA expression plasmids (pU6 si II) (Lee et al. 2002) produced siRNAs that localized in the nucleus, yet still elicited sequence-specific RNAi, in 293 (Figs. 1C, 4A) and HeLa cells (Supplemental Fig. 1). We observed that tandem Pol III-expressed siRNAs were somewhat less effective in triggering RNAi than siRNAs derived from expressed shRNAs (Fig. 4B,E), despite their massive accumulation in the nucleus. This could be due to a number of factors, including rapid export of the unspliced rev–eGFP transcripts from the nucleus to the cytoplasm. When expressed siRNAs and an expressed shRNA targeting the same sequence in a spliced transcript were compared, their levels of target knockdown were virtually identical (A. Ehsani and J. Rossi, unpubl.). Importantly, when sense and antisense 29-mer RNAs were transcribed in 293 cells, no RNAi was elicited, despite their accumulation in discrete nuclear foci. This result suggests that cytoplasmic export of non-covalently linked duplexes does not occur, or takes place inefficiently, since 29 mers would be processed into 21-mer siRNAs by Dicer if they were cytoplasmically exported. These results suggest the existence of a nuclear RISC that requires Dicer-processed siRNAs to function. Since Dicer is largely cytoplasmic (Provost et al. 2002), 29 mers that do not leave the nucleus might not be processed for entry into RISC, whereas 21 mers form functional siRNAs in the nucleus. This may not be true, however, if the RNAi machinery also localized to the nucleus. If a nuclear RNAi mechanism is physiologically relevant, there must be a mechanism for importing the processed 21-mer duplexes into the nucleus, perhaps via one or more components of the RNAi machinery. The possibility exists that siRNAs produced from either pU6 sh II or pVA1 sh II constructs can also be transported to the nucleus following cytoplasmic Dicer processing. One intriguing possibility is that RISC and/or other Argonaute-containing complexes may shuttle between the cytoplasm and nucleus. Of note, mature miR-29b has been shown to possess a 3′ nuclear-localization motif which confines this miRNA largely to the nucleus (Hwang et al. 2007), and other mature miRNAs have also been shown to be localized in the nucleolus (Politz et al. 2006; Marcon et al. 2008).
A different line of evidence for nuclear RNAi comes from our studies of the nuclear-retained DMPK transcript (Langlois et al. 2005). Myotonic dystrophy type 1 (DM1) is a neuromuscular disorder caused by a large unstable CTG expansion in the 3′ UTR of the DMPK gene (Brook et al. 1992). Mutant transcripts (mt) harboring the large CUG repeats are fully transcribed and polyadenylated but are not exported from the nucleus (Davis et al. 1997). We have observed that lentivirus-delivered shRNAs targeting DMPK can simultaneously reduce both nuclear-retained mt DMPK, as well as cytoplasmically exported wild-type DMPK mRNA (Langlois et al. 2005). In addition to our data, a nuclear RNAi pathway is suggested by studies demonstrating that nuclear-retained transcripts, such as those transcribed from the intergenic region of the β-globin gene cluster, are RNAi substrates (Haussecker and Proudfoot 2005), and additional studies indicating that siRNAs may colocalize with their targets in either the cytoplasm or nucleus (Berezhna et al. 2006). The findings of Ago1 and Ago2 association with nuclear proteins (Hock et al. 2007), including proteins involved in cell cycle regulation (Stoica et al. 2006), suggest that the machinery for post-transcriptional RNAi can localize in the nucleus. It is intriguing to speculate that the role for RNAi in the nucleus may differ from that in the cytoplasm, and a nuclear RNAi pathway in human cells might provide a link between post-transcriptional and transcriptional gene silencing mediated by the RNAi machinery.
MATERIALS AND METHODS
Construction of shRNAs
For rapid cloning of shRNAs into the pTZU6+1 DNA vector (pU6 sh II) two primers for PCR amplification were used: The 5′ primer is complementary to sequences of the 5′ end of the U6 promoter, and the 3′ primer contains sequences complementary to the 3′ end of the U6 promoter as well as sequences complementary to the shRNA, and six As. A percentage of the resulting Taq polymerase PCR products have a 3′ A overhang that can be ligated into the pCR2.1 TA cloning vector (Invitrogen) containing a 3′ T overhang. Clones containing the insertion were readily identified as white colonies on X-gal plates. The shRNA-containing plasmids were confirmed by DNA sequencing. The controls for each shRNA contain four-point mutations in the middle of the antisense sequence.
For cloning the shRNAs into the VA1 gene (pVA1 sh II), the small hairpin RNA targeting site II of HIV-1 rev was constructed by enzymatic filling in one of two DNA oligonucleotides with overlapping 3′ termini. ShRNA sequences containing flanking BstEII restriction enzyme sites were prepared from two synthetic oligonucleotides, which share 12 bases of complementary sequence at their 3′ end. The double-stranded DNA sequences were amplified using several rounds of PCR and cloned into the unique BstEII site of pVA1 (Fig. 1A).
Cell culture, transfection, and ponasterone induction conditions
293/EcR cells were grown, transfected, and treated with Ponasteron A as described previously (Lee et al. 2002).
Northern hybridization
RNA sample preparation from 293/EcR cells transiently cotransfected with target and siRNA expression vectors or control constructs and Northern blotting were performed as described previously (Lee et al. 2002). Quantification of rev–eGFP mRNAs was normalized to GAPDH using QuantamOne (Bio-Rad) in α- phosphor images, and was based on two independent experiments.
Nuclear and cytoplasmic fractions
293/EcR cells were separated into nuclear and cytoplasmic fractions using PARIS (Ambion). The nuclear and cytoplasmic RNAs were used for Northern hybridization assay as described previously (Lee et al. 2002; Langlois et al. 2005; Kim et al. 2006).
Quantitative PCR for interferon response
Quantitative PCR was performed using a BIORAD iCycler with an iQ SYBR green PCR supermix (BIORAD no. 170-8880) for GAPDH and OAS1. OAS1 values were normalized to GAPDH. Primers used for GAPDH amplification were 5′-CATTGACCTCAACTACATG-3′ and 5′-TCTCCATGGTGGTGAAGAC-3′ and for OAS1 were 5′-CGAGGGAGCATGAAAACACATTT-3′ and 5′-GCAGAGTTGCTGGTAGTTTATGAC-3′.
Fluorescent in situ hybridization (FISH)
Cells were grown on individual circular gelatin-coated coverslips in six-well tissue culture plates and fixed with 4% formaldehyde as described (http://singerlab.aecom.yu.edu/protocols). 21-mer sense II and 50-mer antisense VA1 DNA-modified oligomers were synthesized in the City of Hope Oligonucleotide DNA synthesis facility. These probes incorporate modified thymidines that were labeled by Cy3 (Amersham) or Oregon Green (Molecular Probes). Coverslips were mounted upside down on a slideglass in an anti-fading solution consisting of nine parts glycerol, one part 1 M Tris-HCl, pH 7.5, 2% 1,4-Diaza-bicyclo-{2,2,2}-octane (Sigma) and DAPI (75 ng/μL) (Sigma).
Microscopy and imaging
Initial observations of Cy3 and DAPI fluorescence were made using wide field microscopy (Olympus AX70). Confocal image stacks were obtained using a Zeiss LSM5 inverted or upright microscope. Cy3 fluorescence was imaged using a 543 HeNe laser and a LP560 emmision filter. DAPI was excited with a TiSaphire 2 photon laser (790 nm) and fluorescence was observed using a LP560 emmision filter. In most cases the spacing of individual confocal sections was set at ½ airy disc overlap between adjacent sections. Maximum intensity projections were generated using the LSM 5 software. Amira (V3) was used to construct 3D surface and voxel images from the 2D confocal stack data. The 3D models were then rotated into favorable orientations and particular nuclear surfaces were made partially transparent or removed using display functions available in Amira.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank L. Brown for help with FACS analysis and M. Lee in the microscope facility for aid in the use of the two-photon Confocal microscope. We also thank M. Miyagishi (University of Tokyo) for Renilla constructs. This work was supported by grants from GSK and MDA to N.S.L, a COH graduate school internal fellowship to D.H.K., and grants from the NIH (NIAID and NHLB) to J.J.R.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1014008.
REFERENCES
- Berezhna S.Y., Supekova L., Supek F., Schultz P.G., Deniz A.A. siRNA in human cells selectively localizes to target RNA sites. Proc. Natl. Acad. Sci. 2006;103:7682–7687. doi: 10.1073/pnas.0600148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- Cagnon L., Rossi J.J. Downregulation of the CCR5 beta-chemokine receptor and inhibition of HIV-1 infection by stable VA1-ribozyme chimeric transcripts. Antisense Nucleic Acid Drug Dev. 2000;10:251–261. doi: 10.1089/108729000421439. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., van Wezel R., Stanley J., Hong Y. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 2003;77:7026–7033. doi: 10.1128/JVI.77.12.7026-7033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley N.R., Labbe J.C., Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Gwizdek C., Ossareh-Nazari B., Brownawell A.M., Doglio A., Bertrand E., Macara I.G., Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 2003;278:5505–5508. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- Haussecker D., Proudfoot N.J. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol. Cell. Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M., Urlaub H., Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Langlois M.A., Boniface C., Wang G., Alluin J., Salvaterra P.M., Puymirat J., Rossi J.J., Lee N.S. Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J. Biol. Chem. 2005;280:16949–16954. doi: 10.1074/jbc.M501591200. [DOI] [PubMed] [Google Scholar]
- Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Marcon E., Babak T., Chua G., Hughes T., Moens P.B. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Miyagishi M., Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Hannon G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J.C., Zhang F., Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica C., Carmichael J.B., Parker H., Pare J., Hobman T.C. Interactions between the RNA interference effector protein Ago1 and 14-3-3 proteins: Consequences for cell cycle progression. J. Biol. Chem. 2006;281:37646–37651. doi: 10.1074/jbc.M604476200. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans . Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]