Abstract
Group II introns are a major class of ribozymes found in bacteria, mitochondria, and plastids. Many introns contain reverse transcriptase open reading frames (ORFs) that confer mobility to the introns and allow them to persist as selfish DNAs. Here, we report an updated compilation of group II introns in Eubacteria and Archaea comprising 234 introns. One new phylogenetic class is identified, as well as several specialized lineages. In addition, we undertake a detailed search for ORF-less group II introns in bacterial genomes in order to find undiscovered introns that either entirely lack an ORF or encode a novel ORF. Unlike organellar group II introns, we find only a handful of ORF-less introns in bacteria, suggesting that if a substantial number exist, they must be divergent from known introns. Together, these results highlight the retroelement character of bacterial group II introns, and suggest that their long-term survival is dependent upon retromobility.
Keywords: reverse transcriptase, retroelement, intron-encoded protein, evolution
INTRODUCTION
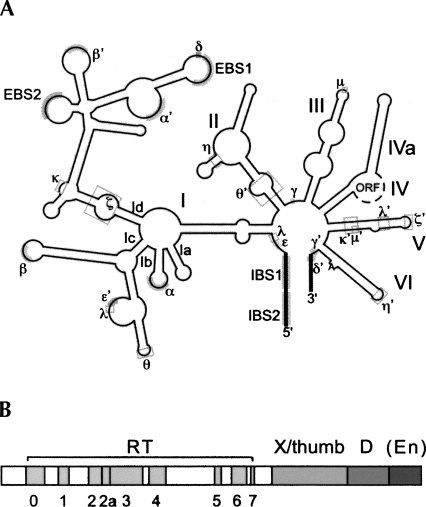
Group II introns are ribozymes and retroelements found in the genomes of organelles and bacteria (i.e., Eubacteria and Archaea) (Michel and Ferat 1995; Dai and Zimmerly 2003; Lambowitz and Zimmerly 2004; Toro et al. 2007). Group II introns in bacteria primarily act as retroelements and consist of a catalytic RNA structure and an intron-encoded protein (IEP). In contrast, introns in mitochondria and plastids frequently lack the IEP and are mobile. The RNA structures of group II introns are comprised of six domains (DI–DVI) that fold into a conserved secondary structure (Fig. 1A). The RNAs are capable of catalyzing intron splicing, although in vivo this activity is usually dependent on protein assistance. The IEPs are multifunctional and encode domains for reverse transcriptase activity (RT), splicing/maturase function (X/thumb), DNA binding (D), and sometimes endonuclease activity (En) (Fig. 1B).
FIGURE 1.
Structure of group II intron RNAs and ORFs. (A) Secondary structure of a group IIA intron RNA showing exons (thick black lines) and ORF (dotted black line). Roman letters (sometimes followed by a lowercase letter) indicate the six RNA domains and their subdomains. EBS and IBS refer to the exon and intron binding sites, respectively. Tertiary interactions are shown with Greek letters and either thick gray lines (Watson–Crick base pairs) or gray boxes (nonpairing interactions). (B) Intron-encoded ORF structure, consisting of the RT domain (subdomains 0–7), maturase (X/thumb), DNA-binding (D), and endonuclease (En) domains. The endonuclease domain is absent in some group II introns, as symbolized by parentheses in the figure.
Mobility of group II introns occurs by a well-defined mechanism that is carried out by a ribonucleoprotein (RNP) composed of the intron lariat and two subunits of the IEP (Lambowitz and Zimmerly 2004). The RNP recognizes a DNA target site, and the intron RNA reverse splices into the top strand of the duplex. The IEP cleaves the bottom strand with the En domain and reverse transcribes the integrated intron using the RT domain (Lambowitz and Zimmerly 2004). Many IEPs in bacteria lack the En domain, and mobility of these introns requires a primer provided by the DNA replication fork (Ichiyanagi et al. 2002; Zhong and Lambowitz 2003). In general, mobility of group II introns is highly site-specific to a ∼35-base-pair (bp) target and is known as retrohoming. At a much lower frequency, introns are able to invade noncognate sites through retrotransposition (Cousineau et al. 2000; Martínez-Abarca and Toro 2000b; Ichiyanagi et al. 2002). A third substrate specificity is seen for a phylogenetic subclass (class C, below) in which the introns insert after diverse intrinsic terminator motifs (Granlund et al. 2001; Dai and Zimmerly 2002; Robart et al. 2007).
The IEPs of bacterial group II introns are more diverse phylogenetically than those in organelles, although organellar introns can be more degenerate in RNA structure (e.g., Choquet et al. 1988; Copertino et al. 1994). Based on phylogenies constructed from ORF sequences, several classes of group II introns have been established (chloroplast-like [CL], mitochondrial-like [ML], and bacterial classes A, B, C, D, E) (Zimmerly et al. 2001; Toro et al. 2002). While introns from all phylogenetic classes are present in bacteria, only classes CL and ML are found in organelles. In addition, each ORF phylogenetic class is associated with a distinct RNA secondary structure (Toor et al. 2001; Ferat et al. 2003; Toro 2003). For example, classes ML and CL have IIA and IIB secondary structures, respectively (Michel et al. 1989), while class C introns have IIC structures that exhibit a number of unique features (Martínez-Abarca and Toro 2000a; Granlund et al. 2001; Toor et al. 2001; Robart et al. 2007).
Based on the general concordance of RNA structural types and ORF phylogenetic classes, as well as the phylogenetic distribution of intron types, we previously proposed an evolutionary scenario called the “retroelement ancestor hypothesis.” This hypothesis predicts that group II ribozymes differentiated into their known structural forms (IIA, IIB, IIC) as components of retroelements in bacteria and then migrated to organelles where many types of degeneration occurred (Toor et al. 2001). A similar idea was proposed by Fontaine et al. (1997). Alternative scenarios are also possible, such as an organellar origin of group II introns or multiple introductions of RT ORFs into different ribozyme subtypes, but these appear less parsimonious.
Given the diversity of known group II introns, continued exploration of sequence data is likely to reveal additional structural variations and specialized lineages. One limitation to fully understanding group II introns in bacteria is that ORF-less introns are very difficult to identify, due to the lack of sequence conservation outside of the intron-encoded protein. In the past, we have identified group II introns on the basis of their RT ORFs. While this method is quite successful in finding diverse introns, it obviously would miss those that either do not encode RT ORFs or encode ORFs of a different family (e.g., a homing endonuclease, as in Toor and Zimmerly [2002]). This methodological weakness is exacerbated by the fact that bacterial group II introns frequently inhabit intergenic regions and mobile DNAs (Dai and Zimmerly 2002; Klein and Dunny 2002). It is therefore important to conduct a general search for group II introns in bacterial genomes based on RNA structural features, independently of the ORF.
In this paper, we update our knowledge of group II intron diversity in Eubacteria and Archaea. We expand our previous compilation (Dai and Zimmerly 2002) by an additional 195 full-length introns, and identify one new ORF phylogenetic class that has a characteristic RNA structure. In addition, we highlight a number of specialized intron lineages that appear to have adapted unusual features for either splicing or mobility. Finally, the results of our search suggest that there is not a significant number of overlooked ORF-less group II introns in bacteria, thus reinforcing the retroelement nature of group II introns in bacteria.
RESULTS AND DISCUSSION
Identification and classification of group II introns in bacteria
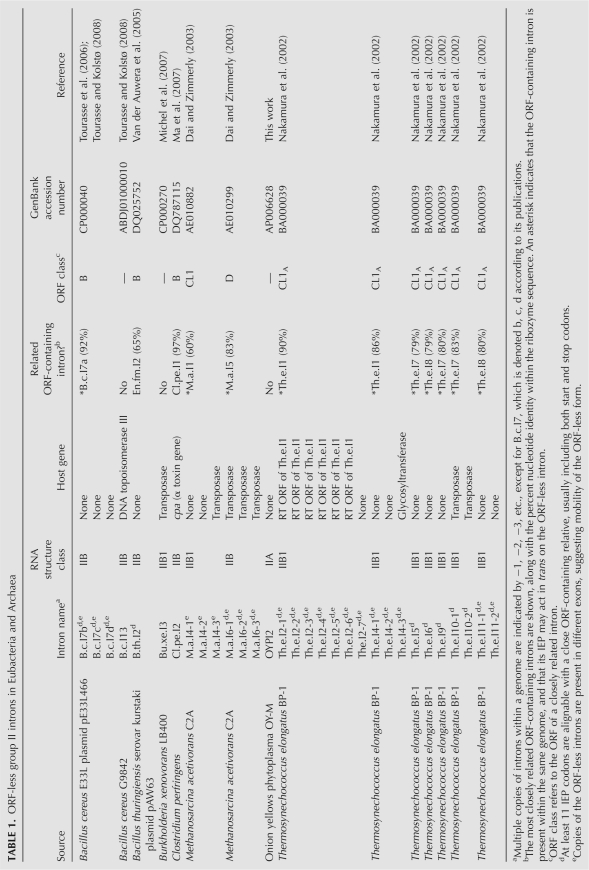
Group II introns were identified in GenBank by a strategy previously described (Dai and Zimmerly 2002), in which RT ORFs are first located by BLAST searches and then the surrounding intron RNAs are identified and folded (see Materials and Methods). Using this method, 180 new ORF-containing introns were identified and added to our database (http://www.fw.ucalgary.ca/group2introns/), making a collection of 219 full-length ORF-containing group II introns in bacteria. The database also includes 15 ORF-less introns (see Table 1 and below) and a selection of organellar ORF-encoding introns. This collection of raw information forms the basis for the analyses presented here.
TABLE 1.
ORF-less group II introns in Eubacteria and Archaea
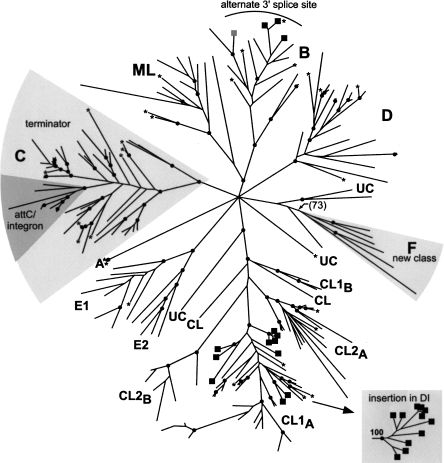
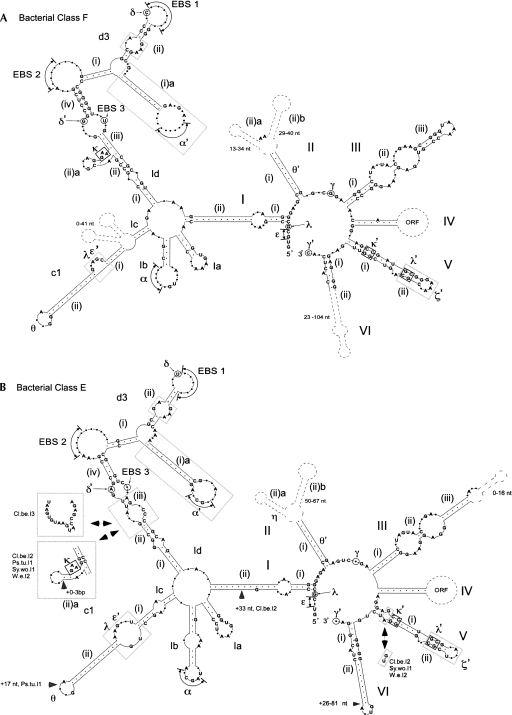
A phylogenetic tree of the bacterial IEPs in our data set (Fig. 2) shows each previously established ORF class (A, B, C, D, E, CL, ML) to be a monophyletic group (Zimmerly et al. 2001; Toro et al. 2002) with one additional class (F). Class F is defined conservatively as a small group that is corroborated by a clearly shared secondary structure (Fig. 3A). Although there is not bootstrap support for class F in Figure 2, a bootstrap value of 73% is obtained when the unclassified introns basal to this group are excluded (see Fig. 2 legend). The RNA secondary structures of class F are type IIB (Fig. 3A), and the characteristic features include a unique ε′ motif, a bulge loop in subdomain Id3(ii) (shared with classes B, C, and E), the presence of α′ at the end of a stem–loop (shared with classes D and E), and a DV(ii) stem–loop of 4 bp with a 5-nucleotide (nt) loop (shared with class E).
FIGURE 2.
Phylogenetic tree of group II intron-encoded proteins. ORFs were subjected to phylogenetic analyses using RAxML (see Materials and Methods). Asterisks indicate introns present in our previous compilation (Dai and Zimmerly 2002). The major groups are labeled classes A-F, mitochondrial-like (ML), and chloroplast-like (CL), with five ORFs remaining unclassified (UC). The proposed new class of introns (F) and specialized lineages (as discussed in the text) are shown with gray sectors or squares at the tips. The inset “insertion in DI” indicates monophyly of the 11 introns in separate analysis of CL1 introns containing a greater number of characters (see Materials and Methods). Black dots indicate nodes with bootstrap support of ≥70%. Nodes uniting multiple classes were collapsed if the bootstrap value was <50%. Although the node uniting ML and B has a high bootstrap value, it is not supported in other analyses (not shown) and should be interpreted cautiously. The number in parentheses at the node uniting class F is the bootstrap value when the analysis was repeated excluding the three unclassified introns at its base.
FIGURE 3.
Secondary structure models for classes E and F. Consensus structures are based on 80% identity, with helices accurate to ±1 bp and single strands to ±2 nt, and with greater deviations indicated. Small and large dots represent base pairs and unpaired nucleotides, respectively. Gray boxes indicate features that are characteristic of the class. (A) The consensus structure for class F is based on the following introns: D.a.I1 (CP000089), Ge.ur.I1 (NZ_AAON01000001), My.xa.I1 (CP000113), Ni.ha.I1 (CP000320), Pe.th.I2 (AP009389), So.us.I2 (CP000473). (B) The consensus structure for class E is based on the following introns: E1: A.v.I5 (NZ_AAAD01000084B), B.j.I2-1 (BA000040), Ch.ph.I2-1 (CP000492), P.l.I1 (BX571862), Pr.ae.I1 (NZ_AAIJ01000002), S.ma.I1 (BX664015), Sr.me.I4 (AJ496462), UB.I1 (AY691909); E2: Cl.be.I2 (AALO01000032), Cl.be.I3 (AALO01000024), Ps.tu.I1 (AAOH01000003), Sy.wo.I1 (NZ_AAJG01000003), W.e.I2 (AE017196).
The updated compilation also shows new subdivisions within previously identified phylogenetic classes. Class E is split into two clades, E1 and E2, which might be considered separate classes based on phylogenetic analysis alone; however, their RNA structures are nearly identical (Fig. 3B), and so we consider them a single class. There are two significant differences between their secondary structures. E1 introns appear to lack the κ–κ′ interaction and have a conserved G–A mispair in DV, whereas most E2 introns possess a typical κ–κ′ motif and have a G–U wobble base pair in DV. The κ–κ′ interaction is important to the group II RNA structure, because along with ζ–ζ′ it forms a docking interaction between DI and DV. For the aI5γ intron, this interaction is a “folding control element” for the entire intron (Waldsich and Pyle 2007). The absence of this motif in the E1 subclass suggests that the RNA either has an equivalent or compensating RNA interaction, or that a different folding pathway is used.
In three other classes (B, C, and ML), basally branching introns are identified that are phylogenetically distinct and could be considered independent classes. However, the existence of shared RNA structural features and a shared insertional preference in the case of class C suggests that the creation of additional classes is not warranted.
Finally, it is notable that the ORFs of classes CL1 and CL2 are not monophyletic. Instead, the CL class is split into four clades (CL1A, CL1B, CL2A, CL2B) (Fig. 2; D.M. Simon and S. Zimmerly, unpubl.), whereas their RNA structures fall into two discrete classes of IIB1 (CL1) and IIB2 (CL2) (Toor et al. 2001). Another example of potential conflict between the ORF and RNA classifications is found for classes E and F; their RNA structures are similar in overall organization (Fig. 3, cf. A and B), yet their ORFs do not appear to be closely related (Fig. 2). Both of these examples may be either exceptions to the general pattern of coevolution between ribozyme and IEP, or examples of convergent evolution of RNA structural features (see Conclusions below).
Phylogenetic distribution of introns
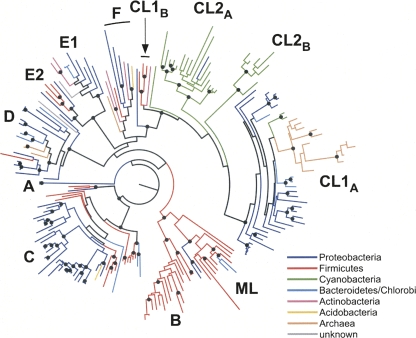
To gain insight into intron dispersal, we examined the distribution of group II introns in Eubacteria and Archaea. Overall, group II introns are found in six subdivisions of Eubacteria (Acidobacteria, Actinobacteria, Bacteroidetes/Chlorobi, Cyanobacteria, Firmicutes, and Proteobacteria) and one of Archaea (Euryarchaeota). The groups for which introns have not yet been found are also the groups with the fewest number of complete genome sequences. Likewise, the largest proportions of introns are found in the groups with the most completed genome sequences (i.e., Proteobacteria, Firmicutes).
The distribution of group II introns in bacteria can be visualized by mapping the host bacterial groups onto an intron phylogeny (Fig. 4). Because the analysis includes all bacterial introns, relatively few characters (230 amino acids) can be aligned reliably across classes, resulting in a tree of low resolution. Nonetheless, a few broad inferences are possible. We find that class B introns are restricted to Firmicutes, as are most of the ML introns. Likewise, the majority of both class C and CL1A introns are found in Proteobacteria. Such clustering would be expected if horizontal transfers of these introns are relatively infrequent among distantly related bacteria.
FIGURE 4.
Distribution of group II introns in bacteria. The intron-encoded ORF phylogeny is presented as a radial phylogram with the groups of host bacteria coded by color. Black dots indicate nodes with ≥70% bootstrap support. The phylogeny is identical to the one shown in Figure 2.
A second general set of observations can be made regarding the introduction of group II introns into specific bacterial lineages. It is clear that group II introns were introduced into Archaea a small number of times (three to four times based on this set of introns). Most archaeal introns are found in a single clade within CL1A, with an additional three introns in class D and one that is still unclassified. Similarly, cyanobacterial introns are restricted to CL2A and CL2B, and virtually all CL2B introns are cyanobacterial, suggesting limited long-distance horizontal transfers involving these introns.
It is notable that the majority of the cyanobacterial introns are found in only a few genomes, with five introns in Crocosphaera watsonii WH8501, four in the Nostoc plasmid pCC7120α, nine in Trichodesmium erythraeum IMS101, and 28 in Thermosynechococcus elongatus BP-1. Cyanobacterial introns are distinctive because of their high copy number in some genomes, the presence of many introns in housekeeping genes (especially in T. erythraeum), and the frequency of ORF-less introns (below and Table 1). Because many cyanobacterial genomes lack group II introns altogether, the pattern suggests that intragenomic mobility and subsequent retention have played a large role in the evolution of group II introns in Cyanobacteria. In contrast, introns of other bacterial groups are widely distributed throughout the tree, indicating multiple introductions of different intron types into these bacterial lineages.
Together, these data clearly indicate that group II introns have undergone extensive horizontal transfers, but also suggest that there are barriers to their spread. Such barriers may include either inherently low frequencies of horizontal transfer between certain groups of bacteria, or adaptations of the intron retroelements to specific groups of bacteria.
Genomic locations of introns
We also examined the distribution of group II introns within genomes. Overall, slightly more than half of the introns in our database are found in predicted intergenic regions, with approximately half of these belonging to class C. This is expected as class C introns target transcriptional terminators (Granlund et al. 2001; Dai and Zimmerly 2002; Robart et al. 2007). The remainder of introns are found in mobile DNA ORFs (19%), hypothetical genes (18%), or housekeeping genes (8%). Putatively intergenic introns are found in nearly every group II intron class, with the exception of the ML class. Likewise, introns in mobile DNA ORFs are present in a large number of classes (B, D, E1, F, CL1A, CL2B, and ML), but are particularly concentrated in class D. The few introns in housekeeping genes are similarly dispersed throughout the intron classes, being found in classes B, E2, CL1A, CL2A, ML, and an additional intron (S.ag.I2 from Streptococcus agalactiae) that is currently unclassified. We find no evidence in our data set of long-term vertical inheritance of introns in housekeeping genes, such as is observed for group II introns in plant organelles (Kelchner 2002). It seems likely that selective pressure inhibits the insertion and/or maintenance of introns in these genes in bacteria.
Specialized lineages
It has been observed that some lineages of group II introns have unique properties. The most well-established example is class C, whose introns insert directly after terminator motifs (Granlund et al. 2001; Dai and Zimmerly 2002; Robart et al. 2007). Interestingly, within class C is a lineage of eight introns with a different insertion pattern: they insert into the inverted repeats of attC sites of integrons rather than the inverted repeats of terminators (Fig. 2; Centrón and Roy 2002; Quiroga et al. 2008). It is perplexing that these introns insert in the orientation opposite to transcription of the integron unit, which would seem to limit mobility of the intron. Nevertheless, the lineage appears to be adapted to survive with this feature.
Another reported example of specialization is a lineage within CL1A, which has a large insertion (∼300–600 nt) near the 5′ end of its RNA structure. These introns are located near the boundaries of genes, either intergenically or at the very beginning or end of genes (Michel et al. 2007). The first reported example was an intron interrupting the groEL gene of Azotobacter vinelandii (Adamidi et al. 2003; Ferat et al. 2003), but the group has now grown to over a dozen members (Fig. 2; Michel et al. 2007). Although the introns are not grouped together in the tree shown in Figure 2, monophyly was confirmed by constructing a tree of CL1 introns using a larger number of characters (Fig. 2, gray inset; Michel et al. 2007). It is unclear how this lineage of introns targets gene boundaries, or how expression of the surrounding genes is affected by the insertion of this highly unusual type of intron. A possible function has been speculated to involve regulation of splicing by competing RNA pairings at the 5′ terminus of the intron (Michel et al. 2007).
Three introns within class B are known to have altered 3′ splice sites. In two instances the splice site is shifted only modestly, by one nucleotide from the expected position (B.a.I2, B.c.I2). However, two other splicing events in vivo (B.c.I4, B.a.I2) involve more substantial shifts of 56 nt and 4 nt downstream from the expected sites, respectively (Robart et al. 2004; Tourasse et al. 2005; Stabell et al. 2007). These three introns form a monophyletic group (B.a.I2, B.c.I2, B.c.I4) and represent a lineage that appears to have flexibility in specifying the 3′ splice sites (black squares within class B in Fig. 2). Other class B introns appear to have a standard 3′ splice site location, with the exception of B.th.I1 (gray square within class B in Fig. 2), which is also likely to splice one nucleotide downstream of the usual position, based on potential γ–γ′ and IBS3–EBS3 pairings.
A final observation is that in the subclass E1, seven of eight IEPs have start codons in domain II or domain III rather than in DIV, the exception being A.v.I5 in Azotobacter vinelandii. The start site is notable because for the Ll.LtrB intron of Lactococcus lactis, the start codon lies within the high-affinity IEP binding site, allowing for feedback inhibition of IEP translation (Wank et al. 1999). It is possible that feedback regulation may not be universal to all group II IEPs, or that the IEP binding site may be in a different location for some introns.
ORF-less group II introns in bacteria
To address the issue of whether there are undetected group II introns in bacteria that do not encode a reverse transcriptase ORF, we searched for introns based on RNA structural criteria. The program RNAMotif (Macke et al. 2001) was used to screen completed bacterial genome sequences, using descriptors of combined sequence and secondary structure information. The descriptors were derived from consensus structures of DV for each phylogenetic class of intron. Descriptors were tested at different stringencies, and ultimately a set of eight descriptors was chosen (Supplemental Fig. 1). Using these descriptors, RNAMotif was able to correctly identify all 17 introns in seven sample genomes (combined size of 31.3 Mb), while generating only two readily identifiable false positives (see Materials and Methods).
The descriptors were then used to individually scan 225 complete bacterial genomes (735 Mb) (Supplemental Fig. 1; Supplemental Table 1; see Materials and Methods). The screen yielded 224 unique hits, which were evaluated as potential DV motifs using criteria outlined in the Materials and Methods. In total, 172 hits were judged to represent ORF-containing introns that are either full-length or truncated, or known ORF-less introns, while 46 were considered false positives, based on genomic locations, quality of the hits, and RNA foldings (about one false positive per 16 Mb). Six hits were considered to be new candidate ORF-less introns; however, we were unable to fold four of them into complete RNA secondary structures, and these were concluded to be likely truncated forms containing DV and DVI structures (Supplemental Fig. 2).
The remaining two introns were folded into complete secondary structures. The first intron, OYPI2, is found in the Onion yellows phytoplasma strain OY-M (AP006628), and its RNA secondary structure is IIA, similar to the RNA structures of class ML IEPs (Supplemental Fig. 3). This genome also contains an ML intron that encodes an ORF (OYPI1), raising the possibility that the OYPI1 ORF may act in trans on OYPI2. A precedent exists for a maturase acting in trans on other group II introns in a genome, in the cyanobacterium Trichodesmium (Meng et al. 2005). In the case of the OYP introns, the introns are not closely related to each other, and OYPI2 does not appear to have derived directly from OYPI1.
The second intron (O.i.I2) was found in Oceanobacillus iheyensis HTE831 (NC_004193), and can also be folded into a IIA structure. Although the intron initially appeared to be ORF-less, a closer examination revealed a highly degenerated ORF in domain IV, which is partially alignable with other IEPs for the entire length of the RT and X domains, yet it lacks many conserved RT motifs (e.g., YADD). It seems likely that the intron-encoded protein in O.i.I2 no longer possesses RT activity, but it may continue to have a maturase function, which is suggested by the maintenance of the reading frame. Interestingly, it is one of the few introns that interrupt a housekeeping gene, radC (involved in DNA repair), thus giving a rationale for retention of splicing function. The closest relative of O.i.I2 is B.c.I9, which also interrupts radC but has a standard RT ORF. To our knowledge, O.i.I2 is the first bacterial group II intron known to encode a degenerated ORF, although there are many examples in mitochondria and plastids (Michel and Ferat 1995; Zimmerly et al. 2001; Barkan 2004).
It thus appears that ORF-less introns in bacteria are indeed rare. Our results indicate that in 225 genomes, which contain 163 ORF-encoding group II introns and nine known ORF-less introns, there is only one new truly ORF-less intron (OYPI2). Table 1 lists additional known ORF-less group II introns. It is striking that 10 of the 15 known ORF-less introns have small but detectable remnants of RTs within the intron RNAs, usually containing both the start and stop codons for IEPs of related introns. Most of these introns reside in genomes that encode related full-length introns, whose IEPs may interact with the ORF-less introns in trans (Meng et al. 2005). In addition, six of the 15 ORF-less introns have evidence for mobility, as suggested by the presence of multiple copies of the ORF-less intron in different exons (Table 1 and footnote). Nevertheless, overall there is a dearth of ORF-less group II introns in bacteria, with only 6% of the introns being ORF-less. The introns are suggested to have lost the ORFs relatively recently, due to the presence of ORF remnants. This combined with the limited number of ORF-less introns suggest that ORF-less forms do not survive independently for long periods in bacteria. We hypothesize that this reflects selective pressure on bacterial introns to maintain their mobility, in sharp contrast with group II introns in organelles, where some introns have evolved with their host gene for several hundred million years (e.g., vertical inheritance of chloroplast introns in plants) (Kelchner 2002).
Conclusions
This compilation of group II introns has increased the number of known introns in bacteria, and examination of this data set in a phylogenetic context has allowed a number of conclusions to be made. Specifically, we identified a novel phylogenetic class of group II introns (class F) and distinguished two RNA subtypes within class E. We have increased our knowledge of specialized lineages, bringing to five the number of bacterial forms that vary in splicing and/or mobility properties compared with canonical group II introns. These include introns that insert at terminators, attC sites, or gene boundaries, introns that have altered 3′ splice sites, and an intron encoding an ORF with degenerated RT motifs. In addition, the data set provides insight into underlying patterns of intron dispersal. While horizontal transfer is clearly rampant, there appear to be barriers to horizontal transfers involving distantly related bacteria. Finally, the results of a thorough search for ORF-less introns suggest that this form is rare in bacteria. Thus, if a significant number of ORF-less group II introns exist in bacteria, they must have DV RNA structures different from known introns.
Together, these data support the “retroelement ancestor hypothesis,” which posits that all extant group II introns are derived from a bacterial retroelement (Toor et al. 2001). We find no evidence for a set of diverse, ancient ORF-less introns into which RTs might have inserted multiple times to generate independent lineages of group II intron retroelements. Rather, it appears that ORF-less introns do not persist independently over long time periods in bacteria. We did identify apparent exceptions to coevolution of the ribozyme and IEP, which is a principle that underlies our evolutionary hypothesis. These exceptions may be attributed to either convergent evolution of a functionally constrained RNA structure, or reshuffling of RNAs and IEPs at the DNA level. The latter possibility is supported by the existence of twintrons in bacteria, in which one group II intron has inserted into another (Nakamura et al. 2002; Dai and Zimmerly 2003), thus offering a plausible mechanism for ORF reshuffling. However, the number of obvious exceptions to coevolution remains low, leaving coevolution as the predominant trend. Taken together, the simplest explanation that accounts for the known structural varieties of group II introns and their phylogenetic distribution continues to be descent from a retroelement ancestor in bacteria. It remains unclear how the reverse transcriptase and ribozyme initially came together to form the putative ancestral group II intron retroelement, but it seems most likely that the current forms of group II ribozymes (IIA, IIB, and IIC) differentiated after that event.
MATERIALS AND METHODS
Identification and analysis of group II introns
Group II introns were identified in GenBank by BLAST searches based on the reverse transcriptase ORF, and then the surrounding RNA was identified and folded into a secondary structure (Dai and Zimmerly 2002). Secondary structures were determined using MFOLD (Zuker 2003) and manual folding constraints, guided by agreement with consensus structures and/or consistency with related intron sequences (Toor et al. 2001). Intron boundaries were determined by RNA folding in combination with examination of potential exon junctions.
Search for ORF-less introns
RNAMotif (v1.7.1) (Macke et al. 2001) was used to search for potential group II introns in bacterial genomes. First, consensus structures were made for the highly conserved and presumably catalytic domain V motif for each ORF phylogenetic class, based on a variety of RNA secondary structures (Supplemental Fig. 1). A set of five to six descriptors was made from each consensus structure, allowing for different degrees of deviation from the consensus. The purpose of this phase of the screen was to identify optimal descriptors that allow the greatest deviation from the consensus without producing an unmanageable number of false hits. The sets of descriptors were evaluated by scanning seven bacterial genomes, which together contain 19 introns representing all seven previously described classes (Escherichia coli pB171, Enterococcus faecalis V583, Bacillus halodurans C-125, Clostridium acetobutylicum ATCC 824, Bacteroides thetaiotaomicron VPI-5482, Bradyrhizobium japonicum USDA 110, Gloeobacter violaceus PCC 7421). One descriptor per class was chosen for B, C, D, E, CL1, and CL2, and two descriptors were used for the ML class (Supplemental Fig. 1). Excluding class C, which has a unique DV length, eight of 11 introns were identified by multiple descriptors, showing redundancy in the search. Two false positives were obtained and readily identified because each lies within the coding sequence of a conserved gene. In an additional negative control, Caenorhabditis elegans chromosome 5 (21 Mb) and Arabidopsis thaliana chromosome 1 (30 Mb) were searched with the descriptors and did not yield any hits. This is expected because group II introns are not known to occur in eukaryotic nuclear DNA, with the exception of recently transferred organellar remnants (Knoop and Brennicke 1994).
A selection of 225 out of >700 sequenced genomes was scanned with these descriptors to determine the frequency of ORF-less group II introns in bacteria. Potential DV motifs identified were then analyzed individually to determine if they were: (1) full-length ORF-encoding introns; (2) truncations of ORF-encoding introns; (3) ORF-less introns; or (4) false positives. ORF-encoding introns, either full-length or truncated, were identified by searching the sequence upstream of the putative DV for an RT ORF. The remaining introns were examined for their genomic location to determine whether they were inside an annotated ORF, or whether sufficient intergenic sequence (>400 bp) was present to accommodate a complete intron sequence. Two additional criteria were the quality of the DV hit and whether it was obtained with more than one descriptor. If there were multiple sequence deviations and/or mispairs in the putative DV, and if it was found with only one descriptor, then the hit was considered less likely to be a group II intron. A final criterion was based on whether the flanking sequence could be folded into a group II intron secondary structure. Potential DVI motifs were easily evaluated by folding the sequences into the simple DVI motif. If a reasonable DVI motif was found, then the entire flanking sequence was folded to obtain a complete group II intron structure.
Phylogenetic analyses
The intron-encoded ORFs were manually aligned to a pre-existing alignment (Zimmerly et al. 2001) using BioEdit v7.0.5.2 (Hall 1999). This alignment included all RT domains, as well as domain X, but not domains D or En because these regions cannot be reliably aligned across classes. After unambiguously aligned regions were removed, 230 amino acids representing 194 unique sequences were used in the analyses. The final alignment contains a small number of gaps (<2% of all characters) in regions that were unalignable for a few introns. Model choice was aided by the Akaike Information Criterion (AIC) as implemented in ProtTest v10.2 (Posada and Buckley 2004; Abascal et al. 2005). An unrooted tree and 100 bootstraps were produced using the program RAxML v.4.0.0 with the model RtREV+Γ+F (Dimmic et al. 2002; Stamatakis et al. 2005). The same procedure was followed to make a tree of only CL1 introns, except in this case 397 amino acids could be unambiguously aligned.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by CIHR grant MOP-49457 and an Alberta Ingenuity fellowship to D.M.S. Salary support for S.Z. was from the Alberta Heritage Foundation for Medical Research. We thank Nicolas Tourasse and Anne-Brit Kolstø (University of Oslo) for communicating data prior to publication.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1056108.
REFERENCES
- Abascal F., Zardoya R., Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adamidi C., Fedorova O., Pyle A.M. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- Barkan A. Intron splicing in plant organelles. In: Daniell H., Chase C., editors. Molecular biology and biotechnology of plant organelles. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 281–308. [Google Scholar]
- Centrón D., Roy P.H. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Goldschmidt-Clermont M., Girard-Bascou J., Kück U., Bennoun P., Rochaix J.D. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell. 1988;52:903–913. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- Copertino D.W., Hall E.T., Van Hook F.W., Jenkins K.P., Hallick R.B. A group III twintron encoding a maturase-like gene excises through lariat intermediates. Nucleic Acids Res. 1994;22:1029–1036. doi: 10.1093/nar/22.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau B., Lawrence S., Smith D., Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- Dai L., Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: Evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–19. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic M.W., Rest J.S., Mindell D.P., Goldstein R.A. rtREV: An amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- Ferat J.L., Le Gouar M., Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- Fontaine J.M., Goux D., Kloareg B., Loiseaux-de Goër S. The reverse-transcriptase-like proteins encoded by group II introns in the mitochondrial genome of the brown alga Pylaiella littoralis belong to two different lineages which apparently coevolved with the group II ribosyme lineages. J. Mol. Evol. 1997;44:33–42. doi: 10.1007/pl00006119. [DOI] [PubMed] [Google Scholar]
- Granlund M., Michel F., Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B Streptococci. J. Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Ichiyanagi K., Beauregard A., Lawrence S., Smith D., Cousineau B., Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- Kelchner S.A. Group II introns as phylogenetic tools: Structure, function, and evolutionary constraints. Am. J. Bot. 2002;89:1651–1669. doi: 10.3732/ajb.89.10.1651. [DOI] [PubMed] [Google Scholar]
- Klein J.R., Dunny G.M. Bacterial group II introns and their association with mobile genetic elements. Front. Biosci. 2002;7:d1843–d1856. doi: 10.2741/klein1. [DOI] [PubMed] [Google Scholar]
- Knoop V., Brennicke A. Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J. Mol. Evol. 1994;39:144–150. doi: 10.1007/BF00163803. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Ma M., Ohtani K., Shimizu T., Misawa N. Detection of a group II intron without an open reading frame in the α-toxin gene of Clostridium perfringens isolated from a broiler chicken. J. Bacteriol. 2007;189:1633–1640. doi: 10.1128/JB.01210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abarca F., Toro N. Group II introns in the bacterial world. Mol. Microbiol. 2000a;38:917–926. doi: 10.1046/j.1365-2958.2000.02197.x. [DOI] [PubMed] [Google Scholar]
- Martínez-Abarca F., Toro N. RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res. 2000b;28:4397–4402. doi: 10.1093/nar/28.21.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Wang Y., Liu X.Q. An intron-encoded protein assists RNA splicing of multiple similar introns of different bacterial genes. J. Biol. Chem. 2005;280:35085–35088. doi: 10.1074/jbc.C500328200. [DOI] [PubMed] [Google Scholar]
- Michel F., Ferat J.L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns—A review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Michel F., Costa M., Doucet A.J., Ferat J.L. Specialized lineages of bacterial group II introns. Biochimie. 2007;89:542–553. doi: 10.1016/j.biochi.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kaneko T., Sato S., Ikeuchi M., Katoh H., Sasamoto S., Watanabe A., Iriguchi M., Kawashima K., Kimura T., et al. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002;9:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- Posada D., Buckley T.R. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Quiroga C., Roy P.H., Centrón D. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiol. 2008;154:1341–1353. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- Robart A.R., Montgomery N.K., Smith K.L., Zimmerly S. Principles of 3′ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA. 2004;10:854–862. doi: 10.1261/rna.5246804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart A.R., Seo W., Zimmerly S. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc. Natl. Acad. Sci. 2007;104:6620–6625. doi: 10.1073/pnas.0700561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell F.B., Tourasse N.J., Ravnum S., Kolstø A.B. Group II intron in Bacillus cereus has an unusual 3′ extension and splices 56 nucleotides downstream of the predicted site. Nucleic Acids Res. 2007;35:1612–1623. doi: 10.1093/nar/gkm031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Toor N., Zimmerly S. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA. 2002;8:1373–1377. doi: 10.1017/s1355838202023087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N., Hausner G., Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N. Bacteria and archaea group II introns: Additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- Toro N., Molina-Sánchez M.D., Fernández-López M. Identification and characterization of bacterial class E group II introns. Gene. 2002;299:245–250. doi: 10.1016/s0378-1119(02)01079-x. [DOI] [PubMed] [Google Scholar]
- Toro N., Jiménez-Zurdo J.I., García-Rodríguez F.M. Bacterial group II introns: Not just splicing. FEMS Microbiol. Rev. 2007;31:342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- Tourasse N.J., Kolstø A.B. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: Insights into their spread and evolution. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn372. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse N.J., Stabell F.B., Reiter L., Kolstø A.B. Unusual group II introns in bacteria of the Bacillus cereus group. J. Bacteriol. 2005;187:5437–5451. doi: 10.1128/JB.187.15.5437-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse N.J., Helgason E., Okstad O.A., Hegna I.K., Kolstø A.B. The Bacillus cereus group: Novel aspects of population structure and genome dynamics. J. Appl. Microbiol. 2006;101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G.A., Andrup L., Mahillon J. Conjugative plasmid pAW63 brings new insights into the genesis of the Bacillus anthracis virulence plasmid pXO2 and of the Bacillus thuringiensis plasmid pBT9727. BMC Genomics. 2005;6:103. doi: 10.1186/1471-2164-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C., Pyle A.M. A folding control element for tertiary collapse of a group II intron ribozyme. Nat. Struct. Mol. Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- Wank H., San Filippo J., Singh R.N., Matsuura M., Lambowitz A.M. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- Zhong J., Lambowitz A.M. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J. 2003;22:4555–4565. doi: 10.1093/emboj/cdg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S., Hausner G., Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]