Abstract
RNA interference (RNAi) is a gene-silencing mechanism by which a ribonucleoprotein complex, the RNA-induced silencing complex (RISC) and a double-stranded (ds) short-interfering RNA (siRNA), targets a complementary mRNA for site-specific cleavage and subsequent degradation. While longer dsRNA are endogenously processed into 21- to 24-nucleotide (nt) siRNAs or miRNAs to induce gene silencing, RNAi studies in human cells typically use synthetic 19- to 20-nt siRNA duplexes with 2-nt overhangs at the 3′-end of both strands. Here, we report that systematic synthesis and analysis of siRNAs with deletions at the passenger and/or guide strand revealed a short RNAi trigger, 16-nt siRNA, which induces potent RNAi in human cells. Our results indicate that the minimal requirement for dsRNA to trigger RNAi is an ∼42 Å A-form helix with ∼1.5 helical turns. The 16-nt siRNA more effectively knocked down mRNA and protein levels than 19-nt siRNA when targeting the endogenous CDK9 gene, suggesting that 16-nt siRNA is a more potent RNAi trigger. In vitro kinetic analysis of RNA-induced silencing complex (RISC) programmed in HeLa cells indicates that 16-nt siRNA has a higher RISC-loading capacity than 19-nt siRNA. These results suggest that RISC assembly and activation during RNAi does not necessarily require a 19-nt duplex siRNA and that 16-nt duplexes can be designed as more potent triggers to induce RNAi.
Keywords: RNAi, Kinetics of RISC, siRNA
INTRODUCTION
RNAi interference (RNAi) is an evolutionarily conserved process whereby double-stranded RNA (dsRNA) induces the sequence-specific degradation of homologous mRNA (for review, see Rana 2007). To induce target cleavage, short-interfering RNA (siRNA), must be perfectly complementary to its mRNA target because siRNAs with sequences mismatched to nucleotides (nt) at the cleavage site do not induce target cleavage. Another class of short (21–24-nt) dsRNAs called microRNAs (miRNA) also associate with RISC but are not perfectly complementary to target sequences; when loaded into RISC, these miRNAs inhibit translation rather than inducing target cleavage (for review, see Chu and Rana 2007; Filipowicz et al. 2008). Small (19–20-nt) synthetic ds siRNAs with dTdT overhangs have been shown to induce RNAi in numerous mammalian cell lines (Caplen et al. 2001; Elbashir et al. 2001a). Variations in the structure and sequence of siRNA have also been shown to be well tolerated and to not adversely affect RNAi (Rana 2007). However, changes in the A-form helical structure of siRNA duplexes or siRNA–mRNA complexes are not well tolerated (Chiu and Rana 2002, 2003), indicating that the A-form helix is important for mediating RNAi. To determine the essential region of the dsRNA helix needed to induce effective RNAi, we synthesized a series of siRNA duplexes with 3′- or 5′-terminal deletions in the passenger and/or guide strand and examined their RNAi activity. Here, we report that the minimal dsRNA A-form helical structure required to assemble a catalytically active effector RNAi complex is a 16-nt duplex siRNA with ∼1.5 helical turns. Our data indicate that this 16-nt siRNA is sufficient to induce potent RNAi with enhanced RISC-loading efficiency.
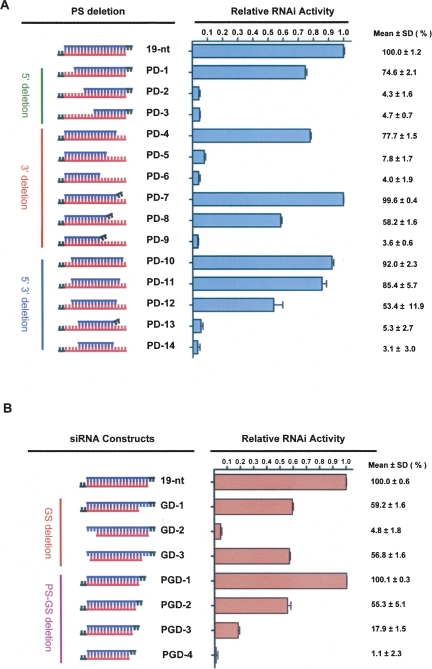
To determine the minimal dsRNA A-form helical structure required to assemble catalytically active RISC, we designed siRNA duplexes to target green fluorescent protein (GFP) and to have a 19-nt guide strand plus dTdT and a passenger strand harboring deletions at the 5′- or 3′-ends (Fig. 1A, PD-1–PD-14; Supplemental Fig. 1). The RNAi activity of these siRNA duplexes was quantitatively analyzed by a dual fluorescence reporter system (Chiu and Rana 2002, 2003). Wild-type 19-nt siRNA (50 nM) silenced 92% of GFP expression in HeLa cells 48 h post-transfection; this RNAi activity is denoted as 100% in Figure 1 for comparison with the activity of other siRNA sequences. Analysis of 19-nt siRNAs with 5′-passenger-strand (PS) deletions showed that a 16-nt siRNA with PS deletions (PD-1) induced RNAi with ∼75% efficiency whereas two other siRNAs with PS deletions, PD-2 and PD-3, did not exhibit RNAi activity.
FIGURE 1.
(A) siRNAs with passenger-strand (PS) deletions as triggers for RNAi. Each GFP siRNA construct shown and reporter plasmids were co-transfected into HeLa cells and RNAi activity was quantified 48 h post-transfection. Relative RNAi activity represents the percentage of GFP knockdown induced by 50 nM siRNA with passenger-strand deletions relative to the inhibition induced by 50 nM 19-nt wild-type siRNA (designated 100%). Each siRNA was tested for knockdown in duplicate in two independent experiments. (B) Effects of guide-strand and two-strand deletions on RNAi activity. The relative RNAi activity of each GFP siRNA construct shown was evaluated as described in (A).
To map the 3′-end boundary required for siRNA function, we systemically deleted the 3′-end of the passenger strand. A 16-nt siRNA with 3′ PS deletions (PD-4) showed efficient RNAi (∼77%). Duplexes with <16-nt passenger strands (PD-5 and PD-6) were inactive. Since 19-nt siRNA duplexes with dTdT overhangs have improved RNAi efficiency, we determined the effect of adding dTdT to the truncated 3′-end of the passenger strand by quantifying the level of GFP knocked down by duplexes PD-7, PD-8, and PD-9 (Fig. 1A). PD-7 exhibited RNAi activity comparable to that of wild-type 19-nt siRNA, whereas adding dTdT to PD-6 (resulting in PD-9) did not increase the RNAi efficiency of this 11-nt duplex.
To address which region of the siRNA must form a duplex structure for efficient RNAi, we synthesized siRNA duplexes with deletions at both the 5′- and 3′-ends of the passenger strand and tested their RNAi activity. PD-10, a 16-nt duplex with a passenger strand at positions 3–18 was highly efficient at knocking down GFP (∼92%), but an 11-nt duplex with a passenger strand at positions 5–15 (PD-14) was nonfunctional, and adding dTdT to the shorter duplex (PD-13) did not improve its RNAi function (Fig. 1A). To determine the minimal length of A-form helix at the central region of the siRNA duplex, we also synthesized siRNAs with a 15- or 14-nt passenger strand. PD-11 with a 15-nt passenger strand exhibited ∼85% RNAi activity, whereas the RNAi activity for PD-12 with a 14-nt passenger strand dropped to ∼53%. These findings demonstrate that efficient RNAi can be accomplished using a 19-nt guide strand and a 16-nt passenger strand. Taken together, these results suggest that a 16-nt duplex RNA structure is required for gene silencing in vivo.
To determine whether this 16-nt rule applied to the guide strand, reciprocal experiments were performed. In these experiments, we used dsRNA duplexes (Fig. 1B, GD-1–GD-3) that harbored a 19-nt plus 3′ dTdT passenger strand and a guide strand truncated from the 5′- and/or 3′-ends. Relative to the wild-type 19-nt siRNA, GD-1 exhibited less efficient RNAi (∼59%). This loss of function may have been due to the 5-nt passenger-strand overhang created by deleting nucleotides 1–3 of the guide strand, since increasing 3′-overhang length is known to negatively affect RNAi (Elbashir et al. 2001b). GD-3 also showed intermediate RNAi activity (∼57%), again likely due to the loss of function contributed by the 4-nt passenger strand 3′-overhang. These results indicate that a 19-nt passenger strand and a 16-nt guide strand can induce RNAi. Surprisingly, GD-2 did not exhibit RNAi activity, suggesting that the 17th to 19th nucleotides from the 5′-end of the guide strand may be important for RISC assembly or target mRNA recognition.
Since our results showed that siRNAs with a 16-nt passenger or 16-nt guide strand exhibited RNAi activity, we next tested whether the 16-nt rule applied to duplexes with both strands truncated (Fig. 1B). Relative to wild-type 19-nt siRNA, 16-nt siRNA (PGD-1) induced RNAi at a high efficiency (∼100%), 15-nt siRNA (PGD-2) induced knockdown at a moderate efficiency (∼55%), whereas 14-nt (PGD-3) and 13-nt (PGD-4) siRNAs induced knockdown at low efficiencies (∼18% and ∼1%, respectively). Collectively, these results demonstrate that the threshold number of nucleotides required to induce highly efficient gene knockdown is 16.
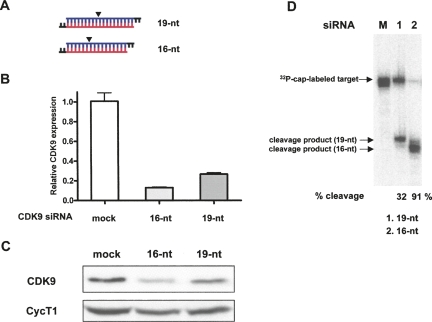
The 16-nt GFP siRNA (PGD-1) exhibited wild-type (WT) levels of GFP knockdown (Fig. 1B), indicating that a 16-nt siRNA is as efficient as a 19-nt siRNA at triggering RNAi in vivo. To show that 16-nt siRNA can be generally used as an RNAi trigger targeting cellular protein, we synthesized the 16-nt siRNA targeting CDK9 based on its published 19-nt siRNA sequence (Fig. 2A; Brown et al. 2005). This 16-nt CDK9 siRNA was evaluated for RNAi efficacy by transfecting it into HeLa cells in parallel with 19-nt WT siRNA and measuring CDK9 mRNA and protein levels at 48 h post-transfection. The 16-nt siRNA was shown by quantitative PCR to knock down CDK9 mRNA with higher efficiency (∼90%) than 19-nt siRNA (∼75%) (Fig. 2B). Consistent with this result, endogenous CDK9 protein was shown by immunoblot analysis to be reduced in HeLa cells transfected with 16-nt or 19-nt siRNA (Fig. 2C).
FIGURE 2.
The 16-nt siRNA targeting CDK9 efficiently induces endogenous gene silencing in HeLa cells. (A) The wild-type and 16-nt CDK9 siRNAs used to program RISC are shown. Arrowheads mark target cleavage sites defined by the 5′-end of the guide strand. siRNA sequences are shown in Supplemental Figure 1. (B) The 16-nt CDK9 siRNA more efficiently knocks down CDK9 mRNA than 19-nt WT siRNA. HeLa cells were transfected with 50 nM CDK9 siRNA (19-nt or 16-nt), harvested 48 h post-transfection, and 1 μg total RNA was reverse-transcribed. CDK9 mRNA levels quantified by quantitative PCR and normalized to GAPDH mRNA are presented relative to mRNA levels in mock-transfected cells. Data represent three independent experiments. (C) The 16-nt CDK9 siRNA more efficiently decreases CDK9 protein expression than 19-nt WT siRNA. Immunoblot analysis of CDK9 knockdown by 19-nt and 16-nt siRNAs. HeLa cells were transfected and harvested as in (B), and 120 μg total protein was analyzed by immunoblot using anti-CDK9 and anti-CycT1 antibodies. (D) The 16-nt CDK9 siRNA more efficiently programs RISC to cleave target CDK9 mRNA than 19-nt WT siRNA. CDK9 siRISC was programmed by transfecting HeLa cells with 19-nt or 16-nt siRNA. Target cleavage reaction was performed at 37°C for 90 min. See Materials and Methods for details. The arrow designated “32P-cap-labeled target” points to full-length 32P-cap-labeled CDK9 target mRNA. The arrows designated “cleavage product (19-nt)” and “cleavage product (16-nt)” point to products of target mRNA cleavage by CDK9 siRISC programmed with 19-nt and 16-nt siRNA, respectively.
To determine if 16-nt siRNA enters the RNAi pathway by the same mechanism as 19-nt siRNA, we compared the extent to which RISC programmed with each siRNA cleaved its target mRNA in vitro. RISC was programmed by transfecting HeLa cells with 19-nt or 16-nt CDK9 siRNA, preparing cell extracts, and measuring the ability of activated siRNA-programmed RISCs (siRISCs) to cleave added 150-nt 32P-cap-labeled CDK9 mRNA target. The 19-nt and 16-nt siRISC enzyme complexes cleaved target mRNA ∼32%, and ∼91%, respectively (Fig. 2D). Thus, the 16-nt siRNA RISC exhibited a much higher cleavage activity with equal amounts of cell extracts, suggesting that 16-nt siRNA is a more potent RNAi trigger. The cleavage product of CDK9 16-nt siRNA RISC revealed that the cleavage site had shifted 3 nt (Fig. 2D; cf. lanes 1,2 and arrows), consistent with previous studies (Elbashir et al. 2001b) and reflecting the new position of the 5′-end of the guide strand after truncating 3 nt.
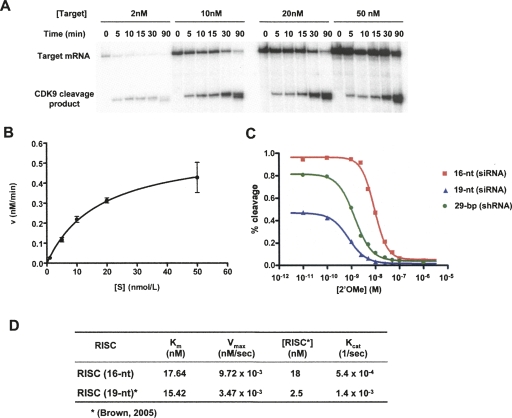
Our results show that 16-nt siRNAs targeting GFP or CDK9 are sufficient to trigger RNAi. In addition, we tested a number of other target mRNA sequences spanning various regions of CDK9 and luciferase reporter constructs, and compared the potency of 19-nt and 16-nt siRNAs which showed that 16-nt siRNA was a potent RNAi trigger (data not shown). The different in vitro cleavage efficiencies of the CDK9 16-nt RISC and 19-nt RISC (Fig. 2D) prompted us to further explore the enzymatic activity of 16-nt RISC. The substrate concentration dependence of CDK9 16-nt RISC cleavage activity was examined by varying the amount of target mRNA (2 nM, 10 nM, 20 nM, and 50 nM) in a fixed-volume (20 μL) reaction with a constant amount of cell extract programmed with 16-nt RISC. The efficiency of RISC target cleavage increased with target mRNA concentration, saturating at higher concentrations (Fig. 3A,B). The K m and V max of CDK9 16-nt RISC target cleavage were determined by nonlinear fitting of substrate concentration versus initial velocity to the Michaelis–Menton equation (Fig. 3B). The concentration of 16-nt RISC was determined by blocking the cleavage activity of RISC with varying concentrations of 2′-O-methyl oligonucleotides complementary to the guide strand 16-nt siRNA and measuring the IC50 (Fig. 3C; Supplemental Fig. 2; Brown et al. 2005).
FIGURE 3.
Kinetic analysis of CDK9 16-nt RISC. (A) CDK9 RISC cleavage of target mRNA depends on substrate concentration. Efficiency of target mRNA cleavage by CDK9 RISC programmed by 16-nt siRNA at various substrate concentrations. (B) CDK9 RISC cleavage of target mRNA shows Michaelis–Menten kinetics. Michaelis–Menten analysis of 16-nt RISC. The initial velocity of cleavage product formation was determined from the slope of a curve fit to the cleavage reactions at 0–30 min as shown in (A). (C) The 16-nt siRNA programs more RISC than 19-nt siRNA or 29-nt shRNA. Concentrations of active CDK9 RISC programmed by 16-nt RNA, 19-nt siRNA, or 29-nt shRNA were determined by 2′-O-Me inhibition in vitro cleavage assay. (D) Kinetic analysis of 16-nt CDK9 RISC. K m and V max were determined by Michaelis–Menten analysis as shown in (B).
Our results indicate that the 16-nt RISC is a multiple-turnover enzyme that recognizes and cleaves its target with classic Michaelis–Menton kinetics. The K m for 16-nt RISC is 17.64 nM and its V max is 9.72 × 10−3 nM/sec (Fig. 3D). Remarkably, the concentration of 16-nt RISC programmed in HeLa cells was 18.26 nM, indicating that 16-nt siRNA programmed ∼ 7 × more RISC than 19-nt siRNA (2.5 nM) (Fig. 3C; Brown et al. 2005). Interestingly, rate constant (K cat) determination showed that RISC programmed with 16-nt RNA was not catalytically more efficient than the 19-nt RISC (Fig. 3D). Overall, these results indicate that the reason why CDK9 16-nt RISC cleaves its target with greater efficiency at steady state is its greater capacity to program a higher concentration of RISC. These findings suggest that increases in RNAi potency correspond to an increased amount of RISC formed by a given siRNA, such as the 16-nt RNA.
In conclusion, our findings demonstrate that 16-nt siRNA duplexes are potent RNAi triggers and can now be considered for use in reverse genetic studies. Further studies on the detailed mechanisms by which 16-nt siRNA is incorporated into RISC and triggers RNAi will provide insight into the principles of RNAi pathways in human cells, particularly those involved in RISC loading and duplex unwinding. Since the current algorithms for designing siRNAs have been specifically developed for 19-nt duplexes, new algorithms should include 16-nt duplexes to create short siRNAs with even higher RNAi efficiencies. Given the advantages of using 16-nt siRNA to trigger RNAi, we expect that these shorter RNA duplexes will be widely employed in biomedical studies.
MATERIALS AND METHODS
siRNA preparation
The sequences of GFP target-specific siRNA duplexes were designed as previously described (Chiu and Rana 2002). 2′-bis(acetoxyethoxy)-methyl ether-protected single-stranded RNA of varying lengths were chemically synthesized by Dharmacon. Synthetic oligonucleotides were deprotected, annealed, and purified as described by the manufacturer. Successful duplex formation was confirmed by 20% nondenaturing PAGE.
Culture and transfection of cells
HeLa cells were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Cells were regularly passaged at subconfluence and plated 16 h before transfection at 70% confluence. Lipofectamine (Invitrogen)-mediated transient co-transfection of reporter plasmids and siRNAs were performed in duplicate six-well plates. A transfection mixture containing 0.16 μg of pEGFP-C1 and 0.33 μg of pDsRed2-N1 reporter plasmids (Clontech), 50 nM of each siRNA deletions, and 7 μL of lipofectamine in 1 mL of serum-reduced OPTI-MEM (Invitrogen) were added to each well. Cells were incubated in the transfection mixture for 4 h and further cultured in antibiotic-free DMEM. Cells were treated under the same conditions without siRNA for mock experiments.
Dual fluorescence assay
The function of GFP siRNAs was evaluated by a dual fluorescence assay as described (Chiu and Rana 2003). RNAi activity was quantified in total cell lysates (150 μg in 200 μL reporter lysis buffer) 48 h after transfecting cells with siRNA. GFP fluorescence was detected using a Safire plate reader (TECAN) by exciting at 488 nm and recording emissions from 504 to 514 nm. The spectrum peak at 509 nm represents the fluorescence intensity of GFP. RFP fluorescence was detected in the same cell lysates by exciting at 558 nm and recording emissions from 578 to 588 nm. The spectrum peak at 583 nm represents the fluorescence intensity of RFP. The fluorescence intensity ratio of target (GFP) to control (RFP) fluorophores was determined in the presence of siRNA duplexes and normalized to the emissions measured in mock-treated cells. Normalized ratios <1.0 indicated specific RNA interference. Relative RNAi activity represents the percentage of GFP knockdown induced by 50 nM siRNA with passenger-strand deletions relative to the inhibition induced by 50 nM 19-nt wild-type siRNA (designated 100%).
Cytoplasmic cell extract preparation and in vitro mRNA cleavage assay
To prepare cell extracts, HeLa cells were transfected with 25 nM siRNA, harvested 18 h later with trypsin, and centrifuged at 1000g for 5 min at 4°C. The pellets were washed 3× with ice-cold PBS pH 7.2 and lysed by adding three packed-cell volumes of lysis buffer (20 mM HEPES pH 7.9, 10 mM NaCl, 1 mM MgCl2, 0.5 M sucrose, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, and 0.35% Triton X-100). Lysis was continued for 10 min on ice. Once lysed, nuclei were removed by centrifugation at 2500 rpm for 10 min at 4°C. Cytoplasmic extracts in supernatants were prepared by adding 0.11 volumes of cold Buffer B (20 mM HEPES pH 7.9, 10 mM NaCl, 1 mM MgCl2, 0.35 M sucrose, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF). Extracts were quick frozen in liquid nitrogen and stored at –80°C.
To evaluate target mRNA cleavage in vitro, the CDK9 mRNA target sequence was amplified by PCR with forward and reverse primers 5′-TAATACGACTCACTATAGGCTTGCGGGAGATCAAGATC-3′ and the reverse primer 5′-CAGCCCAGCAAGGTCATG-3′, respectively, for transcription of a 150-nt CDK9 target mRNA. The resulting transcript was 32P-cap-labeled, as described (Chiu and Rana 2003). CDK9 target mRNA (10 nM) was incubated for 90 min at 37°C in the presence of 4 μL cytoplasmic extract, 1 mM ATP, 0.2 mM GTP, 1 U/μL RNasin (Promega), 30 μg/mL creatine kinase, 25 mM creatine phosphate, 2 mM MgCl2, 20 mM NaCl. Buffer D (1 M KCl, 20 mM HEPES pH 7.9, 10% glycerol, 0.2 mM EDTA) was added to a final reaction volume of 20 μL. Cleavage reactions were stopped by adding nine volumes of proteinase K buffer (200 mM Tris-HCl pH 7.5, 25 mM EDTA, 300 mM NaCl, and 2% [w/v] SDS). Proteinase K (Ambion) was added to a final concentration of 0.6 mg/mL. Reactions were incubated for 15 min at 37°C. Cleavage products were isolated by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and ethanol precipitation, and resolved on a 6.5% polyacrylamide-7 M urea gel.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Craig Mello and Rana laboratory members for helpful discussions and support. This work was supported in part by an NIH grant to TMR.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1161908.
REFERENCES
- Brown K.M., Chu C.Y., Rana T.M. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Parrish S., Imani F., Fire A., Morgan R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.L., Rana T.M. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Rana T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.Y., Rana T.M. Small RNAs: Regulators and guardians of the genome. J. Cell. Physiol. 2007;213:412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001b;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Rana T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]