Abstract
Tertiary or higher-order RNA motifs that regulate replication of positive-strand RNA viruses are as yet poorly understood. Using Japanese encephalitis virus (JEV), we now show that a key element in JEV RNA replication is a complex RNA motif that includes a string of three discontinuous complementary sequences (TDCS). The TDCS consists of three 5-nt-long strands, the left (L) strand upstream of the translation initiator AUG adjacent to the 5′-end of the genome, and the middle (M) and right (R) strands corresponding to the base of the Flavivirus-conserved 3′ stem–loop structure near the 3′-end of the RNA. The three strands are arranged in an antiparallel configuration, with two sets of base-pairing interactions creating L-M and M-R duplexes. Disrupting either or both of these duplex regions of TDCS completely abolished RNA replication, whereas reconstructing both duplex regions, albeit with mutated sequences, fully restored RNA replication. Modeling of replication-competent genomes recovered from a large pool of pseudorevertants originating from six replication-incompetent TDCS mutants suggests that both duplex base-pairing potentials of TDCS are required for RNA replication. In all cases, acquisition of novel sequences within the 3′M-R duplex facilitated a long-range RNA–RNA interaction of its 3′M strand with either the authentic 5′L strand or its alternative (invariably located upstream of the 5′ initiator), thereby restoring replicability. We also found that a TDCS homolog is conserved in other flaviviruses. These data suggest that two duplex base-pairings defined by the TDCS play an essential regulatory role in a key step(s) of Flavivirus RNA replication.
Keywords: Japanese encephalitis virus, Flavivirus, RNA replication, cis-acting elements, base-pairings
INTRODUCTION
The positive-strand RNA viruses include a large number of important human, animal, and plant pathogens (Buck 1996), and their replication is a critical element in the pathogenesis of a variety of acute and chronic diseases in humans, such as severe acute respiratory syndrome (Stadler et al. 2003), encephalitis (Mackenzie et al. 2004), and hepatitis and liver carcinogenesis (Lindenbach and Rice 2005). Although the precise details of their replication vary for each virus, diverse groups of positive-strand RNA viruses in plants and animals share remarkably common replication strategies (Pogue et al. 1994; Ahlquist 2006). Following viral infection, positive-strand genomic RNA is released from endocytosed virions into the cytoplasm of the infected cell, initiating the synthesis of viral proteins, which then direct the replication of multiple positive-strand genomic RNA copies and their encapsidation into new progeny virions. The replication of genomic RNAs requires a complex of cis-acting RNA elements within the viral genome. These elements possess considerable secondary and tertiary structure that is specifically recognized by a complex of viral replicases and cellular proteins. Substantial progress has been made in identifying and characterizing the conserved primary sequences and secondary structures that are characteristic of cis-acting RNA elements in positive-strand RNA viruses; how these RNA elements participate in and coordinate the replication of the genomic RNA is, however, a still challenging question to be addressed. Therefore, in this study, we have analyzed a complex RNA motif within the genome of the Japanese encephalitis virus (JEV) that plays an essential regulatory role in RNA replication.
JEV is a member of the Flavivirus genus of the Flaviviridae family of positive-strand RNA viruses. It is a mosquito-borne virus that is closely related to the human epidemic-causing yellow fever, dengue, West Nile, and Kunjin (a West Nile subtype) flaviviruses (Heinz et al. 2000; Calisher and Gould 2003). JEV is responsible for epidemics of viral encephalitis in many Asian countries, including India, China, Korea, Japan, and Indonesia. The disease mainly affects children and young adults, with a mortality rate of ≈25% and permanent neuropsychiatric sequelae observed in ≈50% of the survivors (Burke and Monath 2001; Solomon 2003). This pathogen has recently emerged in Australia as well, posing a major public health threat worldwide (Mackenzie et al. 2002). Like other flaviviruses, JEV is a small, enveloped virus primarily composed of two viral surface proteins, membrane (M) and envelope (E), which are embedded in a host-derived lipid membrane surrounding a core shell of capsid (C) proteins. Within this shell is a single copy of the positive-strand RNA genome of ≈11 kb, which contains a 5′-cap structure but lacks a 3′-poly(A) tail. The genomic RNA encodes a single long open reading frame (ORF) flanked by 5′- and 3′-nontranslated regions (NTRs) containing most of the cis-acting RNA elements required for viral replication, transcription, and translation (Lindenbach and Rice 2001). The ORF produces a polyprotein that is cleaved by cellular and viral proteases into three structural proteins (C; prM, which is further processed into pr and M; and E) and at least seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, as arranged in the genome), which form a viral replicase complex that mediates replication of the genomic RNA in the cytoplasm of the infected cell (Chambers et al. 1990; Westaway et al. 2003).
Bioinformatic, thermodynamic, chemical, and genetic analyses have identified three common features of the short conserved primary RNA sequence and secondary RNA structure that are required for flavivirus RNA replication (Fig. 2A, below; Markoff 2003):
FIGURE 2.
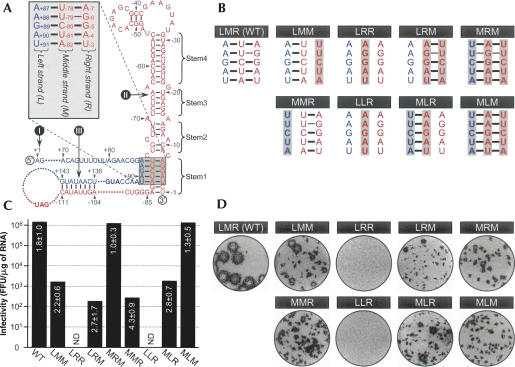
A TDCS is required for the replication of JEV genomic RNA. (A) Schematic presentation of the putative JEV TDCS (shaded box). The 5′-end nucleotide of the genome is numbered +1, and bases extending downstream are assigned (blue) positive numbers; the 3′-end nucleotide is numbered −1, and bases extending upstream are assigned (red) negative numbers. The translation initiation codon (AUG) and stop codon (UAG) are shown in boldface type. The three RNA elements of the flavivirus-conserved primary sequence and the secondary structure required for RNA replication are indicated (Markoff 2003): (I) 5′-AG and CU-3′; (II) the 3′ stem–loop (3′SL), subdivided into four stem regions (Stem1 to Stem4); (III) a pair of cyclization sequences (5′CS, 5′-UCAAUAUG; 3′CS, 5′-CAUAUUGA). (Inset) Relative locations of the three strands of the TDCS, left (L), middle (M), and right (R), and their potential base-pairing patterns are highlighted. (B) Eight mutations introduced into the TDCS of JEV, and the base-pairing potentials disrupted by each mutation. (Shaded boxes) The altered nucleotides are indicated in boldface type. (C,D) (C) Specific infectivities of the synthetic RNA transcripts derived from JEV wild-type (WT) and eight TDCS-mutant cDNAs and (D) representative focus morphologies. The WT and eight TDCS-mutant cDNA templates, as indicated, were used for SP6 polymerase runoff transcription. (C) The specific infectivities of the synthetic RNA transcripts were estimated by infectious center assays, in which naive BHK-21 cells were electroporated with 2 μg of the synthetic RNA transcripts, as described in Materials and Methods. (ND) Not detected. (D) For focus morphology, the cells were immunostained with a mouse JEV-specific hyperimmune antiserum and a peroxidase-conjugated goat anti-mouse IgG.
The genomic RNA begins with the dinucleotide 5′-AG and ends with the dinucleotide CU-3′ (Rice et al. 1985); these dinucleotide sequences are absolutely conserved in all flaviviruses (Wengler and Wengler 1981; Rice et al. 1985; Brinton et al. 1986; Wengler and Castle 1986; Brinton and Dispoto 1988; Mandl et al. 1993; Khromykh and Westaway 1994; Shi et al. 1996), except the cell-fusing agent virus (Cammisa-Parks et al. 1992). In the Kunjin virus, substitution mutations of the 5′-penultimate G or 3′-penultimate C have been shown to block RNA replication, and mutation of the 5′-terminal A or 3′-terminal U blocks or restricts RNA replication, respectively (Khromykh et al. 2003).
The 3′-terminal ≈90 nt is predicted to form a highly conserved stem–loop (3′SL) structure, with a long stem consisting of ≈30 base-pairings that includes several interrupting bulges and a loop of unpaired pentanucleotides (5′-CACAG-3′) at its top. This 3′SL structure was initially noted at the 3′-end of yellow fever genomic RNA (Grange et al. 1985; Rice et al. 1985), was predicted to be undisturbed by base-pairings with upstream sequences (Proutski et al. 1997; Rauscher et al. 1997; Khromykh et al. 2003), and was later shown to be required for the replication of many flaviviruses (Brinton et al. 1986; Men et al. 1996; Shi et al. 1996; Zeng et al. 1998; Yu and Markoff 2005).
The 3′-proximal conserved sequence upstream of the 3′SL is complementary to another conserved sequence located in the 5′ coding region of the C protein in all mosquito-borne flavivirus genomes (Hahn et al. 1987), suggesting that flavivirus RNAs can cyclize. The activity of these two cyclization sequences (5′CS and 3′CS) has been demonstrated in vivo in many mosquito-borne flaviviruses, including Kunjin (Khromykh et al. 2001), yellow fever (Bredenbeek et al. 2003; Corver et al. 2003), West Nile (Lo et al. 2003), and dengue viruses (Men et al. 1996; Alvarez et al. 2005; Filomatori et al. 2006), and in vitro in dengue virus (You and Padmanabhan 1999; You et al. 2001; Alvarez et al. 2005). However, although these three common cis-acting RNA elements have been identified as essential for flavivirus RNA replication, how these RNA elements are organized into tertiary or higher-order structures in the context of the viral genome and how they coordinate RNA synthesis remain unclear.
Here, we describe for the first time a complex RNA motif consisting of a string of three discontinuous complementary sequences (TDCS) that is essential for JEV RNA replication. By using systematic mutagenesis of the genomic RNA, recovery of a large pool of pseudorevertants bearing a variety of compensatory mutations, and characterization of several novel sequences that are capable of restoring JEV RNA replication, we provide strong evidence that the TDCS is required for the formation of the intact 3′SL structure, long-distance communication between the 5′ and 3′ termini of the genome, and the complete replication of the genome. Multiple interactions involving both Watson–Crick and non-Watson–Crick base pairs, rather than the specific nucleotide sequences of this motif per se, are apparently critical not only for the formation and maintenance of this long-range RNA–RNA interaction but also for its biological functions. We further report that a homolog of the TDCS is present in other flaviviruses, suggesting a general requirement for this RNA motif during flavivirus replication. Thus, our data suggest that a complex RNA motif defined by the TDCS is a key regulatory component of flavivirus RNA replication, ensuring long-distance communication of the flavivirus-conserved 3′SL with the flavivirus-conserved 5′SL that precedes the translation initiation codon. This unique sequence-specific RNA motif offers an attractive target for prophylactic or therapeutic interventions that would block the replication of these clinically significant viruses.
RESULTS
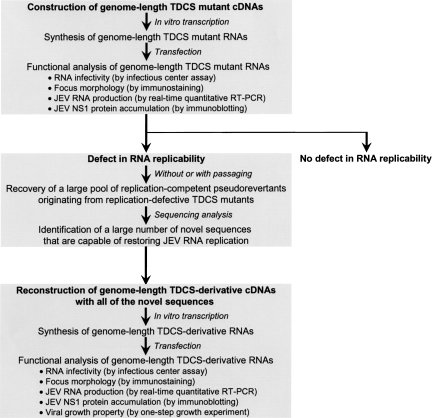
The primary sequence and potential base-pairing interactions of a complex RNA motif required for JEV genome replication were initially identified by systematic mutagenesis of JEV genomic RNA (Fig. 1). Using an infectious JEV cDNA molecular clone, we generated genome-length mutant JEV RNAs by in vitro transcription from recombinant cDNA templates, and then analyzed the replicability of each mutant RNA after transfection into JEV-susceptible BHK-21 cells. In particular, we measured RNA infectivity using an infectious center assay as an indication of RNA replicability and analyzed focus morphology by immunostaining with JEV-specific antibody, assessing the size and homogeneity of infectious foci as an indication of genetic stability. We considered an RNA motif to be required for replication if (1) disrupting the primary sequences and/or base-pairing interactions by mutations led to a loss of RNA infectivity and an absence of the mutated element in new progeny virions recovered from RNA-transfected cells; or (2) restoring the base-pairing interaction by compensatory mutations led to a gain in RNA infectivity to a level similar to that of wild type (WT).
FIGURE 1.
Schematic diagram outlining the identification and characterization of a complex RNA motif, the TDCS, required for the replication of JEV genomic RNA.
A complex RNA motif, the TDCS, is required for viral replication
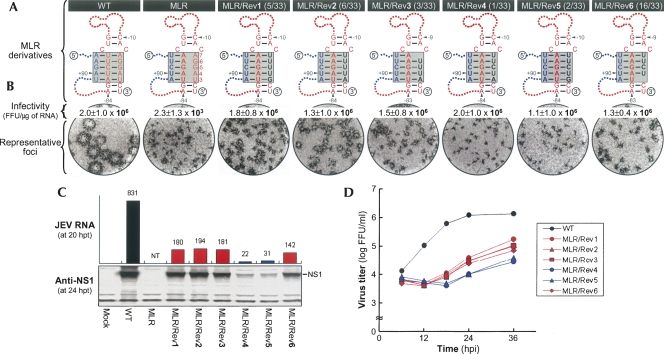
To assess the functional importance of the conserved primary sequence and/or secondary structure of the 3′SL in JEV genome replication, we first subdivided the 3′SL into four stem regions (Fig. 2A, Stem1 to Stem4) and performed a mutagenetic analysis in which each nucleotide of one strand of each stem region was replaced by its complementary base, in order to disrupt base-pairing interactions in the context of the genome. In parallel experiments, we simultaneously exchanged both strands for their complements, in order to maintain the secondary structure but not the primary sequence. Of the four stem regions, three (Stem2, Stem3, and Stem4) showed a clear correlation between disruption/restoration of duplex base-pairing and loss/gain of JEV RNA infectivity, indicating that the duplex stem structures, rather than the primary sequences, in this area were, indeed, essential for JEV genome replication (B.H. Song and Y.M. Lee, in prep.). However, because no such correlation was observed for Stem1, no evidence of Stem1 duplex formation could be inferred. That is, substitution of either strand of Stem1 had a drastic negative effect on infectivity, and replacement of both strands failed to restore the infectivity to near wild-type (WT) levels (Fig. 2, mutants LMM, LRR, and LRM). This led us to hypothesize that Stem1, corresponding to the base of the 3′SL, might be involved in forming a complex RNA motif with tertiary or higher-order RNA structure, as opposed to simple duplex base-pairing. This complex RNA motif might include an additional base-pairing interaction with sequences elsewhere in the JEV genome, with one of the two strands forming Stem1.
Based on computer-based predictions and visual inspection of the nucleotide sequence of the JEV genome, we therefore hypothesized the existence of a complex RNA motif that includes a string of three discontinuous complementary sequences (Fig. 2A, TDCS). For discussion purposes, the 5′-end nucleotide of the genome is numbered +1, and bases extending in the downstream direction are assigned positive numbers; the 3′-end nucleotide is numbered −1, and bases continuing in the upstream direction are assigned negative numbers. The TDCS consists of three 5-nt-long strands, the left (L, 5′-A+87A+88G+89A+90U+91) strand upstream of the translation initiator A+96U+97G+98 adjacent to the 5′-end of the genome, and the middle (M, 5′-A−82U−81C−80U−79U−78) and right (R, 5′-A−7G−6G−5A−4U−3) strands corresponding to the two strands of a 5-nt-long Stem1 duplex proximal to its own 3′-end (Fig. 2A, inset). The three strands are arranged in an antiparallel configuration, with two sets of base-pairing interactions creating L-M and M-R duplexes. Given this configuration, the TDCS could function either by forming one of the two competing duplexes (L-M and M-R) or as a potential intramolecular triple helix (L-M-R).
To examine the functional importance of two potential duplex base-pairings between the L and M strands and the M and R strands of the TDCS, eight mutant constructs differing only in their ability to form the TDCS were individually cloned into an infectious JEV cDNA, pBACSP6/JVFLx/XbaI (Fig. 2B; Yun et al. 2003). For these mutants, the position of the three letters representing the L, M, and R strands of JEV TDCS indicates which strand of the TDCS was substituted by or exchanged with which strand. The parent, denoted LMR, is the WT reference. The functionality of the eight TDCS mutations was analyzed by determining the specific infectivities of the synthetic RNA transcripts derived from each mutant cDNA template after RNA transfection into BHK-21 cells (Fig. 2C). The WT construct gave a specific infectivity of 1.8 ± 1.0 × 106 FFU/μg, in the expected range. In contrast, disruption of the potential for both L-M and M-R duplex base-pairings simultaneously by introducing either an M → R strand substitution (single-strand mutant LRR) or an M → L strand substitution (single-strand mutant LLR) entirely abolished the infectivity. Disruption of the potential for M-R duplex base-pairing alone by introducing either an R → M strand substitution (single-strand mutant LMM) or an L ↔ M strand exchange (double-strand mutant MLR) led to a severe (approximately three orders of magnitude) decrease in the infectivity, to either 2.2 ± 0.6 × 103 or 2.8 ± 0.7 × 103 FFU/μg, respectively, even though L-M duplex base-pairing was still intact (LMM) or restored (MLR). Also, disruption of the potential for L-M duplex base pairing alone by introducing either an L → M strand substitution (single-strand mutant MMR) or an M ↔ R strand exchange (double-strand mutant LRM) reduced the infectivity by approximately four orders of magnitude, to either 4.3 ± 0.9 × 102 or 2.7 ± 1.7 × 102 FFU/μg, respectively, even though M-R duplex base-pairing was still intact (MMR) or restored (LRM). Finally, restoration of the potential for both L-M and M-R duplex base-pairings simultaneously, but not its primary sequence, by introducing either an additional L → M strand substitution in the double-strand mutant LRM (triple-strand mutant MRM, 1.0 ± 0.3 × 106 FFU/μg) or an additional R → M strand substitution in the double-strand mutant MLR (triple-strand mutant MLM, 1.3 ± 0.5 × 106 FFU/μg), completely restored the infectivity to a level similar to WT (1.8 ± 1.0 × 106 FFU/μg). These results demonstrate that disrupting either or both of the L-M and M-R duplex base-pairings leads to a loss of JEV genome replication, and only restoring both of these duplex base-pairings simultaneously with appropriately mutated sequences (but not with either duplex base-pairing alone) can completely restore replication.
Immunostaining of RNA-transfected BHK-21 cells with a mouse JEV-specific hyperimmune antiserum (Fig. 2D) showed that, as expected, the WT reference produced a homogeneous population of large infectious foci. Of the eight TDCS mutants, only the two triple-strand mutants (MRM and MLM), whose potential for both L-M and M-R duplex base-pairings had been simultaneously restored with mutated sequences, produced a population of foci as homogeneous as those of the WT. However, the average focus size was smaller than that of the WT, by ≈4.0-fold for MRM and by ≈5.3-fold for MLM. The size difference was correlated with the levels of JEV RNA production and protein accumulation in RNA-transfected cells (data not shown), suggesting that the nucleotide composition of the TDCS influences the level of RNA replication (see Figs. 3–7 for more experimental data). Four mutants (LMM, MLR, MMR, and LRM), whose potential for either L-M (MMR and LRM) or M-R (LMM and MLR) duplex base-pairing was disrupted, produced foci of heterogeneous sizes. This heterogeneity in focus morphology and the magnitude of the reduction in RNA infectivity indicate some degree of genetic instability, which may reflect a need for compensatory mutations acquired in the TDCS region to restore JEV genome replication.
FIGURE 3.
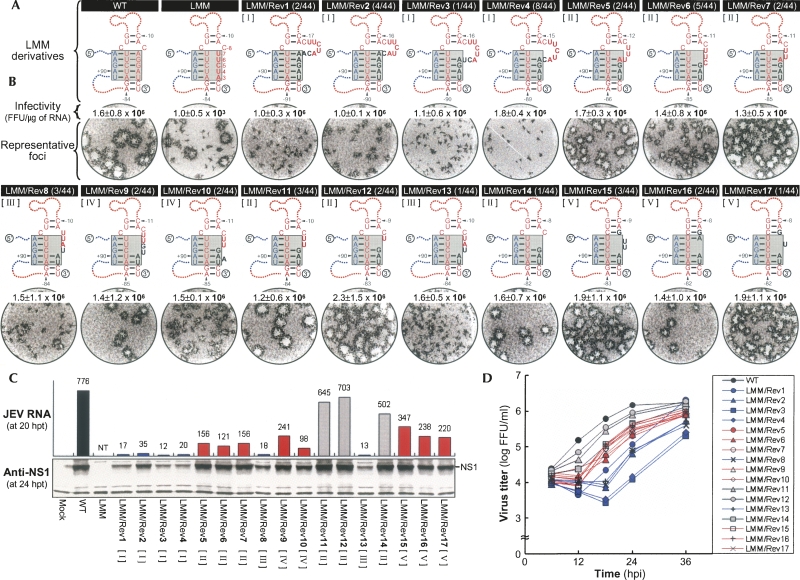
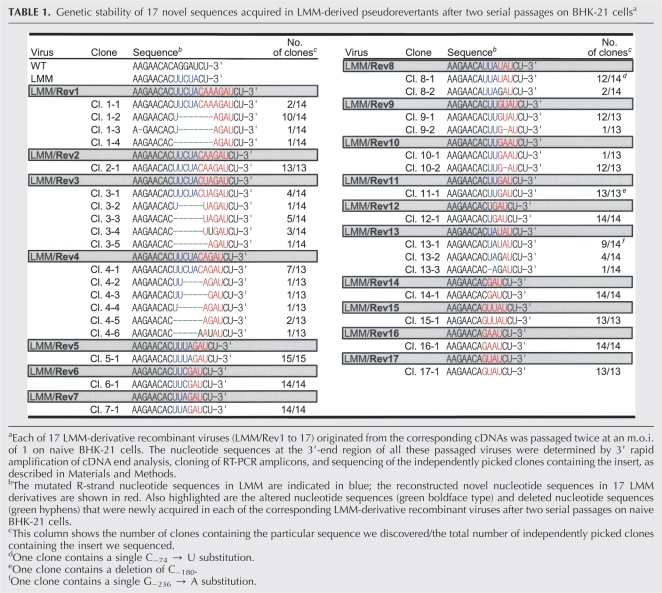
Functional significance of 17 novel sequences discovered in LMM-derived pseudorevertants for M-R duplex base-pairing and genome RNA replication. (A) Novel sequences (LMM/Rev1 to 17) acquired in LMM-derived pseudorevertants, and (shaded boxes) predicted complex motifs of the TDCS formed by WT, LMM, and LMM derivatives containing one of the 17 novel sequences. Nucleotides that were acquired in LMM-derived pseudorevertants are shown in black boldface type. Fractions in parentheses are the number of clones discovered that contain the particular sequence/the total number of independently picked clones that were sequenced. The 17 LMM derivatives were classified into five groups, I to V, as described in Results. (B,C) Specific infectivities and representative focus morphologies obtained 4 d after transfection of BHK-21 cells with the synthetic RNAs transcribed from the reconstructed JEV cDNAs, each containing (B) one of the 17 novel sequences, and (C) their levels of JEV RNA production and JEV NS1 protein accumulation, estimated at 20 and 24 h post-transfection (hpt), respectively. Naive BHK-21 cells were mock-transfected (Mock) or transfected with 2 μg of WT, LMM, or one of 17 LMM-derivative RNAs transcribed from each cDNA, as indicated. (B) At 4 d after transfection, the specific infectivities of the synthetic RNA transcripts were estimated by infectious center assays (Infectivity); the infectious centers of foci were visualized by immunostaining of RNA-transfected cells with anti-JEV antiserum (Representative foci). (C) Changes in JEV RNA levels at 20 h post-transfection, relative to those at 6 h post-transfection, were quantitated by real-time quantitative RT-PCR, using a JEV-specific probe complementary to nucleotides 5837-5856 in NS3 (JEV RNA). JEV NS1 protein accumulation at 24 h post-transfection was analyzed by immunoblotting with anti-JEV NS1 antiserum (Anti-NS1). (NT) Not tested. (D) One-step growth kinetics. BHK-21 cells were infected at a multiplicity of infection of 8 FFU/cell with WT or one of 17 LMM-derivative viruses obtained from the corresponding RNA-transfected cells. Culture supernatants were harvested at the indicated hour post-infection (hpi), and virus titers were determined on naive BHK-21 cells. The curve shows cumulative virus titers.
FIGURE 7.
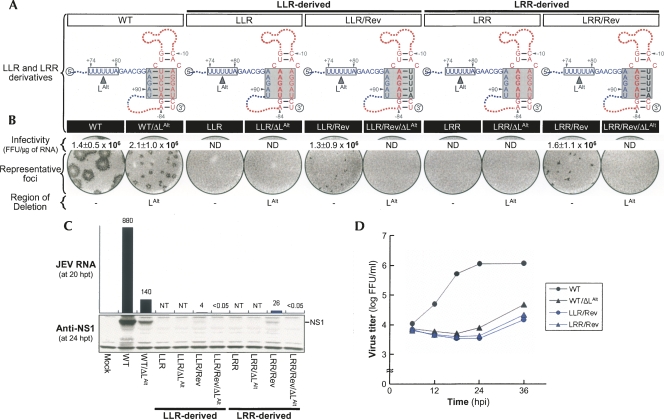
Functional significance of an identical novel 5-nt sequence in the replication of both LLR and LRR genomic RNAs. (A, black boldface type) An identical novel sequence of five nucleotides (U−7U−6U−5U−4A−3) acquired in the R strand of the TDCS in two pseudorevertants originating from LLR (LLR/Rev) and LRR (LRR/Rev). Also presented are predicted complex motifs of the TDCS formed by each RNA in the presence or absence of the 5′LAlt strand (shown as a box outlined with a solid line). (B) Specific infectivities and representative focus morphologies obtained 4 d after transfection of BHK-21 cells with the synthetic RNAs transcribed from each cDNA, as indicated, and (C) their levels of JEV RNA production and JEV NS1 protein accumulation at 20 and 24 h post-transfection, respectively. (D) One-step growth kinetics of the recombinant viruses derived from the corresponding synthetic RNAs, as indicated. Experiments were performed as described for Figure 3. (ND) Not detected; (NT) not tested.
In order to obtain a large pool of various compensatory mutations rather than only a few dominant reversions, culture supernatants collected from RNA-transfected cells, without passaging, were directly used for sequencing analysis. The nucleotide sequence of the TDCS and its flanking regions was determined by cDNA amplification of the 5′- and 3′-terminal regions of the genome (≈650 nt each), cloning of RT-PCR amplicons, and sequencing of a number of the independently picked clones containing the insert. As discussed below, sequence analysis showed that in all four cases (LMM, MLR, MMR, and LRM), the recovered replicating RNA species did not correspond to the original transfected RNA. Instead, a large number of compensatory mutations that varied in size and nucleotide composition were identified (for a detailed description, see Figs. 3–7 and Supplemental Figs. S1–S7). We further characterized these compensatory mutations by using real-time quantitative RT-PCR and immunoblotting to assess the levels of JEV RNA production and NS1 protein accumulation in BHK-21 cells transfected with WT or the mutant JEV RNAs, and by using one-step growth assays to measure the yield of progeny virions in BHK-21 cells infected with WT or the mutant JEV viruses during a single round of infection.
Base-pairing of the M-R duplex in JEV TDCS is a prerequisite for genomic RNA replication
We first examined a large pool of compensatory mutations found in the replicating RNA species that originated from two mutants, LMM and MLR.
Mutant LMM-derived compensatory mutations
Sequencing 25 and 44 independent clones for the 5′- and 3′-terminal regions of LMM-derived pseudorevertants, respectively, identified 17 novel sequences of various sizes that had been acquired immediately upstream of the 3′-end CU, in the presence or absence of a deletion of various sizes in the mutated R strand of LMM (Supplemental Table S1). All of these sequences are predicted to restore the base-pairing potential of the M-R duplex, which had been disrupted in LMM (Fig. 3A). Based on the nucleotide composition and pattern of the base-pairing predicted for the M-R duplex, we classified the novel sequences into five groups, I to V (Fig. 3A; Supplemental Table S1): Group I (LMM/Rev1, 2, 3, and 4) had an insertion of 5–7 nt, thereby restoring the potential for four or five Watson–Crick base-pairings of the M-R duplex and simultaneously converting a bulge of C−8 into a large loop of 7 or 8 nt. Group II (LMM/Rev5, 6, 7, 11, 12, and 14) had a 3-nt insertion of GAU and a deletion of 1–5 nt, restoring the potential for three or four Watson–Crick base-pairings of the M-R duplex and producing a protruding loop of 1–4 nt instead of a C−8 bulge. Group III (LMM/Rev8 and 13) had another 3-nt insertion of UAU and a 3-nt deletion (either C−8C−5U−4 or U−7U−6C−5), restoring the potential for two U-A base-pairings of the M-R duplex and leaving four nucleotides unpaired at the site of a C−8 bulge. Group IV (LMM/Rev9 and 10) had a 3-nt deletion of C−5U−4A−3 and a 4-nt insertion (either GUAU or GAAU) at the site of the deletion, restoring the potential for two or three (interrupted by a new bulge of A−4) Watson–Crick base-pairings of the M-R duplex and creating a loop of 3 or 5 nt above it. Finally, group V (LMM/Rev15, 16, and 17) had a 6-nt deletion (C−8U−7U−6C−5U−4A−3) and an insertion of 4 or 5 nt at the site of the deletion, thereby restoring the potential for two or three U-A base-pairings of the M-R duplex, leaving zero to two nucleotides unpaired within the M-R duplex, and creating a new C-G base-pairing above it.
To address the roles of these 17 novel sequences in LMM RNA replication, we constructed 17 derivatives of LMM with all of these novel sequences and estimated the specific infectivities of their RNAs. In all cases, the infectivities were drastically increased (by approximately three orders of magnitude) to levels resembling WT (Fig. 3B, infectivity). This result indicated that the acquisition of one of these 17 novel R-strand sequences was sufficient for LMM RNA replication. However, the five groups of LMM derivatives displayed a spectrum of focus morphology (Fig. 3B, representative foci) that was correlated with their levels of JEV RNA production and NS1 protein accumulation in RNA-transfected cells (Fig. 3C). We found that all six members of groups I and III displayed the lowest level of RNA replication, three (LMM/Rev5, 6, and 7) of the six members of group II and all five members of groups IV and V showed an intermediate level, and the remaining three (LMM/Rev11, 12, and 14) members of group II displayed the highest level of replication. Pairwise comparisons between groups or between members within a single group revealed that two factors other than the nature of the TDCS motif, the length of the loop immediately upstream of the R strand and the nucleotide composition of the R strand, were critical for achieving maximal RNA replication.
We then characterized these 17 LMM derivatives further by using one-step growth assays to assess the yield of progeny virions during a single round of infection (Fig. 3D). The results of these assays indicated that in all 17 cases, the rate of viral one-step growth was consistent with the level of RNA replication we had just observed. However, the LMM/Rev1 harboring the largest insertion of 7 nt immediately upstream of the 3′-end CU exhibited a sharp increase in the yield of progeny virions at 12 h post-infection and over the next 36 h (Fig. 3D, LMM/Rev1), indicating some degree of genetic instability. This effect was more evident when all 17 LMM derivatives were passaged twice on naive BHK-21 cells. Of the 17 passaged LMM derivatives, 10 (LMM/Rev2, 5, 6, 7, 11, 12, 14, 15, 16, and 17) appeared to be genetically stable, and the remaining seven (LMM/Rev1, 3, 4, 8, 9, 10, and 13) were unstable to varying degrees; nevertheless, the potential for three or four base-pairings of their restored M-R duplex was still maintained (Table 1). In agreement with our observations regarding phenotypic reversion to rapid viral growth, LMM/Rev1 was the most unstable derivative, in which a large loop of 7 or 8 nt had been removed, but the potential for four base-pairings of the restored M-R duplex was still maintained.
TABLE 1.
Genetic stability of 17 novel sequences acquired in LMM-derived pseudorevertants after two serial passages on BHK-21 cellsa
Mutant MLR-derived compensatory mutations
Sequencing of 26 and 33 independent clones for the 5′- and 3′-terminal regions of the MLR-derived pseudorevertants revealed a striking feature (Fig. 4A): No mutation was found in the reconstructed L-M duplex region of the TDCS, although both strands were mutated by exchanging with their complements to maintain its base-pairing potential. Instead, to our surprise, the unaltered R strand of five nucleotides (A−7G−6G−5A−4U−3) was completely replaced by one of six novel sequences immediately upstream of the 3′-end CU, which have the ability to restore the potential for four to five base-pairings of the M-R duplex that had been disrupted in MLR. Four of the six novel sequences were 5 nt long: UUUUG (MLR/Rev1, 5/33 clones), UUUUA (MLR/Rev2, 6/33 clones), UUCUG (MLR/Rev5, 2/33 clones), and UUCUA (MLR/Rev4, 1/33 clones). The remaining two were 4 nt long: UUUA (MLR/Rev3, 3/33 clones) and UUCU (MLR/Rev6, 16/33 clones).
FIGURE 4.
Functional significance of six novel sequences discovered in MLR-derived pseudorevertants for M-R duplex base-pairing and genome RNA replication. (A) Novel sequences (MLR/Rev1 to 6) acquired in MLR-derived pseudorevertants, and predicted complex motifs of the TDCS formed by WT, MLR, and MLR derivatives containing one of the six novel sequences. The nucleotides acquired in MLR-derived pseudorevertants are shown in black boldface type. Fractions in parentheses are the number of clones discovered that contain the particular sequence/the total number of independently picked clones that were sequenced. (B) Specific infectivities and representative focus morphologies obtained 4 d after transfection of BHK-21 cells with the synthetic RNAs transcribed from the reconstructed JEV cDNAs, each containing one of the six novel sequences; and (C) their levels of JEV RNA production and JEV NS1 protein accumulation at 20 and 24 h post-transfection, respectively. (D) One-step growth kinetics of the recombinant viruses originating from the corresponding synthetic RNAs, as indicated. Experimental procedures were performed as described for Figure 3. (NT) Not tested.
The functional importance of each of these six novel R-strand sequences in MLR RNA replication was analyzed by generating six derivatives of MLR reconstructed with all of these novel sequences, and then measuring their specific infectivities. In all cases, the specific infectivities were dramatically increased (by approximately three orders of magnitude) to levels resembling WT (Fig. 4B, infectivity). Again, this result clearly indicated that acquisition of one of these six novel sequences was sufficient for MLR RNA replication. As observed previously for the 17 LMM derivatives (Fig. 3), differences were noted in the sizes of the infectious foci (Fig. 4B, representative foci), the levels of JEV RNA production and NS1 protein accumulation (Fig. 4C), and the rates of viral one-step growth (Fig. 4D); all of which were proportional to one another. In particular, two (MLR/Rev4 and 5) of the six MLR derivatives had a replication efficiency that was lower than that of the other four; in both cases, the efficiency was enhanced by a single C−5 → U−5 substitution in the middle of the R strand, converting the G−80–C−5 base-pairing of the M-R duplex into G−80–U−5 base-pairing (cf. MLR/Rev4 with 2 and MLR/Rev5 with 1). A similar increase was also observed after deleting C−5 (cf. MLR/Rev4 with 3), A−3 (cf. MLR/Rev4 with 6), or G−3 (cf. MLR/Rev5 with 6) of the R strand, thereby leaving either A−78 or U−82 of the M strand unpaired.
In conclusion, all the replicating RNAs derived from a pair of JEV TDCS mutants, LMM and MLR, were able to restore their RNA replication by acquiring one of a large number of novel sequences of various sizes. In all the cases we sequenced here, these novel sequences were introduced into the R strand, restoring the base-pairing potential of the M-R duplex that had been disrupted by introducing either an R → M strand substitution (LMM) or an L ↔ M strand exchange (MLR), in the presence of the L-M duplex base-pairing that was still intact (LMM) or restored (MLR), respectively. Overall, these results support our hypothesis that in accordance with the importance of the 3′SL structure in RNA replication of other flaviviruses (Brinton et al. 1986; Men et al. 1996; Shi et al. 1996; Zeng et al. 1998; Yu and Markoff 2005), base-pairing of the M-R duplex in JEV TDCS is essential for genomic RNA replication. In addition, the nucleotide composition of the R strand had a significant impact on the level of JEV RNA replication.
A 5′–3′ long-range base-pairing of the L-M duplex in JEV TDCS is a prerequisite for genomic RNA replication
We next analyzed a panel of compensatory mutations found in the replicating RNAs derived from two other mutants, LRM and MMR.
Mutant LRM-derived compensatory mutations
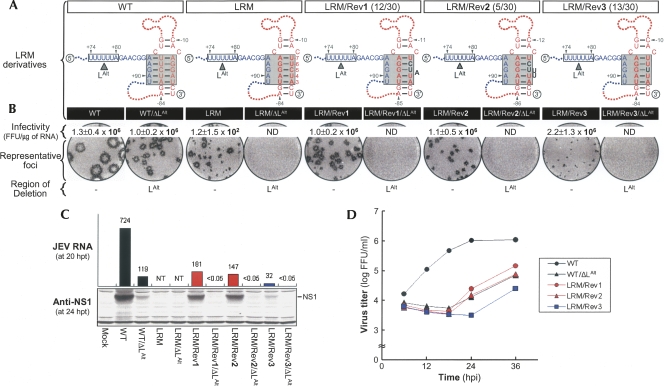
No mutation was found in a pool of 20 independent clones containing the 5′-terminal region of LRM-derived pseudorevertants (Fig. 5A). However, three genetic changes were identified in 30 independent clones containing the 3′-teminal region; to our surprise, all were localized to the R strand of the reconstructed 3′M-R duplex that had been restored by an M ↔ R strand exchange, not to the 5′L strand that had lost its base-pairing potential with the 3′M strand (Fig. 5A). Specifically, in 12 of 30 independent clones (LRM/Rev1), U−6C−5 was replaced with CUA, thereby exchanging the position of two base-pairings (G-C for G−79–U−6 and G-U for G−80–C−5) within the reconstructed 3′M-R duplex and simultaneously creating a new bulge of A below those two base-pairings. Similarly, five of 30 clones (LRM/Rev2) had a U−6C−5 → CUUU replacement, thereby exchanging the position of two base-pairings (G-C for G−79–U−6 and G-U for G−80–C−5) within the reconstructed 3′M-R duplex and simultaneously creating a new loop of UU below those two base-pairings. The remaining 13 clones (LRM/Rev3) had a C−5 → U substitution, converting G−80–C−5 base-pairing of the reconstructed 3′M-R duplex into G-U base-pairing.
FIGURE 5.
Functional significance of three novel sequences discovered in LRM-derived pseudorevertants for base-pairing between the M strand of 3′M-R duplex and an alternative 5′L (5′LAlt) strand and for genome RNA replication. (A) Novel sequences (black boldface type) acquired in LRM-derived pseudorevertants. Also illustrated schematically are predicted complex motifs of the TDCS formed by each RNA in the presence (WT, LRM, LRM/Rev1, LRM/Rev2, and LRM/Rev3) or absence (WT/ΔLAlt, LRM/ΔLAlt, LRM/Rev1/ΔLAlt, LRM/Rev2/ΔLAlt, and LRM/Rev3/ΔLAlt) of the 5′LAlt strand (shown as a box outlined by a solid line). Fractions in parentheses are the number of clones discovered that contain the particular sequence/the total number of independently picked clones that were sequenced. (B) Specific infectivities and representative focus morphologies obtained 4 d after transfection of BHK-21 cells with the synthetic RNAs transcribed from each cDNA, as indicated, and (C) their levels of JEV RNA production and JEV NS1 protein accumulation at 20 and 24 h post-transfection, respectively. (D) One-step growth kinetics of the recombinant viruses derived from the corresponding synthetic RNAs, as indicated. Experimental procedures were performed as described for Figure 3. (ND) Not detected; (NT) not tested.
To assess the functional significance of these three genetic changes in LRM RNA replication, we generated three derivatives of LRM with each of these genetic changes and determined the specific infectivities of their RNAs. In all cases, the specific infectivities were strikingly increased (by approximately four orders of magnitude) to levels resembling WT (Fig. 5B, infectivity). These results indicated that acquisition of one of these three genetic changes in the R strand of LRM was sufficient to restore LRM RNA replication. The most consistently observed of the three changes was a single C−5 → U substitution, converting the potential for G−80–C−5 base-pairing in the middle of the reconstructed 3′M-R duplex to G−80–U−5 (Fig. 5A). We used site-directed mutagenesis to assess the importance of U−5 in LRM RNA replication: Substitution with either A (LRM/C−5A) or G (LRM/C−5G), disrupting the potential for the G−80–C−5 base-pairing, failed to restore WT infectivity (Supplemental Fig. S1). On the other hand, analyses of focus morphology (Fig. 5B, representative foci) and the levels of JEV RNA production and NS1 protein accumulation (Fig. 5C) showed that the RNA replication efficiency of the three LRM derivatives was variable. LRM/Rev3, harboring a single C−5 → U substitution, displayed a lower level of RNA replication than did the other two LRM derivatives bearing additional genetic changes (Fig. 5C, cf. LRM/Rev3 with 1 or 2). In all three cases, the levels of RNA replication were proportional to their rates of viral one-step growth (Fig. 5D).
In seeking to understand how these three LRM-derived compensatory mutations contribute to JEV TDCS organization and genomic RNA replication, we postulated that such modifications might be able to alter the position (or angle) of the reconstructed 3′M-R duplex that had been restored by introducing an M ↔ R strand exchange, so that an alternative L (LAlt) strand of five nucleotides (U+76U+77U+78U+79A+80), located 6 nt upstream of the original 5′L strand, could base-pair with its complements in the M strand (U−82A−81G−80G−79A−78) of the adapted 3′M-R duplex, thereby restoring the 5′–3′ long-range interaction of the 5′LAlt strand with the M strand of the adapted 3′M-R duplex (Supplemental Fig. S2). To test this possibility, we constructed an additional panel of five mutants (WT/ΔLAlt, LRM/ΔLAlt, LRM/Rev1/ΔLAlt, LRM/Rev2/ΔLAlt, and LRM/Rev3/ΔLAlt) from which U+74U+75U+76U+77U+78U+79A+80 was deleted, eliminating the base-pairing potential between the 5′LAlt strand and the M strand of the adapted 3′M-R duplex in each construct (Fig. 5A; Supplemental Fig. S2). We included two additional nucleotides (U+74U+75) immediately upstream of the 5-nt LAlt strand because three (U+74U+75A+83) of the five nucleotides (U+74U+75G+81A+82A+83) present after a deletion of the 5-nt LAlt strand were still predicted to be able to base-pair with their complements (U−82G−79A−78) within the M strand of the adapted 3′M-R duplex in LRM and its three derivatives.
The functionality of the 5′LAlt strand in LRM RNA replication was analyzed by determining the specific infectivities of the synthetic RNAs transcribed from each corresponding deletion mutant (Fig. 5B, infectivity). WT/ΔLAlt had a specific infectivity of 1.0 ± 0.2 × 106 FFU/μg, indistinguishable from WT infectivity (1.3 ± 0.4 × 106 FFU/μg), indicating that a 7-nt deletion containing the 5′LAlt strand did not alter the replication competence. In contrast, the remaining four ΔLAlt-harboring mutants (LRM/ΔLAlt, LRM/Rev1/ΔLAlt, LRM/Rev2/ΔLAlt, and LRM/Rev3/ΔLAlt) had no detectable infectivity, indicating that the 5′LAlt strand in LRM and its three derivatives is essential for RNA replication, probably replacing the role of the original 5′L strand in a 5′–3′ long-range interaction with the M strand of the adapted 3′M-R duplex. In addition, the average size of the foci produced by WT/ΔLAlt was smaller than that of WT (Fig. 5B, representative foci), paralleling its levels of JEV RNA production and NS1 protein accumulation (Fig. 5C) and its rates of viral one-step growth (Fig. 5D) and indicating a role for the 5′LAlt strand in regulating the level of RNA replication.
Mutant MMR-derived compensatory mutations
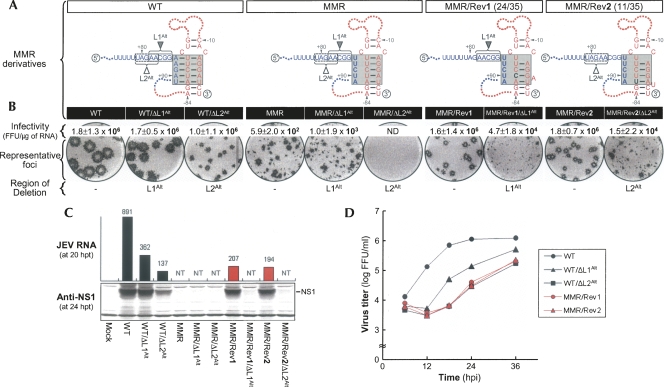
We sequenced 21 and 35 independent clones for the 5′- and 3′-terminal regions of MMR-derived pseudorevertants, respectively. Surprisingly, we found a set of two point mutations in the M strand of the 3′M-R duplex, despite the fact that only the L strand had been mutated by substitution of the M strand (Fig. 6A). Specifically, 24 of 35 independent clones (MMR/Rev1) contained a U−81 → C substitution, maintaining the potential for four Watson–Crick base-pairings of the 3′M-R duplex and creating a new bulge of A−4. The remaining 11 clones (MMR/Rev2) had an A−82 → U substitution, maintaining the potential for one G-U plus three Watson–Crick base-pairings of the 3′M-R duplex and creating a new bulge of U−3. When we reconstructed two derivatives of MMR with either of these point mutations and measured the specific infectivities of their RNAs, we found that the specific infectivities were remarkably increased (by approximately four orders of magnitude) to levels resembling WT (Fig. 6B, infectivity), showing that either a U−81 → C or A−82 → U substitution was required for MMR RNA replication. Based on additional site-directed mutagenetic analyses, we found that a substitution of U−81 with either A (MMR/U−81A) or G (MMR/U−81G) abolished or failed to restore WT infectivity, respectively (Supplemental Fig. S3). In addition, an A−82 → G substitution (MMR/A−82G) also failed to restore WT infectivity, whereas an A−82 → C substitution (MMR/A−82C) restored WT infectivity, but with reduced RNA replication efficiency (Supplemental Fig. S4). Thus, there was a strict requirement in terms of the nucleotide composition, particularly at the base of the M strand of the 3′M-R duplex in MMR.
FIGURE 6.
Functional significance of two point mutations discovered in MMR-derived pseudorevertants for base-pairing between the M strand of 3′M-R duplex and an alternative 5′L (5′L1Alt or 5′L2Alt) strand and for genome RNA replication. (A) Point mutations (black boldface type) acquired in MMR-derived pseudorevertants. Also illustrated schematically are predicted complex motifs of the TDCS formed by each RNA in the presence (WT, MMR, MMR/Rev1, and MMR/Rev2) or absence (two sets of three: WT/ΔL1Alt, MMR/ΔL1Alt, and MMR/Rev1/ΔL1Alt, and WT/ΔL2Alt, MMR/ΔL2Alt, and MMR/Rev2/ΔL2Alt) of either the (box outlined with a solid line) 5′L1Alt or (box outlined with a dotted line) 5′L2Alt strand. Fractions in parentheses are the number of clones discovered that contain the particular sequence/the total number of independently picked clones that were sequenced. (B) Specific infectivities and representative focus morphologies obtained 4 d after transfection of BHK-21 cells with the synthetic RNAs transcribed from each cDNA, as indicated, and (C) their levels of JEV RNA production and JEV NS1 protein accumulation at 20 and 24 h post-transfection, respectively. (D) One-step growth kinetics of the recombinant viruses derived from the corresponding synthetic RNAs, as indicated. Experimental procedures were performed as described for Figure 3. (ND) Not detected; (NT) not tested.
Next, we sought to analyze the functional role of two MMR-derived compensatory mutations, U−81 → C and A−82 → U. As indicated by the results for the three LRM derivatives (Fig. 5), a likely explanation for our results is that either a single C−81 or U−82 nucleotide acquired at the base of the M strand of the 3′M-R duplex might adjust the position (or angle) of the 3′M-R duplex, so that its M strand could base-pair with a 5′LAlt strand available next to the original 5′L strand, at the expense of losing the potential for either a single U−81–A−4 or A−82–U−3 base-pairing of the 3′M-R duplex; this adjustment would restore the 5′–3′ long-range interaction of a 5′LAlt strand with the M strand of the adjusted 3′M-R duplex. Based on computer prediction and visual inspection of the nucleotide sequence adjacent to the original 5′L strand, we predicted one 5-nt 5′L1Alt strand (A+82A+83C+84G+85G+86) for C−81-harboring MMR/Rev1 and another 5-nt 5′L2Alt strand (U+79A+80G+81A+82A+83) for U−82-harboring MMR/Rev2, thereby restoring the potential for at least three or four base-pairings for the L-M duplex (Supplemental Fig. S5). To examine the role of either the 5′L1Alt or 5′L2Alt strand, we generated two sets of six deletion mutants: (1) one set (WT/ΔL1Alt, MMR/ΔL1Alt, and MMR/Rev1/ΔL1Alt), from which A+82A+83C+84G+85G+86 was deleted, eliminating the three base-pairing potential between the 5′L1Alt strand (A+82A+83G+85) and the M strand (C−81U−79U−78) of the adapted 3′M-R duplex in each corresponding construct, and (2) the other (WT/ΔL2Alt, MMR/ΔL2Alt, and MMR/Rev2/ΔL2Alt), from which U+79A+80G+81A+82A+83 was deleted, eliminating the four base-pairing potential between the 5′L2Alt strand (A+80G+81A+82A+83) and the M strand (U−82U−81C−80U−79) of the adapted 3′M-R duplex in each corresponding construct (Fig. 6A; Supplemental Fig. S5).
RNA replication was again monitored by estimating the specific infectivity of each RNA transcript derived from the corresponding deletion mutant (Fig. 6B, infectivity). WT infectivity (1.8 ± 1.3 × 106 FFU/μg) was not altered by introducing a deletion of either the 5′L1Alt or 5′L2Alt strand, showing that neither the 5′L1Alt nor the 5′L2Alt strand is essential for WT RNA replication. As expected, MMR infectivity (5.9 ± 2.0 × 102 FFU/μg) was nearly unchanged or completely abolished by deleting either the 5′L1Alt (MMR/ΔL1Alt, 1.0 ± 1.9 × 103 FFU/μg) or 5′L2Alt (MMR/ΔL2Alt, no detectable infectivity) strand. The infectivities of MMR/Rev1/ΔL1Alt and MMR/Rev2/ΔL2Alt, however, were invariably approximately two orders of magnitude lower than those of MMR/Rev1 and MMR/Rev2, respectively; their foci in RNA-transfected cells were noticeably heterogeneous. Based on the magnitude of the reduction in RNA infectivity and the extent of heterogeneity in focus morphology, we concluded that the 5′L1Alt and 5′L2Alt strands were essential for the RNA replication of MMR/Rev1 and MMR/Rev2, respectively, and that the replicating RNAs must contain one or more second-site reverting mutations. This conclusion was further substantiated by introducing the R-strand sequence at the site of either the L1Alt or L2Alt deletion in all of the six ΔL1Alt- or ΔL2Alt-harboring mutants. These alterations were found to be sufficient to produce phenotypic reversion to high RNA infectivity and homogeneous focus morphology (Supplemental Fig. S6). The nucleotide composition of the L-M duplex was also shown to have a significant impact on the level of JEV RNA replication in the cells transfected with the four replication-competent mutant RNAs (WT/ΔL1Alt, WT/ΔL2Alt, MMR/Rev1, and MMR/Rev2), as evidenced by the homogeneity of their foci, which varied in size (Fig. 6B, representative foci), in parallel with their levels of JEV RNA production and NS1 protein accumulation (Fig. 6C) and their rates of viral one-step growth (Fig. 6D).
An identical compensatory mutation derived from both LLR and LRR mutants
We did not observe any direct reversion of replicating JEV RNAs from BHK-21 cells transfected with either LLR or LRR. In both cases, however, revertants were detected after three serial passages of culture supernatants collected from the transfected cells on naive BHK-21 cells. Immunoblotting with a mouse JEV-specific hyperimmune antiserum showed that JEV-specific viral proteins were produced in the cells infected with each of these two revertants at passage 3 (data not shown). Sequencing of more than 20 independent clones for each of the 5′- and 3′-terminal regions of these two revertants at passage 3 revealed one common reversion: Conversion of the pre-existing 5-nt R strand (A−7G−6G−5A−4U−3) to a novel 5-nt R strand (U−7U−6U−5U−4A−3), restoring the potential for all five base-pairings of the 3′M-R duplex (Fig. 7A, LLR/Rev and LRR/Rev). Replacement of A−7G−6G−5A−4U−3 with U−7U−6U−5U−4A−3 in the context of LLR (LLR/Rev) and LRR (LRR/Rev) showed that this replacement was required for RNA replication (Fig. 7B, infectivity). In addition, analyses of focus morphology (Fig. 7B, representative foci) and the levels of JEV RNA production and NS1 protein accumulation (Fig. 7C) suggested that a single A−79 → G substitution might provide a more favorable environment to support a higher level of RNA replication, a conclusion that was further corroborated by a higher rate of viral one-step growth (Fig. 7D). A similar conclusion was reached by comparing MLM with MRM mutants (Fig. 2; data not shown). Also, acquisition of this novel 5-nt R strand (U−7U−6U−5U−4A−3) was previously identified in two other revertants, MLR/Rev2 (Fig. 4) and LRM/Rev3 (Fig. 5). In LRM/Rev3, all three strands are identical to those of LRR/Rev; in MLR/Rev2, two strands are identical to those of LLR/Rev, but the L strand is distinct.
Subsequently, we sought to understand how this novel 5-nt R strand contributes to the RNA replication of LLR and LRR. As indicated previously by the results for the three LRM derivatives (Fig. 5), a likely explanation for our results is that acquisition of this novel 5-nt R strand in LLR and LRR adjusts the position (or angle) of the newly restored 3′M-R duplex, so that its M strand can base-pair with the 5′LAlt strand (U+76U+77U+78U+79A+80) located 6 nt upstream of the original 5′L strand; this adjustment would restore the 5′–3′ long-range interaction of the 5′LAlt strand with the M strand of the adjusted 3′M-R duplex (Supplemental Fig. S7). As observed previously for LRM/Rev3/ΔLAlt (Fig. 5), deletion of the 5′LAlt strand in LLR/Rev (LLR/Rev/ΔLAlt) and LRR/Rev (LRR/Rev/ΔLAlt) completely abolished RNA replication (Fig. 7), substantiating the critical importance of two base-pairing potentials for JEV genome replication: one involving the 3′M-R duplex and the other a 5′–3′ long-range interaction of the 3′M strand with either the 5′L strand or its alternative. Alternatively, it is possible that the loss of RNA replication in LLR/Rev/ΔLAlt and LRR/Rev/ΔLAlt reflects an additive detrimental effect produced by introducing a deletion of the 5′LAlt strand in the context of LLR/Rev and LRR/Rev, since each of the three mutants (namely, WT/ΔLAlt, LLR/Rev, and LRR/Rev) was replication-competent, but less efficient than WT.
In summary, analyses of all the replicating RNAs originating from a panel of four JEV TDCS mutants (namely, LRM, MMR, LLR, and LRR) support our hypothesis that the 5′–3′ long-range interaction of the 5′L (or its alternative) strand with the M strand of the 3′M-R duplex (or its reconstructed version with an appropriate compensatory mutation) in the JEV TDCS is required for genomic RNA replication. Interestingly, all the compensatory mutations derived from one particular TDCS mutant were found in one particular strand of either the M or R of the TDCS, regardless of the strand(s) we mutated, and in each case, they resulted in a modification of the 3′M-R duplex. These results, therefore, indicate that each of the compensatory mutations potentially renders the M strand of the adapted 3′M-R duplex capable of base-pairing with a 5′LAlt strand located upstream of the original 5′L strand, thereby restoring 5′–3′ long-range RNA–RNA interaction and the potential for two duplex base-pairings. The position of the 5′LAlt strand appears to be determined by the nucleotide composition of the adapted 3′M-R duplex, which plays a regulatory role in the level of RNA replication.
The TDCS is conserved in all JEV isolates; a TDCS homolog is present in other flaviviruses
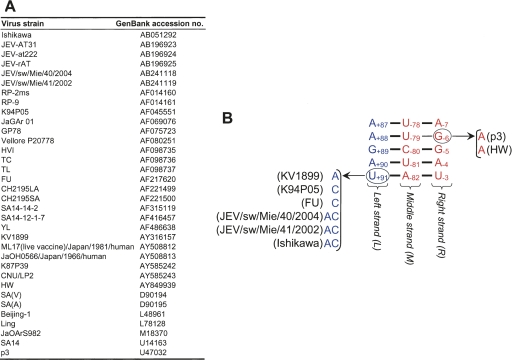
Examination of the 34 full-length JEV genomes currently available from GenBank showed that all 34 sequences were able to form the L-M and M-R duplexes of the TDCS (Fig. 8). Of the 34 genomes, 26 conserved the primary sequences of the TDCS at all 15 nucleotide positions, which are predicted to form nine Watson–Crick base-pairs and one U−79–G−6 base-pair. Two of the JEV genomes had a G−6 → A−6 substitution that would convert the U−79–G−6 base-pairing to U−79–A−6. The remaining six had a single change in U+91 at the end of the L strand, to either A, C, or AC, which would affect the U+91–A−82 base-pairing. Thus, in all 34 full-length JEV genomes in the database, the primary sequences and two duplex base-pairing potentials of the TDCS are conserved. This analysis supports the hypothesis that a complex RNA motif defined by the TDCS has been evolutionally conserved in JEV genome replication.
FIGURE 8.
Phylogenetic conservation in the TDCS of JEV. (A) GenBank accession numbers of 34 full-length JEV genomes used in the sequence alignments. (B) Schematic presentation of the JEV TDCS. The nucleotide sequences and relative locations of three (L, M, and R) strands of the TDCS in the genome are shown. Also presented are potential base-pairings between the L and M strands and between the M and R strands that can occur in other JEV strains. Sequence alignments of the 34 full-length JEV genomes were produced using ClustalX (Thompson et al. 1997), and the resulting output file was analyzed to predict the effect of each variant on TDCS formation. Sequences that differ from the consensus are circled.
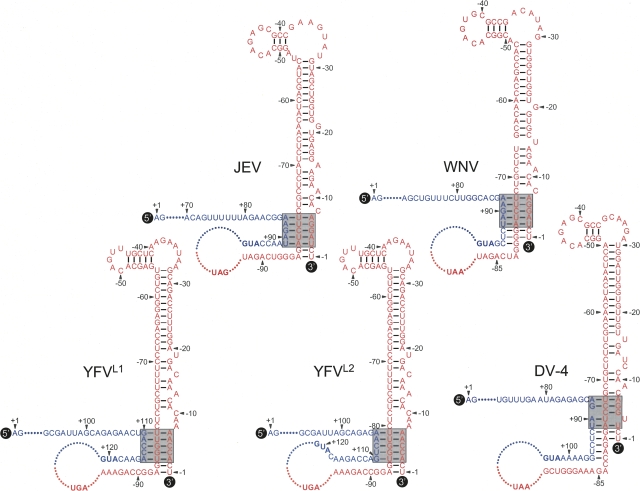
The complex RNA motif of the JEV TDCS is also phylogenetically conserved among other mosquito-borne flaviviruses, including West Nile, yellow fever, and dengue viruses (Fig. 9). Examination of the West Nile virus genome showed a single TDCS identical to that of JEV. For the yellow fever virus genome, we predicted two alternative forms (YFVL1 and YFVL2) of a TDCS homolog analogous to that of JEV, depending on its choice of L strand. For both forms, all 15 nucleotide positions were predicted to form 10 base-pairings, either nine Watson–Crick and one G-U (YFVL1) or eight Watson–Crick and two G-U (YFVL2), but its nucleotide composition was distinct from that of JEV. For the dengue virus genome, all four serotypes shared a single TDCS, homologous to the JEV TDCS in 13 of 15 nucleotide positions, predicting eight Watson–Crick base-pairings. Further analyses suggested that for West Nile and dengue viruses, but not JEV and yellow fever virus, three or five additional base-pairings between the L and M strands below the predicted TDCS might also be involved in its L-M duplex.
FIGURE 9.
JEV TDCS is highly conserved among other flaviviruses, including West Nile virus (WNV), yellow fever virus (YFV), and dengue virus serotype 4 (DV-4). The predicted nucleotide sequences and relative locations of the three (L, M, and R) strands of the TDCS in the various viral genomes are shown, as described in Figure 2. (Shaded boxes) Also highlighted are the potential base-pairing patterns of the TDCSs. The translation initiation codon (AUG) and stop codon (UAG for JEV, UAA for WNV and DV-4, and UGA for YFV) are shown in boldface type. The yellow fever virus genome is predicted to form two alternative forms (YFVL1 and YFVL2) of a TDCS homolog, analogous to that of JEV, depending on the choice of L strand: either L1 (G+110A+111C+112C+113A+114) or L2 (A+106A+107C+108U+109G+110). The GenBank accession numbers of the four flaviviruses analyzed are AY585243 (JEV CNU/LP2), M12294 (WNV), X03700 (YFV 17D), and M14931 (DV-4).
DISCUSSION
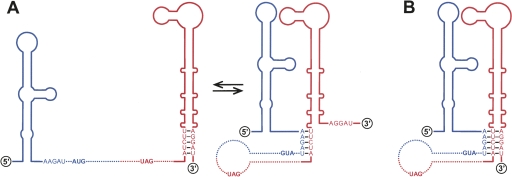
We describe here the first identification of a complex RNA motif containing a TDCS in the positive-strand RNA genome of JEV and demonstrate that this motif plays a central role in viral replication. We have (1) localized three 5-nt strands that participate in the formation of the TDCS; (2) demonstrated that the TDCS has a complicated topology, requiring a 5′L strand or its alternative upstream of the translation initiation codon AUG adjacent to the 5′-end of the genome and a 3′M-R duplex proximal to the 3′-end; (3) provided genetic evidence that complementarity among the three strands of the TDCS, but not the primary sequences, is required for competent RNA replication; (4) shown that the nucleotide composition of the TDCS regulates the level of RNA replication; and (5) predicted that a TDCS homolog is conserved among other mosquito-borne flaviviruses. Based on our findings and previous reports, we propose that the TDCS is a key RNA element participating in the formation of a putative RNA complex that can regulate a key step(s) of the flavivirus replication cycle. This complex (Fig. 10) brings the flavivirus-conserved 3′SL into proximity with the sequence immediately upstream of the translation initiator AUG, and it allows the 3′SL to communicate with a flavivirus-conserved 5′SL, a stem–loop structure predicted at the 5′-end prior to the translation initiator.
FIGURE 10.
Two working hypotheses (A,B) for the TDCS-mediated cyclization and replication of JEV genomic RNA. The predicted stem–loop structures for the (blue) 5′-terminal (5′SL) and (red) 3′-terminal (3′SL) regions of the genome. Also indicated are the relative locations of the L (5′-A+87A+88G+89A+90U+91), M (5′-A−82U−81C−80U−79U−78), and R (5′-A−7G−6G−5A−4U−3) strands of the TDCS and the potential base-pairing patterns. (Boldface type) The translation initiation codon (AUG) and stop codon (UAG). The TDCS brings the flavivirus-conserved 3′SL upstream of the translation initiator AUG adjacent to the 5′-end and renders the 3′SL capable of communicating with the flavivirus-conserved 5′SL. See the text for a detailed explanation.
A striking feature that has emerged from the present study is that the 3′SL of JEV is involved in the formation of a complex RNA motif, the TDCS. An essential component of this motif is three 5-nt single-stranded strands, the L strand (A+87A+88G+89A+90U+91) located 86 nt downstream from the 5′-end of the genome and the M (A−82U−81C−80U−79U−78) and R (A−7G−6G−5A−4U−3) strands located 77 and 2 nt, respectively, upstream from the 3′-end; the M and R strands correspond to the base of the 3′SL. These three strands are predicted to be organized into a RNA complex by base-pairings in an antiparallel configuration, with the L strand being complementary to the M strand, and the same M strand also complementary to the R strand. Overall, our molecular genetic studies offer compelling evidence that JEV RNA replication requires the presence of both L-M and M-R duplexes of the TDCS motif. Furthermore, our sequence analysis of 34 JEV isolates strongly supports the functional importance of this motif. In addition, the high degree of conservation of this motif in the West Nile, yellow fever, and dengue viruses as well as JEV supports its functional importance in flavivirus RNA replication.
Our search for compensatory mutations capable of restoring the replication of each of the six JEV TDCS mutants (LMM, MLR, MMR, LRM, LLR, and LRR) indicated that all the compensatory mutations that originated from a single mutant were found in one particular strand, either M or R, of the TDCS, regardless of the strand(s) we mutated. In the case of LMM and MLR, all 17 (LMM) and six (MLR) compensatory mutations were found in the R strand, as expected, restoring the interaction of the M with R strands that had been disrupted by introducing either an R → M strand substitution (LMM) or an L ↔ M strand exchange (MLR). In the case of MMR and LRM, on the other hand, no compensatory mutations were found at their L strands; instead, to our surprise, all three LRM-derived compensatory mutations containing a single C−5 → U substitution were present in the middle of the R strand, and the two MMR-derived compensatory mutations were localized at the base of the M strand. In these two cases, each complementary mutation rendered the M strand of the modified 3′M-R duplex capable of interacting with one of three 5′LAlt strands. A similar finding was observed for LLR and LRR, replacing the original R strand with a novel R strand of the same length, rendering the M strand of the newly formed 3′M-R duplex capable of interacting with the 5′LAlt strand. Why these compensatory mutations occur in one particular strand of the TDCS is not known; understanding the relevant mechanisms should provide new insight into the functional importance of newly identified TDCS in RNA replication.
The relative location and complementarity of the three strands of the JEV TDCS provide new insights into the molecular events occurring during the process of RNA synthesis. We have now shown that a short stretch of nucleotides in either the 5′L strand or its alternative can interact with its complementary sequence in the M strand of the 3′M-R duplex, mediating the 5′–3′ long-range RNA–RNA interaction and genome cyclization. In many other mosquito-borne flaviviruses, another pair of complementary sequences (5′CS and 3′CS) has previously been shown to contribute to genome cyclization and replication (Hahn et al. 1987; Men et al. 1996; You and Padmanabhan 1999; Khromykh et al. 2001; You et al. 2001; Bredenbeek et al. 2003; Corver et al. 2003; Lo et al. 2003; Alvarez et al. 2005). In JEV, the 5′CS (nucleotides U+136 to G+143) is located 37 nt downstream from the translation initiator AUG, opposite the 5′L strand or its alternative, in the C protein-coding region; the 3′CS (nucleotides C−111 to A−104) is found 19 nt upstream of the 3′M-R duplex (Fig. 2A). Based on our findings and previous reports, at least two pairs of complementary sequences, 5′L/3′M and 5′CS/3′CS, are likely to be involved in the JEV genome cyclization that is a prerequisite to RNA replication, although the requirement for 5′CS/3′CS complementarity in JEV genome replication remains to be demonstrated experimentally. This finding is consistent with a recent report that dengue virus has two pairs of complementary sequences, analogous to those of JEV, that are required for genome cyclization and replication (Alvarez et al. 2005).
The long-distance interaction between the 5′L (or 5′LAlt) strand and the M strand of the 3′M-R duplex brings the flavivirus-conserved 3′SL at the 3′-end of the genome close to the flavivirus-conserved 5′SL at the 5′-end prior to the translation initiator. This juxtaposition could allow the 5′SL and 3′SL to be organized into a putative RNA complex, thereby providing a unique opportunity to facilitate a variety of molecular interactions, such as RNA–RNA, RNA–protein, and protein–protein. Thus, this RNA complex may regulate a key step(s) of the replication cycle, such as RNA synthesis and translation of the genome. Involvement of the TDCS-mediated RNA complex in RNA synthesis is consistent with reports that dengue virus RNA synthesis, both in vitro (You and Padmanabhan 1999; You et al. 2001) and in vivo (Filomatori et al. 2006), is enhanced by the 5′–3′ long-range interaction only in the presence of an RNA element named SLA, equivalent to JEV 5′SL. An explanation for the requirement for SLA in the promotion of dengue virus RNA synthesis has been provided by most recent visual and biochemical analyses of dengue virus cis-acting RNA elements, proposing that the NS5 initially recognizes an intact SLA located at the 5′-end of the genome and then moves to the site of initiation at the 3′-end via 5′–3′ long-range RNA–RNA interactions (Alvarez et al. 2005; Filomatori et al. 2006). Given that the crystal structure of dengue virus NS5 has demonstrated an RNA cap methyltransferase and a novel GTP-binding site at its N terminus (Egloff et al. 2002) and the RNA-dependent RNA polymerase at its C terminus, binding of the NS5 methyltransferase domain to the cap structure at the 5′-end of the genome and of the RNA-dependent RNA polymerase domain to the SLA immediately downstream from the cap structure is thought to be an important step in the initiation of flavivirus RNA synthesis (Filomatori et al. 2006). It is also conceivable that the TDCS-mediated RNA complex regulates the expression of the virus's single ORF. In this study, we have shown that the 5′L strand and its alternative sequence are always found a few nucleotides upstream of the translation initiator AUG adjacent to the 5′-end of the JEV genome. As a consequence of the interaction of the 5′L (or LAlt) strand with the M strand of the 3′M-R duplex, 95 nt of JEV 5′NTR prior to the translation initiator could form two alternative structures in infected cells, and a conformational RNA switch from a linear structure to a complex structure would interfere with the translation of the viral genome.
The mechanism by which a complex RNA motif of the JEV TDCS acts on RNA replication is still unknown. The most likely explanation is that the M strand is only available for duplex base-pairing interaction with one complementary strand (L or R) at a particular stage of RNA replication (Fig. 10A). In this case, the three strands might be involved in the formation of two alternate structures, with the formation of the L-M duplex preventing the formation of the M-R duplex, or vice versa. If this were the case, one might speculate that the strands could act as a molecular switch, regulating a transition occurring during viral replication. One of the two duplexes would play an essential role at one stage of viral replication, whereas the other would provide a regulatory role, turning off that function at another stage of viral replication. Examples of this scenario have been put forward for the replication of several positive-strand RNA viruses (Goebel et al. 2004; Plant et al. 2005; Zhang et al. 2006a,b; Beerens and Snijder 2007). One of the important challenges for future research is to understand the molecular details of the putative function of the TDCS at different stages of viral replication. Given that the genome of positive-strand RNA viruses initially serves as the template for the translation and subsequent synthesis of genome-length negative-strand RNAs in the form of a structurally linear or functionally circular genome, it is possible that the TDCS could act as a switch, helping to regulate genome cyclization versus linearization, genome translation versus replication, and/or negative-strand versus positive-strand RNA synthesis. This motif might also be associated with an as-yet-unknown signal that targets or retains input genomic RNA in the perinuclear membrane compartments in which RNA synthesis generally takes place or that regulates the packaging of nascent genomic RNAs into progeny virions.
Our findings raise interesting possibilities with regard to the identity of the viral and cellular proteins that potentially interact with the TDCS motif and/or its adjacent sequences within the 3′SL. Identifying and characterizing such proteins would provide a broad perspective on how the TDCS mediates a putative molecular switch between the two alternate base-pairings during flavivirus genome replication. From a virology standpoint, the 3′SL of genomic RNA has been shown to interact with a viral replicase protein NS3, which has three enzymatic activities (an NS2B-required serine protease, an RNA-stimulated nucleoside triphosphatase [NTPase]/RNA helicase, and a 5′-RNA triphosphatase [RTPase]), in association with another viral replicase protein NS5 (Chen et al. 1997; Cui et al. 1998). Both the NTPase and RTPase activities of NS3 are enhanced when NS3 is complexed with NS5 (Kapoor et al. 1995; Cui et al. 1998; Johansson et al. 2001; Yon et al. 2005). Also, NS2A has been reported to interact with the 3′NTR of genomic RNA as well as with NS3 and NS5 at the site of RNA replication (Mackenzie et al. 1998).
From a cell biology point of view, the 3′SL of genomic RNA has been shown to possess binding sites for several cellular proteins, including the eukaryotic translation elongation factor eEF1A (Blackwell and Brinton 1995, 1997; De Nova-Ocampo et al. 2002; Davis et al. 2007), human La autoantigen (De Nova-Ocampo et al. 2002; García-Montalvo et al. 2004), and murine Mov34 protein (Ta and Vrati 2000). Of these, the binding of eEF1A to the 3′SL is particularly intriguing, since this interaction has been shown to modulate RNA replication, enhancing viral translation and repressing negative-strand RNA synthesis, in several positive-strand RNA plant viruses (Zeenko et al. 2002; Matsuda and Dreher 2004; Matsuda et al. 2004; Yamaji et al. 2006); also, the prokaryotic homolog of eEF1A (EF-Tu) has been identified as a functional component of the viral replication complex of several positive-strand RNA bacteriophages (Blumenthal and Carmichael 1979; Brown and Gold 1996; Schuppli et al. 1998, 2000). In West Nile virus, however, the binding of eEF1A to the 3′SL has recently been suggested to facilitate viral negative-strand RNA synthesis, switching a viral genome from translation to RNA synthesis (Davis et al. 2007).
Other cellular proteins have also been shown to bind to the 3′NTR of the positive- and/or negative-strand RNAs of several flaviviruses and are thought to play a significant role(s) in the life cycle of the virus: (1) The La protein from mosquito cells inhibits RNA synthesis (Yocupicio-Monroy et al. 2007); (2) the YB-1 protein identified in BHK-21 cells represses viral translation, mediating an antiviral effect (Paranjape and Harris 2007); and (3) the TIA-1/TIAR proteins, also derived from BHK-21 cells, facilitate RNA synthesis and inhibit stress granule formation, which prevents the shutoff of cellular translation (Li et al. 2002; Emara and Brinton 2007). Although these cellular proteins have been shown to interact with cis-acting genomic RNA sequences and viral proteins, the mechanism(s) by which they influence the choice of translation versus RNA synthesis or positive-strand versus negative-strand RNA synthesis remains an under-explored area of research. Given that the TDCS motif is represented by only 15 nucleotides participating in 10 base-pairings, we speculate that a cellular protein(s) is likely to be involved in recognizing and/or operating a putative molecular switch between the two short alternate base-pairings, in association with one or more of the viral replicase proteins. Further investigation is needed to identify a viral replicase component(s) that is (are) recognizing the TDCS motif and/or its adjacent sequences and to explore the RNA–protein and protein–protein interactions that coordinate the putative molecular switch involving the TDCS motif.
Alternatively, it is possible, although less likely, that each of the three strands of the JEV TDCS motif is simultaneously involved in the formation of a triple helix with its consecutive stacked triplets and that this structure plays an essential role in a particular step(s) during the process of RNA replication (Fig. 10B). One might speculate, however, that such a triple helix would more likely be embedded in other more intricate folds, rather than forming an isolated triple helix. Based on our current knowledge of RNA structure, the formation of such a triple helix could involve the first and second strands forming a double helix as an A-form, and the third strand then docking into the minor groove of that double helix and forming productive hydrogen bonds with appropriate functional groups. This possibility is supported by the fact that in addition to forming well-known double helix structures, nucleic acids of either RNA or DNA have been reported to form triple and quadruple helix complexes under some circumstances (Moser and Dervan 1987; Guschlbauer et al. 1990; Sen and Gilbert 1990; Chastain and Tinoco 1992; Kang et al. 1992; Maher 1992; Rettberg et al. 1999; Mills et al. 2002). Under physiological conditions, a short intramolecular triple-helix RNA structure of three to five consecutive nucleotide triplets within a pseudoknot has been proposed to form in the ribosomal frameshift signal sequence of HIV-1 genomic RNA (Dinman et al. 2002) and the telomerase RNAs of the human and budding yeast Kluyveromyces lactis (Theimer et al. 2005; Shefer et al. 2007). In addition, intramolecular triple-helix DNA structures have been reported in the H-DNA structures that are abundant in the eukaryotic genome, suggesting a role in DNA replication and transcription (Htun and Dahlberg 1988; Htun and Dahlberg 1989; Lyamichev et al. 1989). These examples of natural RNAs and DNAs with folded structures suggest that such a complex tertiary structure could play an important role in other biological processes in viral and cellular systems.
In conclusion, our data demonstrate the existence of a long-range TDCS in the JEV genome that plays an essential role in genome cyclization and replication. Although the function of this highly unusual RNA motif in regulating the JEV life cycle is not yet known, the motif may provide a model system for the study of a regulatable replication mechanism that is dependent on the intrinsic nature of the TDCS. Learning more about the fundamental features of this putative RNA complex and its function in viral replication should increase our understanding of the process of flavivirus RNA replication and how it might be controlled. We suggest that it may be worthwhile to screen other positive-strand RNA viruses, whose genomes are organized into a circular form during RNA replication via a 5′–3′ long-range RNA–RNA interaction and/or whose genomes have a 3′SL-like structure at their 5′ or 3′ terminus, in order to identify other such motifs that might be involved in RNA replication. If complex RNA motifs resembling the TDCS are, indeed, present in other positive-strand RNA viruses, this unique sequence-specific structure would offer an attractive target for prophylactic or therapeutic interventions to partially or completely block the replication of these viruses.
MATERIALS AND METHODS
Cells and antisera
Baby hamster kidney BHK-21 cells were maintained and propagated as described (Yun et al. 2003). A mouse hyperimmune antiserum specific for JEV (catalog no. VR-1259AF) was purchased from the American Type Culture Collection. A rabbit polyclonal antiserum (NS1N) specific for JEV NS1 protein was raised against the glutathione S-transferase-fused N-terminal 166 amino acids of JEV CNU/LP2 NS1 protein (nucleotides 2478–2975). Alkaline phosphatase-conjugated goat anti-rabbit IgG and peroxidase-conjugated goat anti-mouse IgG were purchased from Jackson ImmunoResearch Labs Inc.
Construction of recombinant full-length JEV molecular clones
Standard molecular biology protocols (Sambrook et al. 1989) were used to manipulate DNA fragments and construct recombinant full-length molecular clones for JEV. All JEV mutations were introduced in an infectious cDNA molecular clone of JEV CNU/LP2, pBACSP6/JVFLx/XbaI (Yun et al. 2003), designated herein as “WT.” Nucleotide positions refer to the complete nucleotide sequence of JEV CNU/LP2 (GenBank accession no. AY585243). All mutations were introduced by PCR-based site-directed mutagenesis, and plasmids were purified on CsCl/ethidium bromide density gradients. All PCR-derived fragments were sequenced to confirm the desired mutations and exclude further off-site mutations, and the sequences and integrities of all plasmids were checked by extensive restriction analyses. Details of the cloning procedures for the individual mutants and computer-readable sequence files are available upon request.
In vitro transcription
Plasmids containing WT or mutant JEV cDNAs were digested with XbaI, modified with Mung bean nuclease for linearization, and purified by phenol/chloroform extraction and ethanol precipitation. Run-off RNA transcripts were synthesized in vitro using SP6 RNA polymerase as described (Yun et al. 2003). Trace amounts of [3H]UTP were included in the reaction mixtures to calculate yields. The integrity of the full-length RNA transcripts was verified by 1% agarose gel electrophoresis and ethidium bromide staining. Transcripts were used for transfection without any additional purification.
RNA transfection and measurement of the specific infectivity of RNA transcripts
In vitro-transcribed RNAs were transfected by electroporation into naive BHK-21 cells (Yun et al. 2003). Subconfluent cells were trypsinized, washed three times with ice-cold RNase-free phosphate-buffered saline (PBS), and resuspended at 2 × 107 cells/mL in PBS. In vitro-transcribed RNA (2 μg) was mixed with 400 μL of cell suspension (8 × 106 cells) in a 2-mm-gap cuvette (BTX Inc.) and immediately pulsed five times with 99-μsec pulse length at 980 V using a model ECM 830 electroporator (BTX Inc.). After a 10-min recovery at room temperature, the mixture was diluted to 1 mL with fresh complete medium. The specific infectivity of RNA transcripts was determined by infectious center assay and reported as focus-forming units (FFU) per microgram of RNA. A 100-μL aliquot of the 1-mL electroporation mixture was serially diluted by 10-fold with fresh medium and plated on monolayers of naive BHK-21 cells (5 × 105) in a six-well plate. Cells were incubated for 4–6 h at 37°C with 5% CO2, and the medium was then replaced with minimal essential medium containing 10% fetal bovine serum and 0.5% SeaKem LE agarose (FMC BioProducts). Plates were incubated for 4 d at 37°C with 5% CO2. To visualize the infectious centers of foci, cells were immunostained with the mouse anti-JEV antibody and peroxidase-conjugated goat anti-mouse IgG, and then stained with a peroxidase detection kit (Vector Laboratories, Inc.). Under our optimized experimental conditions, the infectious center assay generated RNA infectivity that was directly proportional to the RNA quantity over a range of 2 to 2 × 10−5 μg.
Real-time quantitative RT-PCR
Total RNA from each population of electroporated cells was isolated with TRIzol reagent (Invitrogen Co.). Total cellular RNA (50 ng) was used for first-strand cDNA synthesis with primers specific for the JEV NS3 region, as well as BHK β-actin RNA for normalizing total RNA levels. JEV and BHK β-actin cDNAs were generated by reverse transcription (RT) for 30 min at 45°C, followed by a 10-min inactivation of reverse transcriptase at 95°C. One-tenth of the reaction mixture was used for PCR. Quantitative analysis for JEV-specific and BHK β-actin-specific cDNAs was performed with the iQ Supermix Quantitative PCR System (Bio-Rad Laboratories) and the iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories), for 45 cycles of a two-step PCR amplification (15 sec at 95°C and 1 min at 60°C). Sequences of the PCR primers (forward and reverse, respectively) and fluorogenic TaqMan probes used for quantification were: JEV RNA, 5′-ATCCAACTCAACCGCAAGTC-3′, 5′-TCTAAGATGGTGGGTTTCACG-3′, 5′-6FAM-CATCTCTGAAATGGGGGCTA-BHQ1-3′ (nt 5837–5856); BHK β-actin, 5′-ACTGGCATTGTGATGGACTC-3′, 5′-CATGAGGTAGTCTGTCAGGTC-3′, 5′-HEX-CCAGCCAGGTCCAGACGCAGG-BHQ2-3′ (Yun et al. 2007). Samples were run in duplicate, and a reaction without cDNA was used to establish baseline fluorescence levels. Data are based on a threshold cycle (C T) in which the signal was higher than that of background. No signal was detected when the reverse transcriptase step was omitted. The 2−ΔΔCT method was used to analyze relative changes in JEV RNA levels from real-time quantitative RT-PCR experiments (Winer et al. 1999; Schmittgen et al. 2000).
Immunoblotting
The experimental procedures are described in detail elsewhere (Yun et al. 2003). In brief, a 50-μL aliquot (4 × 105 cells) of the 1-mL electroporation mixture described above was plated on a six-well plate. At 24 h after transfection, cells were lysed with 200 μL of sample loading buffer (80 mM Tri-HCl at pH 6.8, 2.0% SDS, 10% glycerol, 0.1 M DTT, 0.2% bromophenol blue). One-tenth of the lysate was resolved by electrophoresis on glycine-SDS-12% polyacrylamide gels, electroblotted onto methanol-activated polyvinylidene difluoride membranes, and probed with the anti-NS1 antiserum NS1N (1:1000 dilution). The immunoreactive proteins were visualized with the alkaline phosphatase-conjugated goat anti-rabbit IgG (1:5000 dilution), 5-bromo-4-chloro-3-indolyl-phosphate, and nitroblue tetrazolium.
One-step growth experiments
BHK-21 cells were pre-seeded on 35-mm dishes at a density of 3 × 105 cells/dish for 12 h and then infected with viruses at a multiplicity of infection of 8 FFU/cell for 1 h at 5°C with frequent shaking. After incubation, the cell monolayers were washed three times with PBS to remove unabsorbed virus, then incubated in fresh complete medium at 37°C with 5% CO2. At the indicated time points, supernatants were harvested over the first 36 h after infection, and aliquots were stored at −80°C. At the completion of the experiment, the virus titers were determined by focus assays. For these assays, BHK-21 cells were pre-seeded on six-well plates at a density of 3 × 105 cells/well for 12 h prior to infection. The cell monolayers were infected with a series of 10-fold dilutions of virus (1 mL per well) for 1 h. Following removal of the inocula, the cells were overlaid with minimal essential medium (3 mL per well) containing 10% fetal bovine serum and 0.5% SeaKem LE agarose (FMC BioProducts) and incubated for 4 d at 37°C with 5% CO2. The resulting foci were visualized by fixation with 7% formaldehyde, followed by immunostaining with the mouse anti-JEV antibody and peroxidase-conjugated goat anti-mouse IgG and staining with a peroxidase detection kit (Vector Laboratories, Inc.).
Sequence analysis of the 5′- and 3′-terminal regions of the JEV RNA genome
The 5′-terminal region of the JEV RNA genome was sequenced using the sense primer J8 (5′-gatcggaccgAGAAGTTTATCTGTGTGA), complementary to nucleotides 1–18 (uppercase), and antisense primer BsiWR (5′-gatCGTACGTGATAGTGTCCTCACA), complementary to nucleotides 609–630 (uppercase): The ≈661-bp cDNA amplicon was purified, and the ≈649-bp RsrII–BsiWI fragment of the cDNA amplicons was ligated with the 2693-bp pRS2 vector, which had been previously digested with RsrII (underlined) and BsiWI (underlined). About 20 randomly picked independent clones containing the insert were sequenced. To sequence the 3′-terminal region, we used a 3′-RACE protocol (Yun et al. 2003), ligating synthetic oligonucleotides to the 3′-end of the viral RNA to provide a specific primer-binding site during RT-PCR. 5′-Phosphorylated and 3′-blocked oligonucleotide T (5′-ccagtgttgtggcctgcagggcgaatt) was ligated by T4 RNA ligase to the 3′-end of the viral RNA. The ligated RNA was phenol-extracted, ethanol-precipitated, resuspended in RNase-free H2O, and used for cDNA synthesis using Superscript II reverse transcriptase and oligonucleotide TR (5′-gatgaattcgccctgcaggccacaaca), which is complementary to the oligonucleotide T. First-strand cDNA was amplified using a pair of primers, circleF (5′-gatgagctcCACTCAGGAGATACGAAGA), complementary to nucleotides 10,345–10,363 (uppercase), and oligonucleotide TR. The ≈664-bp cDNA amplicon was purified, and the ≈652-bp SacI–EcoRI fragment of the cDNA amplicon was cloned into a 2773-bp fragment of the pRS2 vector, previously digested with SacI (underlined) and EcoRI (underlined). About 30–40 randomly picked independent clones containing the insert were sequenced.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are very grateful to Charles M. Rice (Rockefeller University) for critical reading of the manuscript and helpful discussions and support for this work. Also, we thank Deborah McClellan (Johns Hopkins University) for editorial assistance. This work was supported by a Molecular and Cellular BioDiscovery Research Program grant (M10401000026-06N0100-02610) funded by the Ministry of Science and Technology (Ministry of Education, Science and Technology) and a Korea Research Foundation grant (KRF-2003-015-E00104) funded by the Korean government (MOEHRD), Republic of Korea.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.993608.
REFERENCES
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N., Snijder E.J. An RNA pseudoknot in the 3′ end of the arterivirus genome has a critical role in regulating viral RNA synthesis. J. Virol. 2007;81:9426–9436. doi: 10.1128/JVI.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell J.L., Brinton M.A. BHK cell proteins that bind to the 3′ stem–loop structure of the West Nile virus genome RNA. J. Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell J.L., Brinton M.A. Translation elongation factor-1 α interacts with the 3′ stem–loop region of West Nile virus genomic RNA. J. Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G.G. RNA replication: Function and structure of Qβ-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P.J., Kooi E.A., Lindenbach B., Huijkman N., Rice C.M., Spaan W.J. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 2003;84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Dispoto J.H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Fernandez A.V., Dispoto J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Brown D., Gold L. RNA replication by Q β replicase: A working model. Proc. Natl. Acad. Sci. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K.W. Comparision of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.S., Monath T.P. Flaviviruses. In: Knipe D.M., et al., editors. Fields virology. 4th ed. Lippincott Williams & Wilkins Publishers; Philadelphia, PA: 2001. pp. 1043–1125. [Google Scholar]
- Calisher C.H., Gould E.A. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 2003;59:1–19. doi: 10.1016/s0065-3527(03)59001-7. [DOI] [PubMed] [Google Scholar]
- Cammisa-Parks H., Cisar L.A., Kane A., Stollar V. The complete nucleotide sequence of cell fusing agent (CFA): Homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology. 1992;189:511–524. doi: 10.1016/0042-6822(92)90575-a. [DOI] [PubMed] [Google Scholar]
- Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Poly(rA) binds poly(rG)·poly(rC) to form a triple helix. Nucleic Acids Res. 1992;20:315–318. doi: 10.1093/nar/20.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Kuo M.D., Chien L.J., Hsu S.L., Wang Y.M., Lin J.H. RNA–protein interactions: Involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J., Lenches E., Smith K., Robison R.A., Sando T., Strauss E.G., Strauss J.H. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 2003;77:2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T., Sugrue R.J., Xu Q., Lee A.K., Chan Y.C., Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- Davis W.G., Blackwell J.L., Shi P.Y., Brinton M.A. Interaction between the cellular protein eEF1A and the 3′-terminal stem–loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M., Villegas-Sepúlveda N., del Angel R.M. Translation elongation factor-1α, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Dinman J.D., Richter S., Plant E.P., Taylor R.C., Hammell A.B., Rana T.M. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. 2002;99:5331–5336. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M.M., Brinton M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes & Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Montalvo B.M., Medina F., del Angel R.M. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Res. 2004;102:141–150. doi: 10.1016/j.virusres.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T., Bouloy M., Girard M. Stable secondary structures at the 3′-end of the genome of yellow fever virus (17D vaccine strain) FEBS Lett. 1985;188:159–163. doi: 10.1016/0014-5793(85)80895-4. [DOI] [PubMed] [Google Scholar]
- Guschlbauer W., Chantot J.F., Thiele D. Four-stranded nucleic acid structures 25 years later: From guanosine gels to telomer DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Heinz F.X., Collett M.S., Purcell R.H., Gould E.A., Howard C.R., Houghton M., Moorman R.J.M., Rice C.M., Thiel H.-J. Family Flaviviridae. In: Van Regenmortel M.H.V., et al., editors. Virus taxonomy. Academic Press; San Diego, CA: 2000. pp. 859–878. [Google Scholar]
- Htun H., Dahlberg J.E. Single strands, triple strands, and kinks in H-DNA. Science. 1988;241:1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J.E. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johansson M., Brooks A.J., Jans D.A., Vasudevan S.G. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-β and the viral helicase, NS3. J. Gen. Virol. 2001;82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- Kang C., Zhang X., Ratliff R., Moyzis R., Rich A. Crystal structure of four-stranded Oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- Kapoor M., Zhang L., Ramachandra M., Kusukawa J., Ebner K.E., Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Kondratieva N., Sgro J.Y., Palmenberg A., Westaway E.G. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003;77:10623–10629. doi: 10.1128/JVI.77.19.10623-10629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li Y., Kedersha N., Anderson P., Emara M., Swiderek K.M., Moreno G.T., Brinton M.A. Cell proteins TIA-1 and TIAR interact with the 3′ stem–loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Flaviviridae: The viruses and their replication. In: Knipe D.M., et al., editors. Fields virology. 4th ed. Lippincott Williams & Wilkins Publishers; Philadelphia, PA: 2001. pp. 991–1041. [Google Scholar]
- Lindenbach B.D., Rice C.M. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V.I., Mirkin S.M., Danilevskaya O.N., Voloshin O.N., Balatskaya S.V., Dobrynin V.N., Filippov S.A., Frank-Kamenetskii M.D. An unusual DNA structure detected in a telomeric sequence under superhelical stress and at low pH. Nature. 1989;339:634–637. doi: 10.1038/339634a0. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.M., Khromykh A.A., Jones M.K., Westaway E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.S., Johansen C.A., Ritchie S.A., Van Den Hurk A.F., Hall R.A. Japanese encephalitis as an emerging virus: The emergence and spread of Japanese encephalitis virus in Australasia. Curr. Top. Microbiol. Immunol. 2002;267:49–73. doi: 10.1007/978-3-642-59403-8_3. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Maher L.J. DNA triple-helix formation: An approach to artificial gene repressors? Bioessays. 1992;14:807–815. doi: 10.1002/bies.950141204. [DOI] [PubMed] [Google Scholar]
- Mandl C.W., Holzmann H., Kunz C., Heinz F.X. Complete genomic sequence of Powassan virus: Evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194:173–184. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D., Dreher T.W. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Matsuda D., Yoshinari S., Dreher T.W. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Men R., Bray M., Clark D., Chanock R.M., Lai C.J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: Analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M., Lacroix L., Arimondo P.B., Leroy J.L., François J.C., Klump H., Mergny J.L. Unusual DNA conformations: Implications for telomeres. Curr. Med. Chem. Anticancer Agents. 2002;2:627–644. doi: 10.2174/1568011023353877. [DOI] [PubMed] [Google Scholar]
- Moser H.E., Dervan P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Paranjape S.M., Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Pérez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue G.P., Huntley C.C., Hall T.C. Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses. Arch. Virol. 1994;S9:181–194. doi: 10.1007/978-3-7091-9326-6_18. [DOI] [PubMed] [Google Scholar]
- Proutski V., Gould E.A., Holmes E.C. Secondary structure of the 3′ untranslated region of flaviviruses: Similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S., Flamm C., Mandl C.W., Heinz F.X., Stadler P.F. Secondary structure of the 3′-noncoding region of flavivirus genomes: Comparative analysis of base pairing probabilities. RNA. 1997;3:779–791. [PMC free article] [PubMed] [Google Scholar]
- Rettberg C.C., Prere M.F., Gesteland R.F., Atkins J.F., Fayet O. A three-way junction and constituent stem–loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- Rice C.M., Lenches E.M., Eddy S.R., Shin S.J., Sheets R.L., Strauss J.H. Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schmittgen T.D., Zakrajsek B.A., Mills A.G., Gorn V., Singer M.J., Reed M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- Schuppli D., Miranda G., Qiu S., Weber H. A branched stem–loop structure in the M-site of bacteriophage Qβ RNA is important for template recognition by Qβ replicase holoenzyme. J. Mol. Biol. 1998;283:585–593. doi: 10.1006/jmbi.1998.2123. [DOI] [PubMed] [Google Scholar]
- Schuppli D., Georgijevic J., Weber H. Synergism of mutations in bacteriophage Qβ RNA affecting host factor dependence of Qβ replicase. J. Mol. Biol. 2000;295:149–154. doi: 10.1006/jmbi.1999.3373. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Shefer K., Brown Y., Gorkovoy V., Nussbaum T., Ulyanov N.B., Tzfati Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.Y., Brinton M.A., Veal J.M., Zhong Y.Y., Wilson W.D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Solomon T. Recent advances in Japanese encephalitis. J. Neurovirol. 2003;9:274–283. doi: 10.1080/13550280390194037. [DOI] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS—Beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta M., Vrati S. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 2000;74:5108–5115. doi: 10.1128/jvi.74.11.5108-5115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Blois C.A., Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Castle E. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J. Gen. Virol. 1986;67:1183–1188. doi: 10.1099/0022-1317-67-6-1183. [DOI] [PubMed] [Google Scholar]
- Wengler G., Wengler G. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: Absence of poly(A) and possible role in RNA replication. Virology. 1981;113:544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Westaway E.G., Mackenzie J.M., Khromykh A.A. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- Winer J., Jung C.K., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Yamaji Y., Kobayashi T., Hamada K., Sakurai K., Yoshii A., Suzuki M., Namba S., Hibi T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Yocupicio-Monroy M., Padmanabhan R., Medina F., del Angel R.M. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology. 2007;357:29–40. doi: 10.1016/j.virol.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Yon C., Teramoto T., Mueller N., Phelan J., Ganesh V.K., Murthy K.H., Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:27412–27419. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- You S., Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- You S., Falgout B., Markoff L., Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Yu L., Markoff L. The topology of bulges in the long stem of the flavivirus 3′ stem–loop is a major determinant of RNA replication competence. J. Virol. 2005;79:2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.-I., Kim S.-Y., Rice C.M., Lee Y.-M. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 2003;77:6450–6465. doi: 10.1128/JVI.77.11.6450-6465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.-I., Choi Y.-J., Yu X.-F., Song J.-Y., Shin Y.-H., Ju Y.-R., Kim S.-Y., Lee Y.-M. Engineering the Japanese encephalitis virus RNA genome for the expression of foreign genes of various sizes: Implications for packaging capacity and RNA replication efficiency. J. Neurovirol. 2007;13:522–535. doi: 10.1080/13550280701684651. [DOI] [PubMed] [Google Scholar]
- Zeenko V.V., Ryabova L.A., Spirin A.S., Rothnie H.M., Hess D., Browning K.S., Hohn T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 2002;76:5678–5691. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Falgout B., Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zhang J., George A.T., Baumstark T., Simon A.E. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA. 2006a;12:147–162. doi: 10.1261/rna.2166706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang G., Guo R., Shapiro B.A., Simon A.E. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus-strand synthesis. J. Virol. 2006b;80:9181–9191. doi: 10.1128/JVI.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]