Abstract
RNA levels in a cell are determined by the relative rates of RNA synthesis and decay. State-of-the-art transcriptional analyses only employ total cellular RNA. Therefore, changes in RNA levels cannot be attributed to RNA synthesis or decay, and temporal resolution is poor. Recently, it was reported that newly transcribed RNA can be biosynthetically labeled for 1–2 h using thiolated nucleosides, purified from total cellular RNA and subjected to microarray analysis. However, in order to study signaling events at molecular level, analysis of changes occurring within minutes is required. We developed an improved approach to separate total cellular RNA into newly transcribed and preexisting RNA following 10–15 min of metabolic labeling. Employing new computational tools for array normalization and half-life determination we simultaneously study short-term RNA synthesis and decay as well as their impact on cellular transcript levels. As an example we studied the response of fibroblasts to type I and II interferons (IFN). Analysis of RNA transcribed within 15–30 min at different times during the first three hours of interferon-receptor activation resulted in a >10-fold increase in microarray sensitivity and provided a comprehensive profile of the kinetics of IFN-mediated changes in gene expression. We identify a previously undisclosed highly connected network of short-lived transcripts selectively down-regulated by IFNγ in between 30 and 60 min after IFN treatment showing strong associations with cell cycle and apoptosis, indicating novel mechanisms by which IFNγ affects these pathways.
Keywords: microarray, biosynthetic labeling, half-life, 4-thiouridine, interferon
INTRODUCTION
Multiple regulatory processes govern the flow of genetic information from DNA to protein. Stimulated cells alter transcription rates of individual genes within minutes (Liu et al. 2007). Characteristic and differential mRNA turnover rates either contribute to rapid changes in cellular gene expression profiles or rigidly maintain constant transcript levels. Thus, mRNA levels of genes at a given time are the result of an intensively regulated balance between de novo transcription and mRNA decay (Guhaniyogi and Brewer 2001; Fan et al. 2002; Jing et al. 2005).
Standard microarray analyses on total cellular RNA (total RNA) provide a measure of mRNA abundance but cannot discriminate whether changes are due to alterations in RNA synthesis or decay. Numerous attempts have been undertaken to circumvent this problem. De novo transcription and its contribution to steady-state mRNA levels were determined using nuclear transcription run-on assays (Hirayoshi and Lis 1999; Fan et al. 2002; Garcia-Martinez et al. 2004). However, these assays are not readily compatible with standard microarray formats. Decay rates have been determined by blocking transcription, e.g., using actinomycin-D (act-D), assuming mRNA decay to continue at its normal rate (Frevel et al. 2003; Yang et al. 2003; Bernstein et al. 2004; Raghavan and Bohjanen 2004). However, this method is inherently cell invasive and cannot be combined with assays used to measure de novo transcription.
Low temporal resolution for regulatory changes is another major limitation of standard expression profiling using total RNA. This is particularly true for mammalian cells due to the relatively long half-life of their mRNAs. While the median mRNA half-life (t½m) in prokaryotic cells is in the range of 20–30 min (Bernstein et al. 2002), t½m in human cells was reported to be 600 min (Yang et al. 2003). Hence, even hours after a >100-fold transient down-regulation in transcription rate, this is hardly detectable in total RNA without concordant changes in mRNA decay. In addition, for the majority of transcripts a 10-fold up-regulation in transcription rate requires >2 h to result in a twofold increase in abundance. Therefore, primary changes cannot be differentiated from subsequent events by assays measuring transcript abundance only.
It has been known for 30 years that thiol-group containing nucleosides such as 4-thiouridine (4sU) can be introduced into nucleoside salvage pathways in eukaryotic cells and allow nondisruptive metabolic labeling of newly transcribed RNA (Melvin et al. 1978). These can be used to separate newly transcribed RNA from total RNA using mercury affinity chromatography (Melvin et al. 1978; Woodford et al. 1988; Ussuf et al. 1995; Kenzelmann et al. 2007) or thiol-specific biotinylation and subsequent purification on streptavidin-coated magnetic beads (Cleary et al. 2005) as eukaryotic mRNAs normally do not contain thiol-groups. Here we report on an integrated, improved approach in which the advantages of direct incorporation of 4sU are combined with thiol-specific biotinylation and simple magnetic separation of total RNA into newly transcribed RNA and preexisting unlabeled RNA. This allows microarray analysis on all three obtained RNA subsets in parallel. Thus, for the first time, changes in RNA synthesis and decay as well as their impact on cellular transcript levels can be analyzed in a single experimental setting.
Theoretically, short-term biosynthetic labeling for 15–30 min at different times during cytokine treatment should allow differentiation of the temporal order and kinetics of changes in gene expression at molecular level. We demonstrate this by analyzing the cellular response to type I and II interferons (IFN). IFNs are an intensively studied family of multifunctional cytokines known to dramatically alter transcription rates of hundreds of transcripts (for review, see Platanias 2005). They play an essential role in innate immunity in response to viral, bacterial, and parasitic pathogens (Stetson and Medzhitov 2006). In addition, they exert important anti-proliferative as well as immune modulatory properties (Boxel-Dezaire et al. 2006).
RESULTS
Separation of total cellular RNA into newly transcribed and preexisting RNA
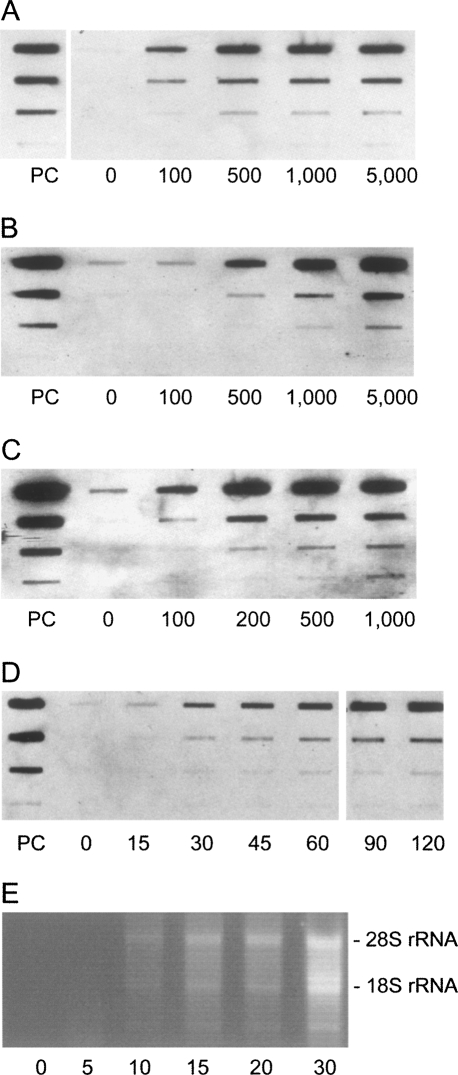
Metabolic labeling of newly transcribed RNA with 4sU has minimal adverse effects on gene expression, RNA decay, protein stability, and cell viability (Melvin et al. 1978; Woodford et al. 1988; Ussuf et al. 1995; Kenzelmann et al. 2007). We compared the global transcriptional profile of three biological replicates of NIH-3T3 cells exposed to 200 μM 4sU for one hour with nontreated cells. No significant differential expression (more than twofold and P < 0.05) attributable to 4sU treatment was detected (see Supplemental Fig. 1). We analyzed 4sU incorporation into newly transcribed RNA by culturing various cell types in the presence of 100 μM to 5 mM 4sU for one hour. Following isolation of total cellular RNA and thiol-specific biotinylation, 4sU-labeled RNA was specifically detected and quantified by dot blot assay. We found 4sU to be efficiently incorporated into RNA by a broad range of cell types of human and murine origin including fibroblasts, endothelial cells, and B-cells (Fig. 1A–C) as well as dendritic cells, macrophages, and T-cells (data not shown). Labeled RNA was detectable by dot blot analysis after 15 min of labeling (Fig. 1D).
FIGURE 1.
Detection and quantification of 4sU incorporation into RNA. 4sU is quantitatively incorporated into newly transcribed RNA by a broad range of cell lines of human and murine origin. Dot blot analyses of thiol-mediated biotinylation of RNA derived from (A) murine NIH-3T3 fibroblasts, (B) SVEC 4-10 endothelial cells, and (C) DG75 human B-cells exposed to 100 μM to 5 mM 4sU for 1 h are shown as examples. Nonlabeled RNA samples (0) were used as controls. All RNA samples were spotted in 10-fold dilutions (top to bottom: 1 μg down to 1 ng). A 5′-biotinylated DNA oligo of 81 nt (PC) was used in 10-fold dilutions (100 ng down to 0.1 ng) to quantify biotin-labeling. (D) Kinetics of 4sU incorporation into cellular RNA were analyzed. NIH-3T3 cells were cultured in the presence of 1 mM 4sU for 0, 15, 30, 45, 60, 90, and 120 min. Dot blot analyses of biotinylated RNA samples are shown as for A–C. (E) Newly transcribed RNA was labeled in NIH-3T3 cells using 500 μM 4sU for 0, 5, 10, 15, 20, and 30 min. Following isolation of total cellular RNA and thiol-specific biotinylation, newly transcribed RNA was purified from 100 μg biotinylated total RNA. Half of the purified RNA was separated on an ethidium bromide-stained agarose gel under nondenaturating conditions.
In cells expressing Toxoplasma gondii uracil-phosphoribosyltransferase (UPRT) newly transcribed RNA can be metabolically labeled using 4-thiouracil (4tU) (Cleary et al. 2005). In this report newly transcribed RNA was isolated by thiol-specific biotinylation followed by affinity purification on streptavidin-coated magnetic beads. We adapted this approach to 4sU labeling, which does not require UPRT expression. We established an improved, column-based protocol for magnetic separation of total RNA into newly transcribed RNA and preexisting RNA (for details, see Materials and Methods). Employing this approach, high-molecular-weight newly transcribed RNA could be detected by agarose gel electrophoresis after as little as 10 min of labeling (Fig. 1E). After one hour of labeling newly transcribed RNA accounted for 3%–6% of total RNA depending on the cell type under study (data not shown). The efficiency of separation was validated by combining 4sU and 3H-cytidine to label newly transcribed RNA for 15, 30, and 60 min. After thiol-specific biotinylation up to 90% of 3H-cytidine-labeled RNA copurified with the newly transcribed RNA fraction (see Supplemental Fig. 2A–D). When 3H-labeled, unbiotinylated RNA was subjected to this separation procedure, the first two of a total of six washing steps already contained the bulk of labeled RNA, indicating preparative recovery of both newly transcribed RNA and preexisting RNA from total RNA. Thus, both RNA subsets, present in the same isolated RNA sample, can be separated with high purity.
Development of an integrative approach to simultaneously analyze RNA synthesis and decay
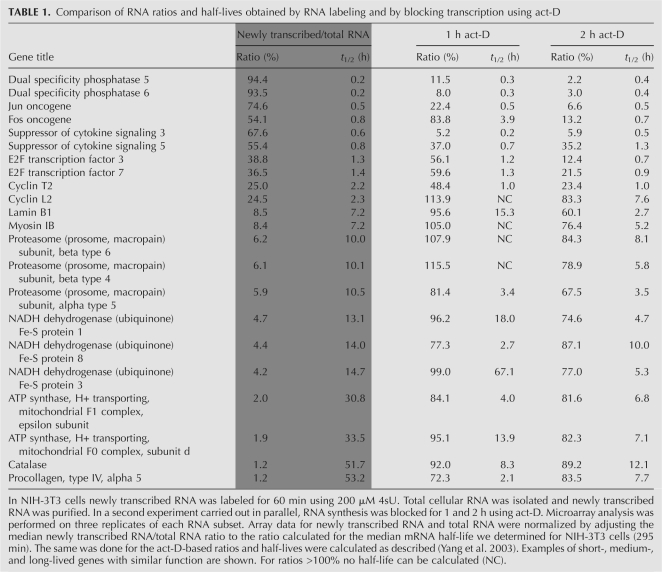
For every transcript total RNA levels are constantly subjected to changes in RNA synthesis and decay. So far it was not possible to analyze both parameters in a systemic approach in a single experimental setting. The standard method used to study RNA decay is to block transcription, e.g., using act-D (Frevel et al. 2003; Yang et al. 2003). However, this approach has a number of limitations. First, blocking transcription provokes a cellular stress response. Some important mechanisms that control mRNA stability, e.g., miRNA-mediated control of gene expression (Jing et al. 2005), are released in cells subjected to stress (Bhattacharyya et al. 2006). In addition, some transcripts have been shown to be rapidly stabilized following act-D treatment (Shyu et al. 1989). Second, for the vast majority of mammalian mRNAs, decay rates determined by blocking transcription are based on very small differences in transcript levels. Only in the case of very short-lived transcripts (t 1/2 < 2 h), RNA levels decrease by >50% upon 2 h of transcriptional blockage. Thus, act-D-based measurements of RNA decay are inherently imprecise for the majority of cellular transcripts.
We developed a novel approach to simultaneously study RNA synthesis and decay for which blocking transcription is not required. Under steady-state conditions, RNA synthesis compensates for RNA decay to maintain stable transcript levels. Therefore, de novo synthesis rates are much higher for short-lived transcripts than for long-lived transcripts. In murine fibroblasts, we found newly transcribed RNA/total RNA ratios to vary from 1% to 2% for long-lived to >95% for very short-lived transcripts (for examples, see Table 1). Based on the newly transcribed RNA/total RNA ratios (R) and the duration of labeling (tL) precise data on mRNA half-life (t 1/2) can be calculated according to the following equation: t 1/2 = −tL × ln(2) / ln(1 − R) (for details, see Supplemental Methods). Thus, assuming steady-state conditions mRNA synthesis and decay rates can be determined by simply analyzing newly transcribed RNA and total RNA derived from the same RNA sample.
TABLE 1.
Comparison of RNA ratios and half-lives obtained by RNA labeling and by blocking transcription using act-D
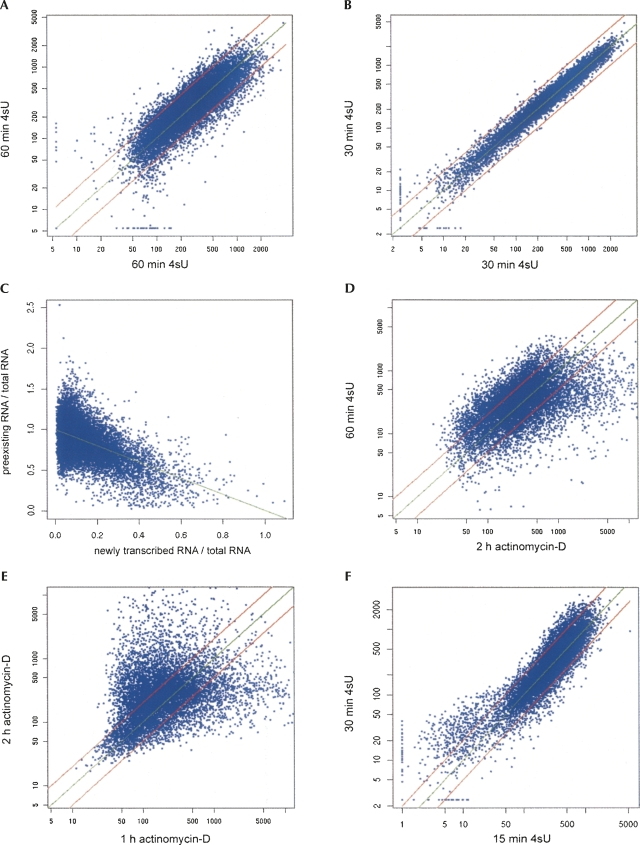
We applied this approach to determine mRNA half-lives for murine fibroblasts. Newly transcribed RNA was labeled in NIH-3T3 cells for 60 and 30 min. After isolation of total cellular RNA newly transcribed RNA was separated from it. Three replicates of both total RNA and newly transcribed RNA were subjected to microarray analysis. Half-life measurements based on newly transcribed RNA/total RNA ratios resulted in precise and reproducible data independent of mRNA half-life ranging from <20 min to >2 d (Fig. 2A,B).
FIGURE 2.
(A,B) Analysis of mRNA half-lives determined based on newly transcribed RNA/total RNA ratios. Newly transcribed RNA was labeled in NIH-3T3 cells for 60 or 30 min. Total cellular RNA was separated into newly transcribed RNA and unlabeled preexisting RNA. For each experiment three biological replicates of each RNA subset were subjected to microarray analysis. For each condition half-lives were determined based on newly transcribed RNA/total RNA ratios obtained from >13,000 probe sets. Scatterplots comparing half-lives (in minutes) obtained by 60 min of labeling in separated experiments (A) and by 30 min of labeling within a single experiment (B) are shown. Red lines indicate deviations from the regression line (green) by twofold in either direction. (C) Normalization of total RNA, newly transcribed RNA, and preexisting RNA microarray data using linear regression analysis. Preexisting RNA/total RNA ratios were plotted against newly transcribed RNA/total RNA ratios. As total RNA is separated into newly transcribed RNA and preexisting RNA, the sum of the two ratios should equal 100% for every transcript. Therefore, a linear correlation exists between the two ratios and linear regression analysis (green line) can be used to determine the parameters required for correct data normalization. Thereby, a median mRNA half-life of 295 min was determined for NIH-3T3 cells. (D–F) Comparison of mRNA half-lives obtained by RNA labeling and actinomycin-D treatment (act-D). In parallel to the RNA labeling experiments, RNA synthesis was blocked in NIH-3T3 cells for 1 and 2 h using act-D. Microarray analyses were performed on three replicates of each RNA subset. (D) Half-lives determined by 2 h act-D treatment and 60 min of labeling correlated reasonably well. (E) Half-lives (in minutes) determined by blocking transcription for 1 and 2 h using act-D only correlated well for very short-lived transcripts (t 1/2 < 2 h) but poor for the remaining transcripts. (F) Half-lives (in minutes) obtained by 15 and 30 min of labeling within a single experiment were compared to identify potential effects caused by 4sU labeling. Significant differences were only observed for very short-lived transcripts (t 1/2 < 50 min).
If steady-state conditions are not given there is no equilibrium in between RNA synthesis and decay. Therefore, only RNA synthesis is analyzed using newly transcribed RNA while RNA decay is determined by comparing preexisting RNA obtained after the application of a stimulus, e.g., a cytokine, with total RNA levels prior to it [ t 1/2 = −tL × ln(2) / ln(P), with P = preexisting RNA/total RNA]. Thereby, the polar effects of act-D treatment on RNA decay are avoided as, instead of blocking RNA synthesis, newly transcribed RNA is physically separated from total RNA.
For act-D-based determination of RNA decay rates, array data from RNA samples obtained after blocking transcription have to be adjusted to data from samples prior to it. This kind of normalization is usually based on the mRNA half-life of a housekeeping gene (e.g., β-actin) independently determined in a separate experiment (Yang et al. 2003). It is a crucial step for comparing data from independent experiments and is also required when RNA half-lives are determined based on newly transcribed RNA/total RNA or preexisting RNA/total RNA ratios. We developed a novel approach to solve this task by statistical means based on the separation of total RNA into newly transcribed and preexisting RNA. For all probe sets the sum of the ratios of newly transcribed RNA/total RNA and preexisting RNA/total RNA must equal 100% as total RNA is separated into newly transcribed RNA and preexisting RNA. Due to the linear relation between the two ratios, normalization parameters simply result from linear regression analysis. Thus, normalization is based on thousands of probe sets and does not require data from separate experiments (for details, see Supplemental Methods). To demonstrate this, we labeled RNA in NIH-3T3 cells for one hour and separated newly transcribed RNA from preexisting RNA. Three replicates of each RNA subset were subjected to microarray analysis. Normalization parameters were determined based on linear regression analysis on array data of >13,000 probe sets with “present” calls in all arrays (Fig. 2C). mRNA half-lives were obtained using both the newly transcribed RNA/total RNA as well as preexisting RNA/total RNA ratios. The median mRNA half-life (t 1/2m) in NIH-3T3 fibroblasts was 295 min. To our knowledge a median mRNA half-life in cells of murine origin has not been described. Once the median mRNA half-life of a given cell type has been determined, array normalization can be achieved by normalizing the median newly transcribed RNA/total RNA ratio to the corresponding ratio predicted for the median mRNA half-life.
In order to validate our new approach we compared mRNA half-lives obtained from NIH-3T3 fibroblasts based on newly transcribed RNA/total RNA ratios with data obtained by the standard method, i.e., by blocking transcription for 1 and 2 h using act-D. Data obtained by 2 h act-D and 60 min of labeling correlated reasonably well (Fig. 2D). In contrast, decay rates obtained by blocking transcription (1 h versus 2 h act-D) only correlated well for transcripts with a half-life of <2 h (Fig. 2E). In order to analyze the effect of time on our new method and show that precise data on mRNA half-life can already be determined after very short labeling, we compared half-lives obtained by 15 and 30 min of labeling. We found half-lives to correlate extremely well (Fig. 2F).
Interestingly, there is deviation from the linear correlation for small values (t 1/2 < 60 min, <5% of all transcripts) and this was also seen when comparing half-lives obtained by 15 and 60 as well as 30 and 60 min of labeling (see Supplemental Fig. 3A,B). This may in part be attributed to the effects of noise in the newly transcribed RNA/total RNA ratios on the logarithmic function used to calculate t 1/2. However, due to their rapid decay, their half-lives can also be determined with high precision based on preexisting RNA/total RNA ratios (see Supplemental Methods).
Monitoring interferon-mediated differential gene expression
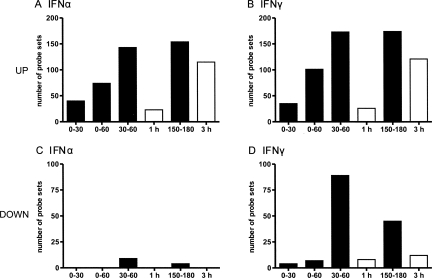
Numerous studies on both type I and II IFN-mediated gene regulation have provided detailed knowledge of their effects on cellular gene expression (for review, see de Veer et al. 2001). For most IFN-regulated genes changes in transcript levels only start to become detectable after 2–3 h. Thus, primary and secondary events are difficult to differentiate. We applied our method to analyze IFN-mediated gene regulation in order to show the advantages of this type of analysis. In a first experiment NIH-3T3 cells were treated for one hour with either 100 U/mL IFNα, 100 U/mL IFNγ, or Φ, adding 4sU at the same time. Total cellular RNA was isolated from cells and newly transcribed RNA was purified. Three biological replicates of each RNA subset were subjected to microarray analysis. In a second larger experiment short-term 4sU labeling was performed at different time intervals during IFN treatment. Specifically, RNA was labeled during the first 15 and 30 min of IFN receptor activation and for 30 min starting 30, 90, and 150 min thereafter. Microarray analyses were performed for three replicates of newly transcribed RNA labeled from 0 to 30, 30 to 60, and 150 to 180 min and from total RNA following 3 h of IFN treatment. In all experiments the number of probe sets showing significant (P ≤ 0.05) (more than fourfold) up-regulation or (less than twofold) down-regulation was substantially higher for newly transcribed RNA than for total RNA (Fig. 3A–D). Analysis of newly transcribed RNA (30–60 min) instead of total RNA (1 h) increased sensitivity for detection of up- and down-regulation by more than five- and >10-fold, respectively. Interestingly, down-regulation during the first 60 min was exclusively seen after IFNγ treatment.
FIGURE 3.
Detection of IFN-mediated differential gene expression in newly transcribed RNA and total RNA. NIH-3T3 cells were treated with 100 U/mL of either IFNα or IFNγ or were left untreated. Newly transcribed RNA was labeled by adding 500 μM 4sU (final concentration) during 0–30, 0–60, 30–60, and 150–180 min of treatment. Microarray analyses were performed on three biological replicates of purified newly transcribed RNA (black bars) as well as of total RNA (empty bars) following 1 and 3 h of treatment. Bar charts depicting the number of probe sets detecting significant (P ≤ 0.05) >4-fold up-regulation (A,B) or >2-fold down-regulation (C,D) by either IFNα or IFNγ are shown.
For further analysis microarray data from all experiments were combined including only probe sets >2-fold regulated and significant (P ≤ 0.01) in any of the 12 statistical comparisons (n = 957, corresponding to 562 unique genes; for a complete list, see Supplemental Table 1). All transcripts which showed significant regulation by >2-fold during the first hour of IFNα or IFNγ treatment were sorted by their half-life (Fig. 4A–C). The median mRNA half-life of genes up-regulated by IFNγ (t 1/2m = 184 min) was significantly shorter than for IFNα (t 1/2m = 346 min; P < 0.0001). We observed a strong correlation between differential gene expression detectable in total RNA at 1 h and transcript half-life. Similar changes for total RNA and newly transcribed RNA were only detectable for short- lived, up-regulated transcripts. In contrast, analysis of newly transcribed RNA revealed substantial differential expression independent of transcript half-life, thereby explaining the superior sensitivity observed when analyzing newly transcribed RNA.
FIGURE 4.
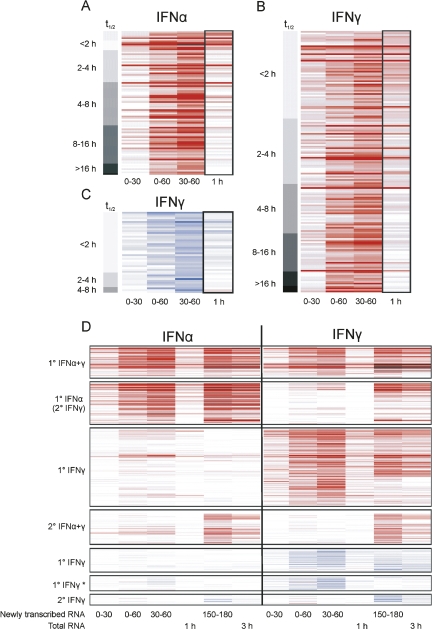
Monitoring IFNα- and IFNγ-mediated differential gene expression. Represented are all genes which were found to be significantly regulated (P ≤ 0.01) within any condition during the first 60 min of IFN treatment (A–C) or in any of the 12 statistical comparisons (D). Each row represents a gene, each column a biological condition. Red-to-black represents up-regulation with respect to mock-treated samples, white represents no change, and blue represents down-regulation. (A–C) Correlation of detectable differential gene expression with mRNA half-life. Heat-maps including genes (A) up-regulated by IFNα, (B) up-regulated by IFNy, and (C) down-regulated by IFNγ by twofold for which half-life could be calculated based on newly transcribed RNA/total RNA ratios are shown. Genes were sorted according to their half-life (top to bottom). Gray scales depicted on the left indicate the half-life of the corresponding group of transcripts ranging from <2 to >16 h. Changes in transcript levels detectable in total RNA after 1 h of IFNα or IFNγ treatment strongly depend on the half-life of the corresponding transcript. In contrast, analysis of newly transcribed RNA revealed substantial differential expression independent of transcript half-life. (D) Temporal order and kinetics of IFNα- and IFNγ-mediated gene regulation. Genes were sorted by similarity (hierarchical clustering) of their regulation across four of the 12 conditions: newly transcribed RNA from 30–60 min (primary) and 150–180 min (secondary) for IFNα and IFNγ. Changes in transcription rates seen already during the first hour were termed “primary” (1°). Additional changes seen first at 150–180 min after stimulation were termed “secondary” (2°). “*” indicates transcripts down-regulated only during the first hour of IFNγ treatment with down-regulation not detectable in total RNA at 1 and 3 h at all.
By quantitative PCR, up-regulation of two major IFN-induced transcription factors, irf1 (interferon regulatory factor 1) and socs3 (suppressor of cytokine signaling 3) by eight- and fivefold, respectively, was already detectable in newly transcribed RNA after 15 min of IFNγ treatment (see Supplemental Fig. 4). By microarray analysis of newly transcribed RNA substantial changes in gene expression were detected after 30 min (data for all conditions visualized in Fig. 4D). Interestingly, virtually all probe sets (IFNα: 86/86; IFNγ 115/119) detecting up-regulation by more than twofold in newly transcribed RNA after 30 min also revealed regulation of more than twofold at 30–60 min. Therefore, analysis of newly transcribed RNA produces highly reliable and reproducible data. Changes in transcription rates seen already during the first hour were termed “primary.” This type of regulation peaked in between 30 and 60 min and mainly remained stable or started to decline thereafter. For a large part (>25%) of these transcripts changes remained undetectable in total RNA at 1 and 3 h. Additional changes seen first at 150–180 min after stimulation were termed “secondary.” These also accounted for ∼25% of changes. Interestingly, a large subset of genes was primary up-regulated by IFNα but secondary by IFNγ. This was remarkable as no induction of any type I IFN gene was observed during IFNγ treatment. Changes in total RNA after 1 and 3 h of IFN treatment could be quantitatively explained by corresponding changes in transcriptional activity when taking into account the half-life of the corresponding transcripts. No effect of IFN treatment on RNA stability and decay was observed.
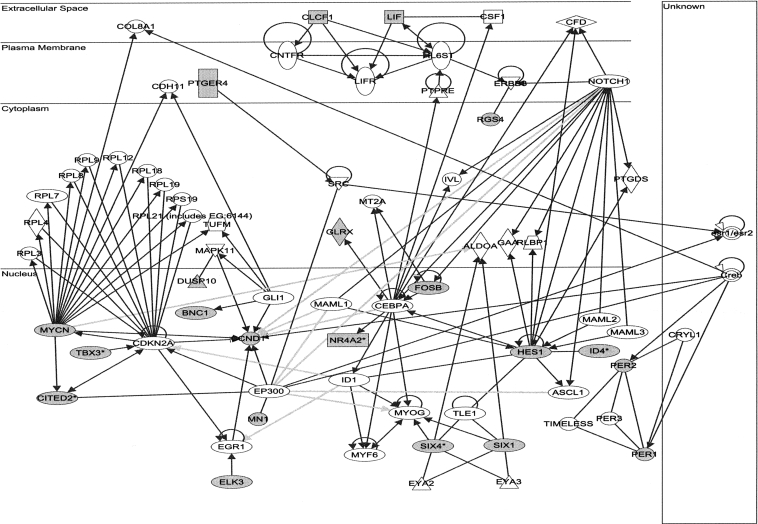
A number of genes have been shown to be down-regulated after 6 h of IFN treatment. Little is known about the mechanisms involved (de Veer et al. 2001; Platanias 2005). Interestingly, IFNα did not lead to any substantial down-regulation of transcription during the first three hours, indicating that down-regulation observed by others following longer treatment (≥6 h; for review, see de Veer et al. 2001) is due to secondary effects. In contrast, we identified a new subset of very short-lived transcripts (t 1/2m = 90 min) significantly down-regulated only by IFNγ following 30–60 min of treatment. These changes were almost undetectable in total RNA at 1 and 3 h when changes seen in newly transcribed RNA already waned. The half-lives of down-regulated genes were much shorter than those of up-regulated genes (t 1/2m = 184 min; P < 10−7). Based on direct interactions described for these genes, comprehensive interaction networks were constructed, two of them showing high connectivity (network scores of 24 and 22, respectively; see Supplemental Fig. 5A,B). It was possible to merge these two networks using direct interactions between nodes to produce a combined network comprising 21 of the 25 “focus genes” used for network analysis (Fig. 5). The majority of “focus genes” present within the networks were located within the nucleus (16 of 21). Networks showed associations with control of gene expression, cellular development, cell death, cellular growth, and proliferation.
FIGURE 5.
Combined gene network obtained based on the genes down-regulated by IFNγ treatment. Network analysis on genes selectively down-regulated at 30–60 min following IFNγ receptor activation suggested two highly connected networks when restricting to direct interactions only (see Supplemental Fig. 5A,B). Network 1 is centered on gene nodes corresponding to cyclin D1 (CDN1) and myc (MYCN) and is comprised of genes associated with protein synthesis, gene expression, and organ development. Network 2 is comprised of genes associated with cellular development, nervous system development, and gene expression functions (based on IPA Functional Analysis; see Materials and Methods for network derivation). Both networks show high connectivity (Network 1 score of 24, Network 2 score of 22) with 11 or 10 of the 35 components of each network derived from the 25 focus genes down-regulated by IFNγ (shown in gray). Interestingly, the two networks could be combined to a single network (shown) based on direct interactions of the involved genes. Direct interactions of genes within a network are shown as dark gray arrows; direct interactions of genes between networks are shown as light gray arrows. The entire list of genes contributing to both networks is provided in Supplemental Table 4.
DISCUSSION
Here, we report on a novel systemic approach to simultaneously analyze short-term changes in RNA synthesis and decay and their impact on cellular transcript levels. Labeling of newly transcribed RNA using 4sU is applicable to a large variety of cell types and organisms including humans, mice (Kenzelmann et al. 2007), and plants (Ussuf et al. 1995), as well as Drosophila (data not shown). It can be used for in vitro and in vivo studies since 4sU is also well tolerated by mice following i.v. injection (Kenzelmann et al. 2007). It was shown that microarray sensitivity for differential gene expression can be substantially increased by the analysis of newly transcribed RNA following two hours of labeling (Kenzelmann et al. 2007). However, in order to be able to study the temporal order of transcriptional regulation at molecular level, monitoring at much shorter time intervals is required. It was shown that incorporation of 4tU can be used to purify newly transcribed RNA from cells expressing toxoplasma gondii UPRT (Cleary et al. 2005) following as little as one hour of labeling. However, we found 4tU incorporation to be dependent on a variety of factors including cell type and UPRT expression levels (data not shown), thereby limiting short-term labeling efficiency. In contrast, incorporation of 4sU into newly transcribed RNA seems to be predominantly concentration dependent. Therefore, labeling efficiency can be adjusted according to the desired duration of labeling and the cell type under study. We were able to achieve a density of thiolation by 4sU sufficient for in vitro biotinylation and subsequent efficient separation on magnetic beads following as little as 10 min of labeling.
We studied the response of fibroblasts to type I and II IFN to test our new approach on a well characterized template of transcriptional regulation. For the first time, we were able to determine the temporal order and kinetics of transcriptional regulation allowing differentiation of secondary signaling effects from primary ones at the time scale of minutes. In fact, >25% of IFN-induced genes differentially expressed at 3 h represented secondary signaling events. However, two major IFN-induced transcription factors (irf1 and socs3) were already up-regulated by five- to eightfold after 15 min of IFNγ treatment, emphasizing the need to study regulation of gene expression within this time scale.
Total cellular transcript levels are constantly subjected to changes in RNA synthesis and decay. Our method allows separation of total cellular RNA into newly transcribed and preexisting RNA. Thus, RNA synthesis and decay can be simultaneously analyzed in a single experimental setting. On the one hand, analysis of newly transcribed RNA allows the quantitative study of regulatory mechanisms governing transcription. This is of particular interest for subsequent promoter analyses. On the other hand, removal of newly transcribed RNA from total RNA offers a simple novel approach to determine RNA decay rates without having to block transcription. If steady-state conditions can be assumed, RNA half-life can even be determined with superior accuracy based on newly transcribed RNA/total RNA ratios alone as these offer a two order of magnitude dynamic range of measurements instead of the twofold dynamic range provided for the majority of transcripts by experiments using act-D. Thereby, precise data on RNA decay are obtained even for medium- to long-lived transcripts. As no cellular stress response is provoked, the regulatory mechanisms that govern mRNA decay, like effects exerted by miRNAs (Bhattacharyya et al. 2006), can be studied.
Calculating half-lives instead of simply attributing changes in total RNA to changes in RNA synthesis or decay as described by Kenzelmann et al. (2007) has several advantages. First, it allows quantification of the actual changes in transcript half-life. Changes in transcript half-life do not simply match changes in newly transcribed RNA/total RNA ratios. In contrast, depending on how high the actual half-life is, it may lead to substantially different results. Second, it allows precise analysis of both changes in RNA synthesis and decay in case both total and newly transcribed RNA levels are altered. Third, functional characteristics can be attributed to genes, e.g., genes with regulatory function are predominantly represented by short-lived transcripts. Finally, it makes data from different experiments comparable.
The most crucial step necessary to be able to compare data on RNA half-life from independent experiments is array normalization. Analysis of all three RNA subsets also provides a new solution to this problem. The standard approach is to use a housekeeping gene for which a half-life was independently determined (Frevel et al. 2003; Yang et al. 2003; Bernstein et al. 2004; Raghavan and Bohjanen 2004). Our approach is based on the concept that total RNA is quantitatively separated into newly transcribed RNA and preexisting RNA. Linear regression analysis can be applied on microarray data of thousands of transcripts. Therefore, array normalization is not dependent on the precision of the measurements of single transcripts any more but can now be based on thousands of probe sets of the actual microarray data under study.
By comparing the regulatory changes detectable for total RNA and newly transcribed RNA after 1 h of IFN treatment with transcript half-life, we show that the low temporal resolution of standard microarray analysis is due to the relatively long half-life of mammalian mRNAs. Thus, standard microarray analyses carried out on total RNA have a bias for picking up changes for short-lived transcripts. These predominantly represent regulatory genes and transcription factors creating further bias in subsequent analysis (Raghavan et al. 2002). With our approach even a transient regulation can now be revealed independent of transcript half-life. We clearly show that changes in gene expression occurring during the first hour following a stimulus are independent of transcript half-life.
Our approach is particularly well suited to detect transient down-regulation in transcription rates which is often impossible to pick up by measuring transcript abundance only. Thus, we found a new subset of short-lived genes selectively down-regulated following 30–60 min of IFNγ treatment, which was not detectable in conventional total RNA at 1 and 3 h. We show that these genes form a high connectivity functional network affecting cell cycle and apoptosis, suggesting that this response is a relevant part of IFNγ signaling. Interestingly, this response was only detectable in the second 30 min of IFN treatment. Therefore, it represents an early but potentially already secondary response specific to interferon γ.
In summary, by combining short-term analysis of RNA synthesis with simultaneous measurements of RNA decay, a comprehensive picture of the regulatory mechanism governing gene expression can be obtained.
MATERIALS AND METHODS
Cell culture and metabolic labeling of cellular RNA
Murine NIH-3T3 fibroblasts (ATCC CRL1658) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum (NCS). Human and murine cell lines including human epithelial cells (HeLa), T-cells (Jurkat), and B-cells (BL41 & DG75), as well as murine macrophages (RAW 264.7) and endothelial cells (SVEC 4-10) were cultured according to ATCC standards. For metabolic labeling of RNA in NIH-3T3 cells 4-thiouridine (Sigma) was added to a 200 μM final concentration into prewarmed, CO2-equilibrated medium in all 60-min labeling experiments (cells used only in between 5. and 15. passage after thawing; split twice weekly and 24 h before start of labeling). For short-term labeling (≤30 min), 500 μM 4sU was used. For most cell types growing in suspension (human B- and T-cells and murine macrophages) 100 μM 4sU was found to be optimal for 60-min labeling experiments (data not shown). Total cellular RNA was isolated from cells using Trizol reagent (Invitrogen) following the modified protocol described by Chomczynski and Mackey (1995).
For combined 4sU and radioactive RNA labeling 5 μM (60-min labeling with 200 μM 4sU) or 12.5 μM (15- and 30-min labeling with 500 μM 4sU) 5-3H-cytidine (25 Ci/mmol, Sigma) was added to the culture media together with the 4sU. For these experiments RNA was extracted using RNeasy kit (Qiagen) to remove unincorporated 5-3H-cytidine.
In order to determine mRNA decay rates by blocking transcription, act-D (Sigma) was used at a final concentration of 5 μg/mL. Both murine IFNα (IFN-αA, PBL Biomedical Laboratories) and murine IFNγ (Chemicon) were used at a final concentration of 100 U/mL in all experiments. In every experiment IFN activity was controlled using the ISRE Luc reporter cell line, which expresses luciferase under an IFN inducible promoter (Zimmermann et al. 2005; data not shown).
Biotinylation and purification of 4sU-labeled RNA
Biotinylation of 4sU-labeled RNA was performed using EZ-Link Biotin-HPDP (Pierce) dissolved in dimethylformamide (DMF) at a concentration of 1 mg/mL and stored at 4°C. Biotinylation was carried out in 10 mM Tris (pH 7.4), 1 mM EDTA, and 0.2 mg/mL Biotin-HPDP at a final RNA concentration of 100 ng/μL for 1.5 h at room temperature. In general 50–100 μg total RNA were used for the biotinylation reaction. By dot blot assay (see below) we noted that simple precipitation of biotinylated RNA using isopropanol/ethanol as described (Cleary et al. 2005) leads to coprecipitation of large amounts of unbound biotin. As this is likely to interfere with capture of biotinylated RNA by streptavidin-coated magnetic beads we efficiently removed unbound Biotin-HPDP by chloroform/isoamylalcohol (24:1) extraction using Phase-lock-gel (Heavy) tubes (Eppendorf). Afterward, a 1/10 volume of 5 M NaCl and an equal volume of isopropanol were added and RNA was precipitated at 20,000g for 20 min. The pellet was washed with an equal volume of 75% ethanol and precipitated again at 20,000g for 10 min. The pellet was resuspended in 50–100 μL RNAse-free water. After denaturation of RNA samples at 65°C for 10 min followed by rapid cooling on ice for 5 min, biotinylated RNA was captured using μMACS streptavidin beads and columns (Miltenyi). Our column-based assay eliminated the risk of carry-over of unlabeled RNA we observed when using the original protocol (Cleary et al. 2005). Up to 100 μg of biotinylated RNA were incubated with 100 μL of μMACS streptavidin beads with rotation for 15 min at room temperature. The beads were transferred and magnetically fixed to the columns. Columns were washed three times with 1 mL 65°C washing buffer (100 mM Tris-HCl, pH 7.4, 10 mM EDTA, 1 M NaCl, 0.1% Tween20) followed by three washes with room temperature washing buffer. To recover the unlabeled preexisting RNA the flow-through of the first two washes was collected and combined. Labeled RNA was eluted by the addition of 100 μL of freshly prepared 100 mM dithiothreitol (DTT) followed by a second elution round 5 min later. RNA was recovered from the washing fractions and eluates using the RNeasy MinElute Spin columns (Qiagen).
Detection of 4sU incorporation by dot blot assay
Dot blot analysis on biotinylated RNA samples was carried out to detect and quantify the amount of RNA-bound biotin residues within a given RNA sample. First, biotinylated RNA samples were denaturated for 10 min at 65°C and subsequently placed on ice for 5 min. RNA samples were diluted in ice-cold alkaline binding buffer (10 mM NaOH, 1 mM EDTA) at a concentration of 1 μg/200 μL and spotted onto a Zeta+-probe blotting membrane (Bio-Rad) in 10-fold dilutions (1 μg down to 1 ng) using the Bio-Dot Apparatus (Bio-Rad). A 5′-biotinylated DNA oligo of 81 nt was used as quantitative positive control and applied to the membrane in 10-fold dilutions (100 ng down to 0.1 ng). After rinsing each well with 500 μL alkaline binding buffer, the blotting apparatus was disassembled and the membrane was incubated in blocking solution (phosphate buffered saline [PBS], pH 7.5, 10% SDS, 1 mM EDTA) for 20 min. The membrane was probed with 1:1000 dilution of 1 mg/mL streptavidin-horseradish peroxidase (Pierce) in blocking solution for 15 min. The membrane was washed six times in PBS containing decreasing concentrations of SDS (10%, 1%, and 0.1% SDS, applied twice each) for 10 min. The signal of biotin-bound horseradish peroxidase was visualized by ECL detection reagent (Amersham).
Reverse transcription and real-time PCR
Reverse transcription of 100 ng purified newly transcribed RNA was carried out in 25 μL reactions using Superscript III (Invitrogen) and oligo-dT primers (Invitrogen) following the manufacturers instructions. PCR was performed on a Light Cycler (Roche Molecular Biochemicals). Each reaction was carried out using 5 μL of cDNA (1:10 dilution) and 15 μL reaction mixtures of Light Cycler Fast Start, DNA MasterPlus Sybr Green I and 0.5 μM of the primers. PCRs were subjected to 10 min of 95°C hot-start, and Sybr Green incorporation was monitored for 45 cycles of 95°C denaturation for 10 sec, 58°C annealing for 3 sec, and 72°C elongation for 10 sec, followed by melting curve analysis to confirm specific amplification. The specific primer pairs for each target are listed in Supplemental Table 2. Standard curves were prepared using cDNA prepared from total RNA of NIH-3T3 cells. Relative quantification was performed based on gapdh normalization.
Microarray sample labeling, hybridization, and preprocessing
Total RNA (1.5 μg) and newly transcribed RNA (280 ng) were amplified and labeled using the Affymetrix One-Cycle Target Labeling Kit according to the manufacturer's recommendations. As newly transcribed RNA mainly consists of mRNA, it was amplified and labeled according to the manufacturer's protocol for mRNA. The amplified and fragmented biotinylated cRNA was hybridized to Affymetrix MG 430 2.0 arrays using standard procedures.
Microarray data processing and statistical analysis for analysis of IFN-mediated gene regulation
Data were processed and analyzed with R and Bioconductor (Gentleman et al. 2004; R Development Core Team 2007). For the analysis of IFN-mediated differential gene expression processing and statistical hypothesis testing was implemented separately for total RNA and newly transcribed RNA data sets. The former contains a total of 24 arrays in seven biological conditions, the latter 36 arrays in 12 biological conditions (for details, see Supplemental Table 3).
Arrays were assessed for quality, GCRMA-normalized, filtered for low and invariant expression, and analyzed using an empirical Bayes moderated t-test for paired samples. Hybridization results for newly transcribed RNA and total RNA were expected and observed to show significant differences.
“Quality assessment” consisted of RNA degradation plots, Affymetrix quality control metrics, sample cross-correlation, data distributions, and probe-level visualizations.
“Normalization” incorporated (separately for each RNA type data set) background correction, normalization, and probe-level summation by GCRMA.
“Nonspecific filters” consisted of a combined intensity-based and variation-based step removing genes that can safely be considered to be biologically irrelevant or nonmeasurable by microarray technology. Filter one only retains genes which are called “present” or “marginal” in at least three out of the total number of arrays in that data set. Filter two only retains genes with a measured Inter-Quartile-Range ≥0.75. In the total RNA data set, n = 4327 genes pass this combined filter; in the newly transcribed RNA data set, this number is n = 10,053. The difference in numbers is due to the higher proportion of “present” probes in the newly transcribed RNA data set.
“Statistical testing” for differential expression was applied separately for the total RNA and newly transcribed RNA data sets. Analysis hypotheses were centered around the effect of IFNα and IFNγ at particular time points, compared to mock-treated samples. The small statistical sample size for individual biological conditions is partly offset by the paired nature of samples, in that the mock and treatment samples are the same sample with the only difference being the treatment received. This is made use of by applying a paired-test version of the empirical Bayes moderated t-test (Smyth 2004), which in itself is the most robust test for small sample sizes. An increased rate of false-positive results due to simultaneously testing on a large number of genes was corrected for by applying a multiple testing correction algorithm to the observed P-values, in this case using the Benjamini and Hochberg method (Benjamini and Hochberg 1995). Fisher's exact test was applied for comparison of the number of down-regulated genes in between IFNα and IFNγ for newly transcribed RNA.
Calculation of mRNA half-life based on newly transcribed/total RNA ratios, preexisting RNA/total RNA ratios, and actinomycin-D data
The different bioinformatical approaches used to determine RNA half-lives are described in detail in the Supplemental Methods. For all experiments three biological replicates of each RNA subset were subjected to microarray analysis.
In total this comprised 45 arrays in 11 biological conditions (for details, see Supplemental Table 3).
To calculate RNA half-lives CEL-files of all samples from all conditions (including total RNA, newly transcribed RNA, and preexisting RNA as well as the act-D RNA samples) were normalized together using the GCRMA algorithm (R Development Core Team 2007). Only probe sets called “present” in all three replicates of all three RNA subsets under study were included in the analysis of transcript half-lives. Statistical comparison of half-life values between groups was performed using the Wilcoxon rank–sum test.
Pathway analysis for genes down-regulated by IFNγ
Network analysis involved upload of gene identifier lists to the ingenuity pathway analysis (IPA) software application. The IPA Knowledge Base, a comprehensive knowledge base of biological findings for genes of human, mouse, and rat, was used to construct pathways and functional modules. The IPA application maps each gene identifier to its corresponding gene object in the IPA Knowledge Base. These focus genes are then overlaid onto a global molecular network developed from information contained in the Knowledge Base. Networks of focus genes are then algorithmically generated based on their connectivity and assigned a significance score (based on P-values) representing the likelihood that the focus genes within the network are found there by chance. A high number of focus genes within a data set leads to a higher network score (equal to the negative exponent of the respective P-value). To identify focus genes down-regulated after IFNγ treatment, the database was queried using the 44 probe IDs present in this list and 25 focus genes were identified as eligible for network analysis. Comprehensive interaction networks were constructed showing high connectivity (Network 1 score of 24 and Network 2 score of 22; for details, see Supplemental Fig. 5A,B).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Angela Servatius for her excellent technical assistance, Klaus Conzelmann for his help and advice with setting up the IFN experiments, and Steven Watterson for proofreading the mathematic sections. This work was funded by NGFN grants 01GS0405 (to U.H.K.) and FKZ 01G0113 TP 37 (to R.H.) as well as grants for a Centre for Integrative Systems Biology supported by BBSRC and EPSRC, FP6 InfoBioMed programme and Wellcome Trust (to P.G.). L.D., Z.R., and U.H.K. designed the study and wrote the paper. L.D. and B.R. performed the experiments. L.D., C.C.F., and R.Z. developed and C.C.F. implemented the new bioinformatics approach used to calculate mRNA half-lives from the experimental results. R.H. and J.M. provided the microarray data at the TUM microarray facility with the help of Angela Servatius and contributed to data analysis. P.D., T.F. and P.G. helped with array study design, analyzed the microarray data, and performed pathway analysis for IFN regulation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1136108.
REFERENCES
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Bernstein J.A., Khodursky A.B., Lin P.H., Lin-Chao S., Cohen S.N. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J.A., Lin P.H., Cohen S.N., Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. 2004;101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Boxel-Dezaire A.H., Rani M.R., Stark G.R. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Mackey K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- Cleary M.D., Meiering C.D., Jan E., Guymon R., Boothroyd J.C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- de Veer M.J., Holko M., Frevel M., Walker E., Der S., Paranjape J.M., Silverman R.H., Williams B.R. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- Fan J., Yang X., Wang W., Wood W.H., III, Becker K.G., Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevel M.A., Bakheet T., Silva A.M., Hissong J.G., Khabar K.S., Williams B.R. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez J., Aranda A., Perez-Ortin J.E. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi J., Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Hirayoshi K., Lis J.T. Nuclear run-on assays: Assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Kenzelmann M., Maertens S., Hergenhahn M., Kueffer S., Hotz-Wagenblatt A., Li L., Wang S., Ittrich C., Lemberger T., Arribas R., et al. Microarray analysis of newly synthesized RNA in cells and animals. Proc. Natl. Acad. Sci. 2007;104:6164–6169. doi: 10.1073/pnas.0610439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Yang Y., Chernishof V., Loo R.R., Jang H., Tahk S., Yang R., Mink S., Shultz D., Bellone C.J., et al. Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- Melvin W.T., Milne H.B., Slater A.A., Allen H.J., Keir H.M. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur. J. Biochem. 1978;92:373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2007. ISBN 3-900051-07-0; 2007. A language and environment for statistical computing.http://www.R-project.org [Google Scholar]
- Raghavan A., Bohjanen P.R. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief. Funct. Genomic. Proteomic. 2004;3:112–124. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- Raghavan A., Ogilvie R.L., Reilly C., Abelson M.L., Raghavan S., Vasdewani J., Krathwohl M., Bohjanen P.R. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Greenberg M.E., Belasco J.G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes & Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Ussuf K.K., Anikumar G., Nair P.M. Newly synthesised mRNA as a probe for identification of wound responsive genes from potatoes. Indian J. Biochem. Biophys. 1995;32:78–83. [PubMed] [Google Scholar]
- Woodford T.A., Schlegel R., Pardee A.B. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal. Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E., Jr Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A., Trilling M., Wagner M., Wilborn M., Bubic I., Jonjic S., Koszinowski U., Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-γ signaling and antiviral responses. J. Exp. Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]