Abstract
The hepatitis delta virus (HDV) ribozyme occurs in the genomic and antigenomic strands of the HDV RNA and within mammalian transcriptomes. Previous kinetic studies suggested that a wobble pair (G•U or A+•C) is preferred at the cleavage site; however, the reasons for this are unclear. We conducted sequence comparisons, which indicated that while G•U is the most prevalent combination at the cleavage site, G-C occurs to a significant extent in genomic HDV isolates, and G•U, G-C, and A-U pairs are present in mammalian ribozymes. We analyzed the folding of genomic HDV ribozymes by free energy minimization and found that variants with purine–pyrimidine combinations at the cleavage site are predicted to form native structures while pyrimidine–purine combinations misfold, consistent with earlier kinetic data and sequence comparisons. To test whether the cleavage site base pair contributes to catalysis, we characterized the pH and Mg2+-dependence of reaction kinetics of fast-folding genomic HDV ribozymes with cleavage site base pair purine–pyrimidine combinations: G•U, A-U, G-C, and A+•C. Rates for these native-folding ribozymes displayed highly similar pH and Mg2+ concentration dependencies, with the exception of the A+•C ribozyme, which deviated at high pH. None of the four ribozymes underwent miscleavage. These observations support the A+•C ribozyme as being more active with a wobble pair at the cleavage site than with no base pair at all. Overall, the data support a model in which the cleavage site base pair provides a structural role in catalysis and does not need to be a wobble pair.
Keywords: ribozyme, RNA folding, GU wobble, catalysis
INTRODUCTION
The hepatitis delta virus (HDV) ribozyme is a small (∼85 nucleotides [nt]), self-cleaving RNA that occurs in both genomic and antigenomic forms and is responsible for processing nascent viral transcripts during double rolling-circle replication (Lai 1995; Wadkins and Been 2002; Been 2006). More recently, a mammalian version of the HDV ribozyme was identified (Salehi-Ashtiani et al. 2006). These ribozymes have similar secondary structures that comprise five pairing regions that form a nested double pseudoknot (Fig. 1A). The ribozyme uses the 2′-hydroxyl of U(−1) as a nucleophile to cleave the phosphodiester bond between nucleotides −1 and +1, yielding products with 5′-hydroxyl and 2′,3′-cyclic phosphate termini. High-resolution crystal structures have been solved for both the self-cleaved (Fig. 1B) and precleaved forms of the genomic ribozyme and reveal compact structures with similar overall folds and a buried active site (Ferre-D'Amare et al. 1998; Ferre-D'Amare and Doudna 2000; Ke et al. 2004; Ke et al. 2007). Notably, these structures show that C75 is well-positioned to participate in general acid–base chemistry (Fig. 1C).
FIGURE 1.
Various structures of the genomic HDV ribozyme. (A) Secondary structure of the ribozyme used in this study. The catalytic core is in uppercase letters and flanking sequences are in lowercase letters. The cleavage site between U−1 and G1 is denoted with an arrow. The G•U wobble pair at bp 1 is in red (throughout) and boxed, while C75 is blue (throughout) and is boxed. The G11C mutation that promotes fast, single-exponential kinetics is shown (Chadalavada et al. 2002). (B) Crystal structure of the self-cleaved form of the ribozyme. Bases highlighted in the secondary structure are also highlighted here. Also shown is the U1A protein (purple), which was used to facilitate crystallization (Ferre-D'Amare and Doudna 2000; Ferre-D'Amare et al. 1998). (C) Crystal structure of the active site of the self-cleaved form of the ribozyme. Bases highlighted in the secondary structure are also highlighted here, and wobble-pairing between G1 and U37 in bp 1 is highlighted. Panels B and C were generated using DS ViewerPro 5.0 (Accelrys) and PDB entry 1drz.
Kinetic and mutagenesis studies on the HDV ribozyme support a cleavage mechanism involving general acid–base catalysis by C75 (Perrotta et al. 1999; Nakano et al. 2000; Oyelere et al. 2002; Das and Piccirilli 2005; Perrotta et al. 2006). Two models have been advanced for how C75 participates in general acid–base catalysis, as recently summarized by Nakano and Bevilacqua (2007). In general acid–base model 1 (GAB model 1), C75 acts as a general base to deprotonate the 2′-hydroxyl of U−1 and an outer-sphere hydrated Mg2+ ion acts as a general acid to protonate the 5′-oxygen leaving group. In general acid–base model 2 (GAB model 2), protonated C75 acts as a general acid and a hydrated Mg2+ hydroxide ion acts as a general base. Crystal structures of a precleaved ribozyme, inactivated by a C75U change, show a Mg2+ ion in a position consistent with GAB model 1 but are ambiguous with respect to the role of C75 (Ke et al. 2004, 2007). In contrast, the crystal structure of the self-cleaved form of the ribozyme shows C75 in a position consistent with GAB model 2 (Ferre-D'Amare et al. 1998; Ferre-D'Amare and Doudna 2000; Fig. 1C). The latter structure does not show well-ordered metal ions near the active site, however, perhaps owing to the absence of the scissile phosphate and the 2′-hydroxyl nucleophile. Thus, despite considerable effort, uncertainty about the positioning of catalytic species persists.
Several computational approaches have also been used to probe the mechanism of the HDV ribozyme; however, some results support GAB model 1 (Krasovska et al. 2005, 2006), while others support GAB model 2 (Liu et al. 2007; Wei et al. 2007). In general, biochemical data are more consistent with GAB model 2, in which C75 acts as a general acid and a hydrated Mg2+ hydroxide ion acts as general base (Nakano et al. 2000, 2001; Das and Piccirilli 2005).
In an effort to gain further insight into the catalytic mechanism of the HDV ribozyme, we probed the role of the base pair immediately proximal to the cleavage site.1 This base pair involves residues 1 and 37 (Fig. 1A) and is a G•U wobble (Fig. 2) in most HDV ribozymes (Chadalavada et al. 2007). Since this base pair abuts the scissile phosphate, it could potentially be involved in catalysis. We tested whether bp 1 could play roles in binding the catalytic Mg2+ ion, positioning functional groups for catalysis, and/or influencing the pK a of C75.
FIGURE 2.
Base pairs tested in this study. The major and minor groove faces and numbering for purines and pyrimidines are shown for the G•U wobble pair.
Indeed, G•U wobble pairs have been shown to perform two of these functions in another ribozyme, the self-splicing group I intron. This large catalytic RNA leaves products with termini opposite to the HDV ribozyme and it has been shown that both the geometry and minor groove of the G•U wobble pair at the active site are important for positioning functional groups for RNA substrate recognition (Barfod and Cech 1989; Doudna et al. 1989; Downs and Cech 1994; Knitt et al. 1994; Cate and Doudna 1996; Strobel et al. 1998; Adams et al. 2004; Golden et al. 2005). In addition, crystal (Cate and Doudna 1996) and NMR (Allain and Varani 1995; Kieft and Tinoco 1997) structures revealed that the active site G•U wobble pair, as well as tandem G•U wobble pairs in other parts of the ribozyme, form metal ion binding sites (Keel et al. 2007). G•U wobble pairs have also been found to be important in other RNAs, as reviewed by Varani and McClain (2000) and Xu et al. (2007).
In an effort to clarify what roles, if any, the G•U wobble pair plays in the HDV ribozyme reaction, constructs with the purine–pyrimidine combinations G•U, A-U, G-C, and A+•C at bp 1 were studied, and rate-pH and rate-Mg2+ profiles were determined. In order to minimize the impact of alternative folding on activity and therefore study effects on chemistry, we used RNA sequences based on the previously characterized G11C fast-folding ribozymes (Chadalavada et al. 2000, 2002). Ultimately, our data suggest that, in contrast to previous results, bp 1 does not need to be a wobble pair and that it provides a structural role in catalysis.
RESULTS
Rationale for experiments: Sequence comparison data
Characterization of bp 1 was initiated by investigating the degree to which it is conserved in HDV ribozymes. We recently compared sequences of the ribozyme and upstream and downstream flanking sequences (Chadalavada et al. 2007). The −54 to 140 region of the genomic HDV RNA was aligned for 76 isolates using blastn (Altschul et al. 1990). This region encompasses 54 nt upstream of the cleavage site and 56 nt downstream of the 3′ boundary of the ribozyme.
We found that position 1 is a G for all 76 genomic sequences, but that position 37 is a U for only 61 isolates, with the other 15 isolates having a C. Thus, the genomic ribozyme can tolerate either pyrimidine opposite G1. In contrast, sequence comparisons carried out on the antigenomic ribozyme revealed that all isolates (269 available) have a G•U wobble at bp 1. In addition, bp 1 of the mammalian HDV ribozyme has been found to be G•U, G-C, or A-U, depending on the organism (Salehi-Ashtiani et al. 2006). Taken together, these results support two conclusions. First, all ribozymes require a purine–pyrimidine combination at bp 1. Second, a G•U wobble is conserved in the antigenomic ribozyme but not in the genomic or mammalian ribozymes.
Rationale for experiments: Prior biochemical studies on bp 1
Contribution of the G•U wobble at bp 1 to the HDV ribozyme reaction has been studied previously. For the genomic ribozyme, Wu and coworkers found that G•U was preferred at bp 1 over G-C, A-U, and C-G, by at least six- to eightfold, while other base combinations (U•U, C•U, G•G, G•A, U-A, U•G, C•A) were essentially inactive (Wu et al. 1993). Experiments by Nishikawa et al. (1997) on the genomic ribozyme indicated that G was preferred over A at position 1 by three- to eightfold, regardless of the base at position 37 as long as it was a pyrimidine (i.e., G•U = G-C > A-U = A+•C). In addition, other combinations at bp 1 (A•G, U•U, U•G, G•G, C•U, C•C) resulted in miscleavage between G1 and G2 (Nishikawa et al. 1997). The basis for greater tolerance of a Watson–Crick G-C in one study and not the other was unclear.
For the antigenomic ribozyme, experiments from the Wu and Been laboratories indicated that a G•U wobble at bp 1 was approximately 2-fold more reactive than G-C and at least 15-fold more reactive than A-U (Wu and Huang 1992; Perrotta and Been 1996). Been and coworkers also studied an antigenomic ribozyme with an A+•C combination at bp 1 and found that it reacted with a rate intermediate to the G•U and G-C ribozymes (Perrotta and Been 1996; Been and Wickham 1997). Since an A and C can form a wobble pair upon protonation (A+•C), it was proposed that a wobble pair at the active site might be important for cleavage activity.
These studies were important for identifying secondary structural interactions in the HDV ribozyme and for beginning to understand HDV ribozyme catalysis. However, the ribozymes in these studies were only assayed under a single set of conditions, usually pH 7–8 and ∼10 mM MgCl2, and were not especially fast-reacting, native-folding variants. In this study, we investigated the HDV ribozyme mechanism under a wide range of pH and Mg2+ conditions, using a fast-reacting ribozyme variant. Variation of pH is especially important for A+•C wobble pairs, which typically have a pK a near 6.5 (Legault and Pardi 1994; Cai and Tinoco 1996; Bevilacqua et al. 2004).
Predicted folding of ribozymes with different base combinations at bp 1
In designing ribozymes for evaluating the role of bp 1 in the mechanism, we took into account prior studies, which showed that ribozymes with certain bp 1 combinations had exceptionally poor activity. In particular, nearly all pyrimidine–purine (including Watson–Crick base pairs), pyrimidine–pyrimidine, and purine–purine combinations resulted in either slow kinetics, miscleavage, or both (Wu et al. 1993; Nishikawa et al. 1997). Since we are interested in the role of bp 1 in the ribozyme reaction, we first tested whether these changes are predicted to affect folding of the −30/99 genomic HDV ribozyme, which we have used for most other mechanistic investigations (Fig. 1A). Since the HDV ribozyme has two pseudoknots, we chose an algorithm capable of predicting pseudoknots. The iterated loop matching (ILM) algorithm was used, as it is much less time-consuming than other programs such as PKNOTS but has similar accuracy (Ruan et al. 2004a,b). All predictions were carried out in the background of a G11C mutation that, in combination with an antisense oligonucleotide, facilitates fast-folding of the −30/99 ribozyme (see next paragraph and Materials and Methods) (Chadalavada et al. 2000, 2002).
We began by examining folds of −30/99 ribozymes with a G•U, A-U, or G-C at bp 1 (Fig. 3, folds B,C). As expected from experiments on constructs of this length (see next paragraph and Materials and Methods), −30/99 ribozymes were predicted to contain a large fraction of nonnative pairings in the absence of an antisense oligonucleotide (Fig. 3, folds B,C). The structures predicted for the G•U and A-U ribozymes contained two near-native pairings, P1 and P4, and three alternative (Alt) pairings.2 Two of the Alt pairings have been observed experimentally in the genomic ribozyme: Alt 1, which involves upstream flanking sequence and the 3′ portion of P2, and Alt 3, which involves the 5′ portion of P2 and 3′ portion of P3 (Chadalavada et al. 2000). The third Alt pairing, termed “Alt A,” is positioned near Alt 1.3 The structure predicted for the G-C ribozyme (Fig. 3B) was identical to that for the G•U and A-U ribozymes except that it contained another known Alt pairing, Alt P1, which involves a slipped P1, part of the 5′strand of P2, and the entire 3′ strand of P1.1 (Chadalavada et al. 2002).
FIGURE 3.
Folds predicted for the ribozymes (all with G11C) using the ILM algorithm (Ruan et al. 2004a, b). (A) Primary structure representation of the predicted pairings. Positions 1 and 37 are shown in italics and are highlighted with a gray vertical bar. Base-paired regions are underlined, with the 5′ and 3′ strands noted. “A” is the native fold for the −30/99 wild-type sequence. Note that the sequence includes the G11C mutation, but shows the wild-type P2 pairing. “B” is the fold for the −30/99 ribozyme, predicted with either G•U or A-U at bp 1. “C” is the fold for the −30/99 ribozyme, predicted with G-C at bp 1. “D” is the fold for the −6/99 ribozyme, predicted with G•U, A-U, or G-C at bp 1. “E” is the fold for the −6/99 ribozyme, predicted with A+•C at bp 1. “F” is the fold for the −6/99 ribozyme, predicted with UG or CG at bp 1. “G” is the fold for the −6/99 ribozyme, predicted with UA at bp 1. “H” is the fold for the −6/99 ribozyme, predicted with CA at bp 1. Previously described alternative pairings are denoted with numbering (e.g., Alt 1), while new alternative pairings are denoted with lettering (e.g., Alt A). In some of the folds, “R” and “Y” are used to denote purine and pyrimidine. (B) Representative secondary structure representation of predicted pairings. For clarity, sample two-dimensional representations of the secondary structures are shown for folds A (see Fig. 1A), C and E; ribozyme sequence are shown in uppercase and the flanking sequence in lowercase. For folds C and E, the pairings are oriented on the page in a native-like fashion for convenience only, and P4 is represented as dots for simplification. Native pairings are shaded in gray and reveal a greater fraction of native pairings for the −6/99 ribozymes.
Fast-reacting self-cleavage reactions for the −30/99 ribozyme are performed in the presence of an antisense oligonucleotide complementary to nt −30 to −7, referred to as “AS(−30/−7).” Base-pairing of AS(−30/−7) to the ribozyme disrupts Alt 1, which facilitates folding of the ribozyme to the native state (Chadalavada et al. 2000). To mimic these conditions computationally, we folded additional sequences containing only nt −6 to 99 (Fig. 3, folds D–H).
The first −6/99 ribozymes examined had a G•U, A-U, G-C, or A+•C at bp 1. As expected, structures predicted for the −6/99 G•U, A-U, and G-C ribozymes were considerably more native than their −30/99 counterparts. They contained four native or near-native pairings, P1, P2, P3, and P4, along with two short (3 or 4 bp) Alt pairings (Fig. 3, fold D). Results for the A+•C ribozyme were similar except P1 contained one less base pair, which was accompanied by two additional Alt folds (Fig. 3, fold E; Fig. 3B); the program does not predict A+•C wobble pairs, a result that is expected experimentally at higher pH. These data suggest that if the A and C can form an A+•C wobble pair, as expected at lower pH, then the A+•C ribozyme will adopt the same fold as the other purine–pyrimidine ribozymes; otherwise, the ribozyme will be substantially less native in its folding.
Next, we folded 6/99 ribozymes with the four pyrimidine–purine combinations UG, UA, CG, and CA at bp 1. The predicted structures contained a near-native P4 and, for the UA ribozyme, P1, but were otherwise misfolded (Fig. 3, folds F–H). Extensive misfolding of these constructs is consistent with prior observations that ribozymes with pyrimidine–purine combinations are much less active (Wu et al. 1993; Nishikawa et al. 1997). On the basis of the above sequence comparisons, secondary structure predictions, and prior biochemical studies, we chose to focus our experimental studies of bp 1 on the better-folding and biologically relevant purine–pyrimidine combinations: G•U, A-U, G-C, and A+•C (Fig. 2).
G•U, A-U, and G-C ribozyme pH-dependent kinetics can be described with a single-channel model
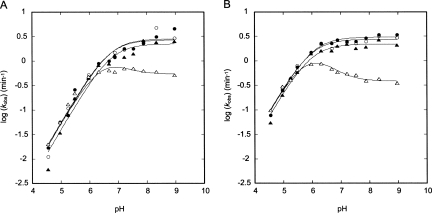
In order to test possible roles of bp 1 in catalysis, we examined pH and Mg2+ concentration dependencies of the reaction rate for the four bp 1 variants. Rate-pH and -Mg2+ profiles have been reported for the G•U ribozyme, wherein anticooperative coupling between H+ and Mg2+ was observed (Nakano et al. 2000; Nakano and Bevilacqua 2007). To see if this behavior is maintained upon mutation of bp 1, rate-pH profiles were obtained for the four ribozymes at 1 and 10 mM Mg2+ (Fig. 4).
FIGURE 4.
Comparison of the pH-dependence for the G•U (●), A-U (▲), G-C (○), and A+•C (△) ribozymes. (A) Experiments performed in 1 mM Mg2+. Fits to the G•U, A-U, and G-C (R2 = 0.98–0.99) ribozymes were according to the logarithm of Equation 2b and gave k max values of 2.7 ± 0.4, 2.2 ± 0.4, and 2.8 ± 0.4 min−1, and pK a values of 6.68 ± 0.09, 6.8 ± 0.1, and 6.73 ± 0.09, respectively. The A+•C ribozyme data were fit using the logarithm of Equation 4b and gave a k max value for the slow channel of 0.57 ± 0.05 min−1 and pK a values of 6.57 ± 0.03 and 6.3 ± 0.1 for C75 and the A+•C pair, respectively. (B) Experiments performed in 10 mM Mg2+. Fits to the G•U, A-U, and G-C (R2 = 0.98–0.99) ribozymes were according to the logarithm of Equation 2b and gave k max values of 3.0 ± 0.1, 2.2 ± 0.2, and 2.8 ± 0.2 min−1, and pK a values of 6.04 ± 0.04, 5.97 ± 0.06, and 5.97 ± 0.05, respectively. The A+•C ribozyme data were fit using the logarithm of Equation 4b and gave a k max value for the slow channel of 0.39 ± 0.02 min−1 and pK a values of 5.95 ± 0.03 and 6.00 ± 0.06 for C75 and the A+•C pair, respectively. In both panels, we set k fast for the A+•C ribozyme equal to k max for the G•U ribozyme, as described in the Results. The average error in k obs was found to be ±10%. Statistical justification for fitting the A+•C ribozyme data to Equation 4b (and Scheme 3) rather than Equation 2b (and Scheme 1) is as follows. In 1 mM Mg2+, the χ2 values for the A+•C ribozyme were 0.084 and 0.037 for Equation 2b (fit not shown) and Equation 4b, respectively, supporting Equation 4b and Scheme 3. In 10 mM Mg2+, the χ2 values for the A+•C ribozyme were 0.26 and 0.018 for Equation 2b (fit not shown) and Equation 4b, respectively, providing even stronger support for Equation 4b and Scheme 3. In addition, attempts to fit the A+•C ribozyme data (at both 1 mM and 10 mM Mg2+) using Equation 2b resulted in sinusoidally distributed residuals, while fits to Equation 4b gave randomly distributed residuals.
Rate-pH profiles for the G•U, A-U, and G-C ribozymes in 1 mM Mg2+ were nearly identical (Fig. 4A). A log-linear increase of k obs occurs between pH 4.5 and 6.3 with a slope of ∼1, while above pH ∼7 k obs is insensitive to pH, as previously observed (Nakano et al. 2000). The profiles were fit to the logarithm of Equation 2b, which is derived from a kinetic model in which a single deprotonation event provides the active ribozyme species (Scheme 1) (see Materials and Methods for alternative, kinetically equivalent schemes). The apparent pK as for G•U, A-U, and G-C ribozymes are experimentally indistinguishable at 6.68 ± 0.09, 6.8 ± 0.1, and 6.73 ± 0.09, respectively (Table 1), and are assigned to C75 on the basis of previous kinetics and mutagenesis studies (Nakano et al. 2000; Nakano and Bevilacqua 2007). In addition, the G•U, A-U, and G-C rate-pH profiles leveled off at similar rate constants in the high-pH regime, with k max values of 2.7 ± 0.4, 2.2 ± 0.4, and 2.8 ± 0.4 min−1, respectively.
SCHEME 1.
Single-channel/single-deprotonation mechanism (GAB Model 1).
TABLE 1.
Rate-pH profile parameters
The rate-pH profiles obtained for the G•U, A-U, and G-C ribozymes in 10 mM Mg2+ were also nearly identical (Fig. 4B). A log-linear increase of k obs occurs between pH 4.5 and 6.0 with a slope of ∼1, and above pH 6 k obs is insensitive to pH. These profiles were fit to the logarithm of Equation 2b. The apparent pK as for G•U, A-U, and G-C ribozymes remained experimentally indistinguishable at 6.04 ± 0.04, 5.97 ± 0.06, and 5.97 ± 0.05, respectively (Table 1), which are ∼0.7 units lower than the values at 1 mM Mg2+. In addition, the G•U, A-U, and G-C rate-pH profiles leveled off at similar rate constants in the high-pH regime, with k max values of 3.0 ± 0.1, 2.2 ± 0.2, and 2.8 ± 0.2 min−1, respectively.
Overall, rate-pH profiles on the G•U, A-U, and G-C ribozymes at 1 and 10 mM Mg2+ reveal that replacing the G•U wobble pair with a Watson–Crick purine–pyrimidine base pair has no effect on rate or the pK a of C75. In addition, the observed decrease in the pK a of C75 with increasing Mg2+ concentration is consistent with the previously established anticooperative coupling between H+ and Mg2+ (Nakano et al. 2000), suggesting that the G•U, A-U, and G-C ribozymes bind Mg2+ and catalyze the reaction in a similar fashion.
A+•C ribozyme pH-dependent kinetics require a multichannel model
For the A+•C ribozyme, the dependence of log k obs on pH at both 1 and 10 mM Mg2+ was more complex and could not be accounted for by Scheme 1 (see caption to Fig. 4 for statistical justification). At 1 mM Mg2+, a log-linear increase of k obs occurs between pH 4.5 and 6.0 with a slope of ∼1. This portion of the rate-pH profile, where the A+•C wobble is likely to be protonated, is indistinguishable from that of the other three ribozymes (Fig. 4A). Above pH 6, however, the rate-pH profile of the A+•C ribozyme deviates from that of the other ribozymes. In particular, between pH 6.0 and 7.2 k obs is insensitive to pH, while above pH 7.2 k obs decreases slightly and then levels off.
Similar behavior for the A+•C ribozyme was observed at 10 mM Mg2+ (Fig. 4B). A log-linear increase of k obs occurs between pH 4.5 and 5.5, with a slope of ∼1. Again, the lower pH portion of the rate-pH profile is indistinguishable from that of the other three ribozymes. At higher pH values, however, the rate-pH profile of the A+•C ribozyme again deviates from that of the other ribozymes. Between pH 6.0 and 6.3, k obs is insensitive to pH, while above pH 6.3 k obs decreases slightly and then levels off. The departure in behavior of the A+•C ribozyme at higher pH is consistent with deprotonation of the predicted A+•C wobble pair (see Discussion).
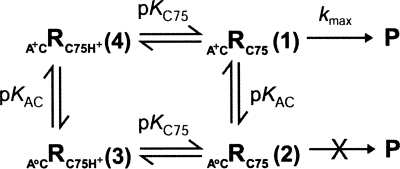
To understand the behavior of the A+•C ribozyme, we developed two kinetic models of increasing complexity and performed simulations (Schemes 2, 3). Scheme 2 shares the characteristic with Scheme 1 that only one reaction channel is reactive. In Scheme 2, we assume that the fold with wobble-pairing at bp 1 is the active species. According to Scheme 2, the rate-pH profile should be bell-shaped (Fig. 5, filled squares), which does not explain the data (Fig. 5, triangles). Thus, a model involving more than one reaction channel is needed.
SCHEME 2.
Single-channel/double-deprotonation mechanism (GAB Model 1).
SCHEME 3.
Double-channel/double-deprotonation mechanism (GAB Model 1).
FIGURE 5.
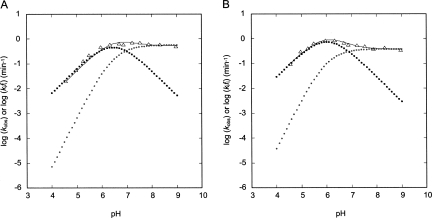
Data and simulations for the A+•C ribozyme. Simulations were performed using Microsoft Excel (not shown) and Scheme 3. (A) Data and fit parameters are the same as in Figure 4A. (Line) log k obs = log (f (1) k fast + f (2) k slow) (Scheme 3), (darker dots) log k obs (Scheme 2) = log (f (1) k fast) (Scheme 3), and (lighter dots) log (f (2) k slow) (Scheme 3). (B) Data and fit parameters are the same as in Figure 4B. (Line) log k obs = log (f (1) k fast + f (2) k slow) (Scheme 3), (darker dots) log k obs (Scheme 2) = log (f (1) k fast) (Scheme 3), and (lighter dots) log (f (2) k slow) (Scheme 3). Only Scheme 3 can account for the observed data.
A more complex, double-channel mechanism in which the A+•C ribozyme retains partial activity in the absence of an A+•C wobble pair was considered next. In Scheme 3, the ribozyme is more active when the A and C form an A+•C wobble pair than when they form no base pair at all (a weak one hydrogen bond base pair might also be possible in the absence of protonation; Allawi and SantaLucia 1998) and reacts with rate constants k fast and k slow, respectively. Multichannel analysis has been used previously to explain kinetics data for HDV and hammerhead ribozymes (Nakano et al. 2001, 2003; Zhou et al. 2002; Nakano and Bevilacqua 2007). It is clear from the simulations that this model can account for the data. The fast channel (A+•C wobble pair present) accounts for the low pH data (Fig. 5, filled squares), the slow channel (A+•C wobble pair absent) accounts for the high pH data (Fig. 5, open squares), and the logarithm of the sum of the channels (Fig. 5, thin line), fits the observed data (Fig. 5, triangles).
To obtain kinetic parameters for the A+•C ribozyme, the rate-pH profiles were fit to the logarithm of Equation 4b, which was derived from the kinetic model in Scheme 3, with no parameter constraints applied. However, unconstrained fitting led to large errors in the values for k fast, pK a,AC, and pK a,C75. We then made the assumption that k fast is equal to k max for the G•U ribozyme at a given Mg2+ concentration (2.7 min−1 at 1 mM Mg2+ and 3.0 min−1 at 10 mM Mg2+), which is reasonable given the indistinguishable rate-pH profiles for the four ribozymes in the lower pH regime where the A+•C wobble pair would be formed (see Fig. 4A,B between pH 4.5 and ∼6). This approach yielded the following values for the 1 mM Mg2+ profile: k slow = 0.57 ± 0.05 min−1, pK a,C75 = 6.57 ± 0.03, and pK a,AC = 6.3 ± 0.1; and the following values for the 10 mM Mg2+ profile: k slow = 0.39 ± 0.02 min−1, pK a,C75 = 5.95 ± 0.03, and pK a,AC = 6.00 ± 0.06 (Table 1).
Overall, the pK a values for C75 in the A+•C ribozyme are similar to those for the other three ribozymes at both Mg2+ concentrations, and the pK a values for the A+•C wobble pair are similar to those observed for A+•C wobble pairs in other RNAs (Legault and Pardi 1994; Cai and Tinoco 1996; Bevilacqua et al. 2004). Scheme 3, having just one more reactive channel than the unsatisfactory Scheme 2, is therefore the minimal kinetic scheme that describes the pH dependence of the A+•C ribozyme reaction. Taken together, the pH profiles of the four ribozymes suggest that any stable purine–pyrimidine interaction at bp 1 accomplishes catalysis in a similar fashion, and that introducing a less stable purine–pyrimidine interaction at bp 1 results in only a modest (approximately five- to eightfold) decrease in rate.
All four ribozymes yield the same cleavage products
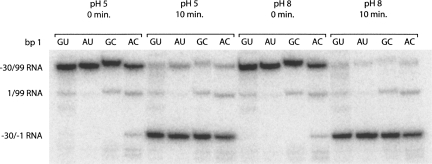
To further probe the role of bp 1, we checked to see if the identity of the purine–pyrimidine combination at bp 1 affects proper cleavage site selection. This was especially important given the aforementioned report of substrate miscleavage (between positions +1 and +2, instead of −1 and +1) from Nishikawa et al. (1997). Representative time points from the pH 5 and 8 (10 mM Mg2+) reactions for each ribozyme were fractionated on a denaturing 20% (w/v) polyacrylamide gel, chosen for its higher resolving power (Fig. 6). Also, these two pH values allow the cleavage product to be evaluated for protonated and unprotonated A•C ribozymes. It is clear from the gel that the 5′ product migrates the same for all four ribozymes at both pH values. Thus, the identity of the purine–pyrimidine combination at bp 1 is not important for proper cleavage site selection. Interestingly, this is true even for the A•C ribozyme at pH 8, where the A and C are not expected to form an A+•C wobble pair.
FIGURE 6.
Self-cleavage product analysis. Representative time points from pH 5 and 8 reaction mixtures (10 mM Mg2+) were fractionated on a 20% (w/v) polyacrylamide gel. Substrate was 5′-end labeled, and the major bands are marked. The slowest migrating band corresponds to the −30/99 RNA and the fastest migrating band corresponds to the −30/−1 RNA. The slight differences in the migration of the −30/99 RNAs are likely due to heterogeneity at the 3′ ends that is common to T7 polymerase transcription reactions and has no effect on the length of the self-cleavage product. When 5′-end-labeled −30/99 RNA self-cleaves, it produces 5′-end-labeled −30/−1 RNA and unlabeled 1/99 RNA. The 1/99 RNA (middle band) that is observed is left over from the 5′-end-labeling reaction. (Mg2+ is present in the 5′-end-labeling reaction, allowing for some ribozyme self-cleavage and 5′-end-labeling of the 1/99 RNA.) Migration of the −30/−1 band is identical for all four ribozymes.
Metal switch experiments suggest that the catalytic ion binding site is the same for all ribozymes
Because similar negative linkage between H+ and Mg2+ was observed for all four ribozymes, we reasoned that changing bp 1 does not alter the mode (outer-sphere-like) by which the catalytic Mg2+ binds or the location of its binding site. To further investigate this issue, we performed metal switch experiments. It was previously shown that the G•U ribozyme reacts nearly identically in Mg2+ and Ca2+ (Nakano et al. 2000). This observation, along with other metal-dependence kinetic data (Nakano et al. 2003), suggested that the ribozyme binds and utilizes Mg2+ and Ca2+ ions in similar ways. To test whether bp 1 identity affects the metal preference of the four ribozymes in this study, we performed reactions for each ribozyme in the presence of 10 mM Ca2+ (pH 7).
The G•U and A+•C ribozymes reacted slightly faster (1.2- and 1.4-fold, respectively) in Ca2+ than Mg2+, while the A-U and the G-C ribozymes reacted at essentially the same rate for the two metal ions (Table 2). The slightly greater rate of the G•U ribozyme in Ca2+ is consistent with previous observations (Nakano et al. 2000, 2003). Overall, switching metal ions does not have a large effect on ribozyme activity, and so it is likely that the four ribozymes bind and utilize the catalytic metal ion in very similar ways.
TABLE 2.
Metal switch experiments (pH 7)
All four ribozymes have similar Mg2+ dependencies at low pH, with A+•C differing at higher pH
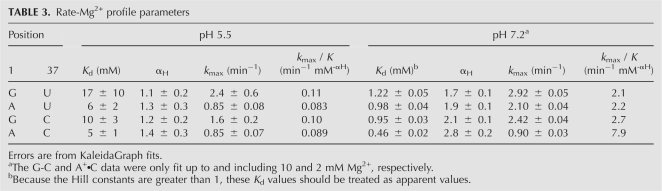
To determine whether changing bp 1 from a G•U wobble to the other base combinations affects Mg2+ binding affinity, we obtained rate-Mg2+ profiles. Such profiles have been reported for the G•U ribozyme (Nakano et al. 2000). In order to further investigate anticooperative coupling between H+ and Mg2+ binding, Mg2+ binding was tested at two pH values. We chose a low pH at which the four ribozymes behave similarly (pH 5.5) and a higher pH at which the A+•C ribozyme has compromised reactivity (pH 7.2). With the exception of the A+•C ribozyme at pH 7.2, the rate-Mg2+ profiles could be fit to a Hill equation, Equation 5 derived from Scheme 4.
SCHEME 4.
Multiple-Mg2+mechanism.
At pH 5.5, the G•U and G-C ribozymes have k max values near 2 min−1 (2.4 ± 0.6 min−1 and 1.6 ± 0.2 min−1, respectively) while the A-U and A+•C ribozymes have k max values of 0.85 min−1, representing a two- to threefold difference (Table 3). It appears that the A-U and A+•C ribozyme profiles reach saturation over the range of Mg2+ concentrations tested, but that the G•U and G-C ribozymes do not (Fig. 7A). The apparent K d values for Mg2+ binding for the G•U, A-U, G-C, and A+•C ribozymes are similar within experimental error at 17 ± 10 mM, 6 ± 2 mM, 10 ± 3 mM, and 5 ± 1 mM, respectively (Table 3). The Hill coefficient, αH, at pH 5.5 is ∼1 for each ribozyme, supporting binding of at least one functional Mg2+ ion, consistent with previous findings (Nakano et al. 2000).
TABLE 3.
Rate-Mg2+ profile parameters
FIGURE 7.
Comparison of the Mg2+-dependence for the G•U (●), A-U (▲), G-C (○), and A+•C (△) ribozymes. (A) Experiments performed at pH 5.5. Fits to the G•U, A-U, G-C, and A+•C (R2 > 0.98) ribozymes were according to Equation 5 and gave k max values of 2.4 ± 0.6, 0.85 ± 0.08, 1.6 ± 0.2, and 0.85 ± 0.07 min−1, αH values of 1.1 ± 0.2, 1.3 ± 0.3, 1.2 ± 0.2, and 1.4 ± 0.3, and K d values of 17 ± 10, 6 ± 2, 10 ± 3, and 5 ± 1 mM, respectively. (B) Experiments performed at pH 7.2. Fits to the G•U, A-U, G-C, and A+•C (R2 > 0.99) ribozymes were according to Equation 5 and gave k max values of 2.92 ± 0.05, 2.10 ± 0.04, 2.42 ± 0.04, and 0.90 ± 0.03 min−1, αH values of 1.7 ± 0.1, 1.9 ± 0.1, 2.1 ± 0.1, and 2.8 ± 0.2, and apparent K d values of 1.22 ± 0.05, 0.98 ± 0.04, 0.95 ± 0.03, and 0.46 ± 0.02 mM, respectively. The G-C and A+•C data were only fit up to and including 10 and 2 mM Mg2+ data, respectively. The average error in k obs was found to be ±10%.
At pH 7.2, the G•U, A-U, and G-C ribozymes have similar k max, K d, and αH values, using data truncated at 10 mM Mg2+ for the G-C ribozyme (Table 3). The G-C ribozyme differs from the G•U and A-U ribozymes above 10 mM Mg2+ in that its k obs decreases and then starts to level off as the concentration of Mg2+ approaches 50 mM (Fig. 7B). The Hill coefficients at pH 7.2 for these three ribozymes are ∼2, suggesting that each ribozyme binds at least two Mg2+ ions at higher pH.
At pH 7.2, the rate-Mg2+ profile for the A+•C ribozyme differs in that the rate decreases above 2 mM Mg2+. The A+•C data were therefore only fit up to and including 2 mM Mg2+. The apparent K d and k max values obtained for the A+•C ribozyme were two- to threefold lower than for the other ribozymes, and the Hill coefficient was 2.8 ± 0.2 (Table 3). Unusual behavior for the A+•C ribozyme at pH 7.2 is not unexpected, given the deviation of its rate-pH profiles near or above pH 6 (Fig. 4) where the A+•C wobble pair is not fully formed.
We note that k max/K d αH values are very similar for all four ribozymes at pH 5.5 (Table 3). This parameter is similar to k cat/K m for Michaelis–Menten enzyme kinetics, which is a measure of enzyme specificity (Fersht 1984). Similarity of this parameter among the four variants suggests little if any influence of the purine–pyrimidine base combinations at bp 1 on the reaction mechanism. Likewise, at pH 7.2, the k max/K d αH values are similar for the G•U, A-U, and G-C ribozymes, although the value for the A+•C ribozyme is three- to fourfold higher.4
DISCUSSION
Both prevailing mechanistic models, GAB1 and GAB2, for phosphodiester bond cleavage by the HDV ribozyme involve proton transfer by C75 and a hydrated Mg2+ ion. The bond that is cleaved during the reaction is located between G1 of bp 1 and U−1. In this study, we kinetically characterized four purine–pyrimidine bp 1 variants. In this section, we consider potential mechanistic roles bp 1 could play in the reaction, including binding a catalytic Mg2+ ion, influencing the pK a of C75, or positioning functional groups for catalysis.
Base pair 1 does not have to be a wobble pair but is required for optimal activity
We predicted secondary structures of genomic ribozymes with purine–pyrimidine and pyrimidine–purine base combinations at bp 1 using an energy minimization program capable of pseudoknot prediction (Ruan et al. 2004a,b). Predicted folds were consistent with sequence comparison data and prior kinetics studies in that ribozymes with purine–pyrimidine combinations at bp 1 were predicted to form largely native structure while ribozymes with pyrimidine–purine combinations were predicted to misfold (Fig. 3). Since viral and mammalian HDV(-like) ribozymes have only been found to have purine–pyrimidine combinations at bp 1 and pyrimidine–purine combinations severely impair activity, we conducted experiments on ribozymes with purine–pyrimidine combinations at bp 1. These ribozymes were evaluated as a function of pH and Mg2+ concentration using a fast-folding RNA sequence in order to focus on issues of chemistry rather than folding.
The four combinations tested at bp 1 are shown in Figure 2. The G•U and A+•C base combinations form wobble pairs, with the A+•C wobble requiring protonation to fold, while the A-U and G-C combinations form Watson–Crick base pairs. Rate-pH profiles for the G•U, A-U, and G-C ribozymes were nearly identical at both 1 and 10 mM Mg2+ concentrations (Fig. 4) and could be fit using a model in which one ionization event, presumably protonation of C75, gives rise to the active species (Scheme 1). The A+•C ribozyme behaved identically to the other three ribozymes up until pH ∼6, above which its activity decreased (Fig. 4). This effect is consistent with deprotonation of the A+•C wobble pair and formation of a different structure at elevated pH. Given that bp 1 is part of the active site, it is reasonable that disrupting it decreases activity. Ultimately, we developed a four-state, two-channel model to account for the kinetic behavior of the A+•C ribozyme (Scheme 3), in which the ribozyme is fully active when the A and C form a protonated wobble pair, and in which the ribozyme retains moderate activity with an unprotonated A•C without miscleaving. These data support the conclusion that a cleavage site base pair is required for optimal activity.
It is typically assumed that A-U and G-C combinations positioned within the context of a helix form Watson–Crick base pairs. However, it was previously shown for the group I intron that when the G•U wobble pair at the active site is mutated to a G-C, the C protonates and the G and C+ form a wobble pair with the same geometry as the G•U wobble (Knitt et al. 1994). This conclusion was made on the basis of a distinct pH dependence of the mutant G-C group I intron reaction (Knitt et al. 1994). In the case of the HDV ribozyme, however, the pH dependence of the G-C ribozyme reaction is identical to that of the G•U and A-U ribozymes (Fig. 4). Thus, it is highly unlikely that the G and C of the HDV ribozyme form a protonated wobble pair at bp 1, which further supports the conclusion that base pair 1 does not need to be a wobble pair for the ribozyme to be fully functional.
Negative linkage occurs between Mg2+ and C75H+, as well as the A+•C wobble pair
The rate-pH profiles indicated that the pK a of C75 at a particular Mg2+ concentration is the same within error for all four ribozymes (Fig. 4; Table 1). The profiles also revealed that the pK a of C75 decreases as the concentration of Mg2+ increases, consistent with previously established anticooperative coupling between H+ and Mg2+ binding (Nakano et al. 2000). Combined with the observation that each ribozyme reacts similarly in Mg2+ or Ca2+ (Table 2) and that the ribozymes have similar Mg2+ binding isotherms (Fig. 7; Table 3), this suggests that the four bp 1 variant ribozymes bind the catalytic metal ion and accomplish the reaction in very similar fashions.
Interestingly, in the case of the A+•C ribozyme, the pK a of the A+•C wobble pair also decreases with increasing Mg2+ concentration (Table 1). The presence of thermodynamic linkage between bp 1 and Mg2+ suggests that the catalytic metal ion may bind near both bp 1 and C75, although we cannot rule out the influence of diffusely bound metal ions.
Unusual effects of Mg2+ on the G-C and A+•C ribozymes suggest weak binding of additional ions
We examined the Mg2+ concentration dependence of the reaction for the four bp 1 variant ribozymes. At pH 5.5, all four ribozymes have similar k max and K d values within error and Hill coefficients of unity (Fig. 7A; Table 3), which suggests that they behave similarly as long as bp 1 is formed.
At pH 7.2, the G•U, A-U, and G-C ribozymes exhibit a similar Mg2+ dependence up to 10 mM Mg2+, above which the rate of the G-C ribozyme decreases and then levels off as the Mg2+ concentration nears 50 mM (Fig. 7B). The origin of this effect is not entirely clear, but it is possible that there is one or more additional, weaker Mg2+ binding sites for the G-C ribozyme, which cause it to adopt a less reactive structure. The G-C ribozyme was also predicted to form an additional alternative pairing, Alt P1, for the −30/99 folding (Fig. 3, fold C), so perhaps high Mg2+ causes this fold to populate. This would support the notion that the purine–pyrimidine combinations tested at bp 1, when properly formed in the context of a native ribozyme, have little if any influence on the reaction mechanism. Overall, under physiological Mg2+ concentrations, the G•U, A-U, and G-C ribozymes have similar k max and K d values at all pH values.
Curiously, the Hill coefficients suggest that the G•U, A-U, and G-C ribozymes bind at least two Mg2+ ions at pH 7.2 (Table 3). There is some precedent for Hill coefficients greater than unity in the HDV ribozyme at higher pH. Previous work from our laboratory revealed that the HDV ribozyme can react by one Mg2+-independent and two Mg2+-dependent channels (Nakano et al. 2001). In particular, channel 1 involves cleavage in the absence of divalent metal ions and uses solvent or hydroxide ion as a catalyst, channel 2 involves cleavage in the presence of a structural Mg2+ ion without participation of a catalytic metal ion, and channel 3 involves both structural and catalytic Mg2+ ions. Under channel 2 conditions (1 M NaCl and micromolar to near-molar concentrations of Mg2+) at higher pH (pH 8 and 9), Hill coefficients close to two were observed, suggesting a linkage between the binding of the structural and catalytic Mg2+ ions (Nakano et al. 2001).
The A+•C ribozyme rate-Mg2+ profile at pH 7.2 differs from those obtained for the other ribozymes (Fig. 7B). The profile is similar to those for the other ribozymes in terms of shape, but only up to 2 mM, above which k obs decreases as the Mg2+ concentration approaches 50 mM. The molecular origin of the rate decrease with higher concentrations of Mg2+ is unclear; however, it may be associated with a C at position 37 since the rate of the G-C ribozyme also decreased at higher Mg2+ concentrations (Fig. 7B). Notably, the aforementioned Alt P1 pairing is stabilized by C37 (Fig. 3, fold C), which is common to the A+•C and G-C ribozymes.
The A+•C ribozyme data up to 2 mM were fit with the same equation (Equation 5) used for the other ribozymes. The K d and k max values are two- to threefold lower than for the other ribozymes, and it appears that the A+•C ribozyme may bind at least three Mg2+ ions (Table 3). Given that the A+•C ribozyme exhibits a different pH dependence than the other ribozymes, it is not surprising that its Mg2+ dependence is also different at high pH.
Possible mechanistic roles for bp 1 in the HDV ribozyme reaction
As illustrated in Figure 2, the 4 bp tested differ in the hydrogen bond donors and acceptors presented to the major and minor grooves, as well as their geometries. Of the 4 bp, the G•U wobble has the widest major groove and is the only pair whose major groove contains only hydrogen bond acceptors. Indeed, the major groove of the G•U wobble pair can be a motif for the binding of metal ions and other positively charged ligands (Varani and McClain 2000; Keel et al. 2007; Xu et al. 2007). A recently compiled database of metal ion binding sites in RNA structures supports this notion (Stefan et al. 2006). Both the A-U and G-C Watson–Crick base pairs as well as the A+•C wobble pair have one or more amino groups in the major groove, which may disrupt the negative electrostatic potential present in the G•U wobble major groove as well as present steric blocks to metal ion binding. In addition, the A+•C wobble pair has a positive charge, which has the potential to disfavor binding of a metal ion.
Our initial hypothesis was that if the G•U wobble pair at bp 1 comprised the catalytic metal ion binding site, then ribozymes with other base combinations would have compromised metal ion binding. However, this turned out to not be the case. As illustrated in Figures 4 and 7A, the four ribozymes behaved similarly under conditions where bp 1 is formed, leading us to conclude that the majority of the bp 1 functionalities are not ligands for metal ion binding, nor do they prevent steric blocks.
At the same time, the data herein provide indirect evidence to suggest that a metal ion may be binding at or near bp 1. The pK a of the A+•C wobble pair decreased by 0.3 unit upon increasing Mg2+ concentration from 1 to 10 mM (Table 1). This negative thermodynamic linkage is similar to that observed between C75H+ and Mg2+, suggesting that the catalytic Mg2+ ion may bind near both C75 and bp 1. In addition, the two ribozymes with wobble pairs at bp 1 showed a slight rate enhancement with Ca2+ (Table 2), supporting proximity of bp 1 and a metal ion. If bp 1 is part of a metal ion-binding site, then the largely similar behavior of the four variants suggests that some feature common to all four ribozymes is involved. Since the 4 bp tested are purine–pyrimidine combinations, they all have N7 and N3 at the purine position (position 1) available as potential ligands. Another possibility is that the metal ion is coordinated to the ribose sugar-phosphate backbone of bp 1.
In addition to influencing metal ion binding, we considered whether bp 1 could contribute to catalysis by influencing the pK a of C75. The pK a values acquired for the four bp 1 variants suggest that this is not the case (Table 1). The pK a of C75 derived from various rate-pH profiles is the same within error for all four ribozymes at both low and high Mg2+ concentrations; this includes a C75 pK a in the presence of a protonated A+•C wobble, which has a different electrostatic potential. Lack of an influence of bp 1 on the pK a of C75 is consistent with distances between N3 of C75 and key atoms within the G•U wobble pair of greater than 8 Å in the product crystal structure (Ferre-D'Amare et al. 1998).
In addition to the major groove, the G•U wobble pair differs from the other base pairs in the minor groove (Fig. 2). Specifically, the exocyclic amine of G is not involved in base-pairing and is shifted into the minor groove, while the A-U and A+•C pairs lack the amino functionality altogether. Although the exocyclic amine of guanine is important for mediating RNA-RNA and RNA–protein interactions in a number of systems (Varani and McClain 2000; Xu et al. 2007), crystal structures (Ferre-D'Amare et al. 1998; Ke et al. 2004) and biochemical data (Been and Wickham 1997) suggest that this functional group is not important for the HDV ribozyme reaction. Observation that base combinations without a minor groove amino group at bp 1 behave similarly supports the minor groove of bp 1 as not being important for catalysis.
We also considered the possibility that the geometry of the wobble pair could be important for the HDV ribozyme reaction. In addition to shifting the exocyclic amine of the G into the minor groove, the geometry of a G•U wobble pair can introduce certain changes into the structure of A-form RNA, such as overtwisting and undertwisting, depending on the sequence context (Varani and McClain 2000; Xu et al. 2007). However, structure prediction and rate-pH and rate-Mg2+ profiles reveal that bp 1 does not have to be a G•U wobble pair or a wobble pair at all. At present, the reason for strict conservation of a G•U wobble pair in the antigenomic ribozyme is unclear. It could reflect other selective pressures on this ribozyme, such as a need to pair with the attenuator or to avoid misfolds unique to the antigenomic ribozyme. Further experiments are needed to distinguish these possibilities.
In conclusion, the data presented herein directly support a model in which bp 1 provides a structural role in catalysis and does not need to be a wobble pair. It remains possible that bp 1 is involved in binding the catalytic metal ion through a common molecular feature such as the N7 or N3 of the purine. This work, in combination with other studies, may provide constraints for future investigations into the mechanism of HDV ribozyme catalysis.
MATERIALS AND METHODS
Preparation of RNA
The −30/99 genomic HDV RNA used here was transcribed from pT7 −30/99 using phage T7 polymerase as described (Chadalavada et al. 2000). The transcript contains 30 nt upstream of the cleavage site and 15 nt downstream of the 3′ end of the ribozyme (Fig. 1A). All transcripts contain the G11C mutation that biases the Alt P1–P1 equilibrium toward the native fold (Chadalavada et al. 2002). Transcripts are Bfa I run-offs, which contain ribozyme and HDV-derived RNA sequence only. RNA was transcribed, purified, and radiolabeled as described (Chadalavada et al. 2000). All mutant plasmids were generated from pT7 −30/99 using the QuikChange kit (Stratagene). Sequences were confirmed by dideoxy sequencing after both minipreps and maxipreps (Qiagen).
Ribozyme kinetics and data fitting
Reactions were performed as described (Nakano et al. 2000). A typical self-cleavage reaction contained 2 nM 5′-32P-end-labeled RNA, 25 mM buffer, 10 μM antisense oligonucleotide, and 0.05–50 mM MgCl2. The RNA was renatured at 55°C for 10 min in either water or 0.5 mM Tris (pH 7.5)/0.05 mM EDTA in the presence of the antisense oligonucleotide AS(−30/−7), which disrupts Alt 1 and facilitates native folding of the ribozyme (Chadalavada et al. 2000), and then cooled at room temperature for 10 min. Buffer was added and the mixture was incubated at 37°C for 2 min. Reactions were limited to a pH range of ∼4–9, outside of which acid- and alkaline-denaturation events interfere (Moody et al. 2005). The buffer was MES for experiments at pH 4.5–6.3 and HEPES for experiments at pH 6.7 to 9.0. Buffers were prepared at room temperature and the pH values were measured at 37°C to determine the experimental pH. A zero time point was removed and self-cleavage was initiated by the addition of MgCl2 or CaCl2, as appropriate. All time points were quenched by mixing with an equal volume of 95% (v/v) formamide loading buffer containing 60 mM EDTA and placing on dry ice. At the end of each reaction, the reaction mixture was checked on pH paper to ensure that the correct pH had been maintained. Time points were fractionated on a denaturing 10% (w/v) polyacrylamide gel. Gels were dried and visualized using a PhosphorImager (Molecular Dynamics). The Mg2+ concentrations were corrected for the small amount of EDTA present in some of the reactions as described (Nakano et al. 2001).
Plots of fraction product versus time were constructed and fit to the single-exponential equation (Equation 1),
where f is the fraction of ribozyme cleaved at time t, A is the fraction of ribozyme cleaved at completion, A + B is the burst fraction (in all cases, A + B ≈ 0), and k obs is the observed first-order rate constant. Kinetic parameters were obtained using nonlinear least-squares fitting by KaleidaGraph (Synergy Software). Typically, 4–6 half-lives of data were collected. Multiple determinations of k obs for the G•U ribozyme were in good agreement, and the same rate-pH profile at 10 mM Mg2+ was obtained as before (Nakano et al. 2000). Rate-pH and rate-Mg2+ profiles were constructed for all ribozymes and fit as described below. The average error in k obs was determined to be ±10% from the covariance/correlation matrices for the time courses (taking all pH and Mg2+ studies into account).
In general, the equations used to fit the rate-pH and rate-Mg2+ profiles were derived from schemes involving the minimal number of protonation and deprotonation events necessary to arrive at the active ribozyme species. Rate-pH profiles for the G•U, A-U, and G-C ribozymes were fit to the logarithm of Equation 2b, which was derived from Equation 2a and the single-channel/single-deprotonation mechanism shown in Scheme 1.
 |
Scheme 1 depicts GAB Model 1, in which C75 acts as an ionizable general base and hydrated Mg2+ acts as a fully functional general acid; a water coordinated to fully hydrated Mg2+ has a pK a of 11.4 (Dahm et al. 1993). However, the kinetically equivalent GAB model 2 in which protonated C75 is the general acid and a hydrated Mg2+ hydroxide ion is the general base leads to the same rate-pH profile, albeit with a slightly more complex mathematical form (Nakano and Bevilacqua 2007). Both approaches provide the kinetic parameters k max and pK a,C75.
The rate-pH profiles for the A+•C ribozyme could not be described by Scheme 1 (see below). We therefore developed two more schemes. Scheme 2 is similar to Scheme 1 in that it is a single-channel mechanism and therefore it is derived beginning with Equation 2a. Equation 3 applies to Scheme 2.
Scheme 3, on the other hand, has two reactive channels, which describes the data better (see Results). Equations 4a and 4b apply to Scheme 3
In these two schemes, we assumed that C75 and the A+•C base pair do not couple thermodynamically, which is consistent with the data (see Discussion).
The A+•C ribozyme data were initially fit using the logarithm of Equation 2b (not shown). In 1 mM Mg2+, the χ2 values for the A+•C ribozyme were 0.084 and 0.037 for Equation 2b and Equation 4b, respectively, supporting Equation 4b and Scheme 3. In 10 mM Mg2+, the χ2 values for the A+•C ribozyme were 0.26 and 0.018 for Equation 2b and Equation 4b, respectively, providing even stronger support for Equation 4b and Scheme 3. In addition, attempts to fit the A+•C ribozyme data (at both 1 mM and 10 mM Mg2+) using Equation 2b resulted in sinusoidally distributed residuals, whereas using Equation 4b resulted in randomly distributed residuals. Based on these observations, the A+•C ribozyme data is better described by Scheme 3 as compared to Scheme 1. In addition, Equation 3 from Scheme 2 does not fit the data well because it gives a bell-shaped curve.
Rate-Mg2+ profiles were fit to Equation 5, which can be derived from the single-channel mechanism shown in Scheme 4 in which Mg2+ ions bind cooperatively to free ribozyme, with a Hill coefficient (αH) and apparent dissociation constant (K d) to provide the active ribozyme species.
 |
Free energy minimization
We folded various ribozymes using the iterated loop matching (ILM) algorithm web server: http://cic.cs.wustl.edu/RNA/ (Ruan et al. 2004a). This algorithm can predict pseudoknot formation, which is important for the HDV ribozyme.
ACKNOWLEDGMENTS
We thank Susan Senchak for conducting pilot kinetics experiments. This work was supported by NIH Grant R01-58709, NSF Grant 0527102, and an NSF graduate research fellowship (to A.L.C.-S.).
Footnotes
As a shorthand, we refer to the cleavage site base pair as bp 1.
The small (2 bp) P1.1 pairing is not predicted correctly. Instead, formation of a base pair between U- 1 and G38, which extends P1 by 1 bp, is predicted. In addition, the base pair between A43 and G74 at the top of P4 is not predicted. Lastly, a terminal base pair in L4 between A56 and U60 is predicted, while experiments coupled with mFold predictions suggest that these bases are single stranded at least part of the time (Chadalavada et al. 2000).
Previously described alternative pairings are denoted with (previously assigned) numbering (e.g., Alt 1), while new alternative pairings are denoted with lettering (e.g., Alt A).
The AC ribozyme kmax/Kdα H value is higher because both Kd and kmax are two- to threefold lower (Table 3) and the ribozyme may bind an additional Mg2+ ion. It is therefore not straightforward to compare the kmax/Kdα H value for the AC ribozyme at higher pH.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1140308.
REFERENCES
- Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Allain F.H., Varani G. Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: Sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barfod E.T., Cech T.R. The conserved U.G pair in the 5′ splice site duplex of a group I intron is required in the first but not the second step of self-splicing. Mol. Cell. Biol. 1989;9:3657–3666. doi: 10.1128/mcb.9.9.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M.D. HDV ribozymes. Curr. Top. Microbiol. Immunol. 2006;307:47–65. doi: 10.1007/3-540-29802-9_3. [DOI] [PubMed] [Google Scholar]
- Been M.D., Wickham G.S. Self-cleaving ribozymes of hepatitis δ virus RNA. Eur. J. Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., Brown T.S., Nakano S., Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Cai Z., Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Doudna J.A. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- Chadalavada D.M., Knudsen S.M., Nakano S., Bevilacqua P.C. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis δ virus ribozyme. J. Mol. Biol. 2000;301:349–367. doi: 10.1006/jmbi.2000.3953. [DOI] [PubMed] [Google Scholar]
- Chadalavada D.M., Senchak S.E., Bevilacqua P.C. The folding pathway of the genomic hepatitis δ virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- Chadalavada D.M., Cerrone-Szakal A.L., Bevilacqua P.C. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA. 2007;13:2189–2201. doi: 10.1261/rna.778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm S.C., Derrick W.B., Uhlenbeck O.C. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- Das S.R., Piccirilli J.A. General acid catalysis by the hepatitis δ virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Cormack B.P., Szostak J.W. RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc. Natl. Acad. Sci. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs W.D., Cech T.R. A tertiary interaction in the Tetrahymena intron contributes to selection of the 5′ splice site. Genes & Dev. 1994;8:1198–1211. doi: 10.1101/gad.8.10.1198. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare A.R., Doudna J.A. Crystallization and structure determination of a hepatitis δ virus ribozyme: Use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme structure and mechanism. W.H. Freeman; New York: 1984. [Google Scholar]
- Golden B.L., Kim H., Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Ke A., Zhou K., Ding F., Cate J.H., Doudna J.A. A conformational switch controls hepatitis δ virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Ke A., Ding F., Batchelor J.D., Doudna J.A. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Keel A.Y., Rambo R.P., Batey R.T., Kieft J.S. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S., Tinoco I., Jr Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure. 1997;5:713–721. doi: 10.1016/s0969-2126(97)00225-6. [DOI] [PubMed] [Google Scholar]
- Knitt D.S., Narlikar G.J., Herschlag D. Dissection of the role of the conserved G.U pair in group I RNA self-splicing. Biochemistry. 1994;33:13864–13879. doi: 10.1021/bi00250a041. [DOI] [PubMed] [Google Scholar]
- Krasovska M.V., Sefcikova J., Spackova N., Sponer J., Walter N.G. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis δ virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Krasovska M.V., Sefcikova J., Reblova K., Schneider B., Walter N.G., Sponer J. Cations and hydration in catalytic RNA: Molecular dynamics of the hepatitis δ virus ribozyme. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. The molecular biology of hepatitis δ virus. Annu. Rev. Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- Legault P., Pardi A. In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 1994;116:8390–8391. [Google Scholar]
- Liu H., Robinet J.J., Ananvoranich S., Gauld J.W. Density functional theory investigation on the mechanism of the hepatitis δ virus ribozyme. J. Phys. Chem. B. 2007;111:439–445. doi: 10.1021/jp064292n. [DOI] [PubMed] [Google Scholar]
- Moody E.M., Lecomte J.T., Bevilacqua P.C. Linkage between proton binding and folding in RNA: A thermodynamic framework and its experimental application for investigating pKa shifting. RNA. 2005;11:157–172. doi: 10.1261/rna.7177505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S., Bevilacqua P.C. Mechanistic characterization of the HDV genomic ribozyme: A mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- Nakano S., Chadalavada D.M., Bevilacqua P.C. General acid–base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Nakano S., Proctor D.J., Bevilacqua P.C. Mechanistic characterization of the HDV genomic ribozyme: Assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- Nakano S., Cerrone A.L., Bevilacqua P.C. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- Nishikawa F., Fauzi H., Nishikawa S. Detailed analysis of base preferences at the cleavage site of a trans-acting HDV ribozyme: A mutation that changes cleavage site specificity. Nucleic Acids Res. 1997;25:1605–1610. doi: 10.1093/nar/25.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelere A.K., Kardon J.R., Strobel S.A. pK(a) perturbation in genomic Hepatitis δ Virus ribozyme catalysis evidenced by nucleotide analogue interference mapping. Biochemistry. 2002;41:3667–3675. doi: 10.1021/bi011816v. [DOI] [PubMed] [Google Scholar]
- Perrotta A.T., Been M.D. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A.T., Shih I., Been M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Perrotta A.T., Wadkins T.S., Been M.D. Chemical rescue, multiple ionizable groups, and general acid–base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Stormo G.D., Zhang W. ILM: A web server for predicting RNA secondary structures with pseudoknots. Nucleic Acids Res. 2004a;32:W146–W149. doi: 10.1093/nar/gkh444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Stormo G.D., Zhang W. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics. 2004b;20:58–66. doi: 10.1093/bioinformatics/btg373. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K., Luptak A., Litovchick A., Szostak J.W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- Stefan L.R., Zhang R., Levitan A.G., Hendrix D.K., Brenner S.E., Holbrook S.R. MeRNA: A database of metal ion binding sites in RNA structures. Nucleic Acids Res. 2006;34:D131–D134. doi: 10.1093/nar/gkj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S.A., Ortoleva-Donnelly L., Ryder S.P., Cate J.H., Moncoeur E. Complementary sets of noncanonical base pairs mediate RNA helix packing in the group I intron active site. Nat. Struct. Biol. 1998;5:60–66. doi: 10.1038/nsb0198-60. [DOI] [PubMed] [Google Scholar]
- Varani G., McClain W.H. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadkins T.S., Been M.D. Ribozyme activity in the genomic and antigenomic RNA strands of hepatitis δ virus. Cell. Mol. Life Sci. 2002;59:112–125. doi: 10.1007/s00018-002-8409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Liu L., Cheng Y.H., Fu Y., Guo Q.X. Theoretical examination of two opposite mechanisms proposed for hepatitis δ virus ribozyme. J. Phys. Chem. B. 2007;111:1514–1516. doi: 10.1021/jp070120u. [DOI] [PubMed] [Google Scholar]
- Wu H.N., Huang Z.S. Mutagenesis analysis of the self-cleavage domain of hepatitis δ virus antigenomic RNA. Nucleic Acids Res. 1992;20:5937–5941. doi: 10.1093/nar/20.22.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.N., Lee J.Y., Huang H.W., Huang Y.S., Hsueh T.G. Mutagenesis analysis of a hepatitis δ virus genomic ribozyme. Nucleic Acids Res. 1993;21:4193–4199. doi: 10.1093/nar/21.18.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Landon T., Greenbaum N.L., Fenley M.O. The electrostatic characteristics of G.U wobble base pairs. Nucleic Acids Res. 2007;35:3836–3847. doi: 10.1093/nar/gkm274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.M., Zhou D.M., Takagi Y., Kasai Y., Inoue A., Baba T., Taira K. Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme. Nucleic Acids Res. 2002;30:2374–2382. doi: 10.1093/nar/30.11.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]