Abstract
Autogenous regulation is a general strategy of balancing ribosomal protein synthesis in bacteria. Control mechanisms have been studied in detail for most of ribosomal protein operons, except for rpsB-tsf encoding essential r-protein S2 and elongation factor Ts, where even the promoter has remained unknown. By using single-copy translational fusions with the chromosomal lacZ gene and Western-blot analysis, we demonstrate here that S2 serves as a negative regulator of both rpsB and tsf expression in vivo, acting at a single target within the rpsB 5′-untranslated region (5′-UTR). As determined by primer extension, transcription of the Escherichia coli rpsB-tsf operon starts 162 nucleotides upstream of the rpsB initiation codon at a single promoter TGTGGTATAAA belonging to the extended −10 promoter class. Both the promoter signature and the 5′-UTR structure of the rpsB gene appear to be highly conserved in γ-proteobacteria. Deletion analysis of the rpsB 5′-UTR within rpsB′-′lacZ fusions has revealed that an operator region involved in the S2 autoregulation comprises conserved structural elements located upstream of the rpsB ribosome binding site. The S2-mediated autogenous control is impaired in rpsB mutants and, more surprisingly, in the rpsA mutant producing decreased amounts of truncated r-protein S1 (rpsA∷IS10), indicating that S2 might act as a repressor in cooperation with S1.
Keywords: rpsB-tsf operon, extended −10 promoter, translation initiation region, autogenous regulation, ribosomal proteins S1 and S2, elongation factor Ts
INTRODUCTION
In bacteria, ribosome biosynthesis is governed by transcriptional and translational regulatory mechanisms that provide a balanced and coordinated production of individual ribosomal components. Transcription of rRNA responds to nutritional cues, while production of ribosomal proteins (r-proteins) is tightly linked to the rRNA level by the feedback inhibition mechanism known as autogenous control. Most of r-protein operons encode a regulatory r-protein that directly binds 16S or 23S rRNA during ribosome assembly, but if synthesized in excess relative to its target on rRNA, serves as an operon-specific translational repressor by binding to its own mRNA to prevent further translation (for reviews, see Nomura et al. 1984; Zengel and Lindahl 1994; Nomura 1999). In the 30S ribosomal subunit, only r-proteins S1 and S2 present exceptional cases, as they do not recognize naked 16S rRNA and participate in the 30S assembly at the very late step (Culver 2003), but nevertheless possess the potential to act as specific autogenous regulators. However, while the autoregulation of a key mRNA-binding protein S1 has been corroborated both in vitro and in vivo by various approaches (Skouv et al. 1990; Boni et al. 2000, 2001), still little is known about how the synthesis of essential r-protein S2 might be controlled.
Ribosomal protein S2 is highly conserved in all forms of life (its counterparts are referred to as S0 in yeast and SA in higher eukaryotes) and is essential for the translational machinery in all prokaryotes, eukaryotes, mitochondria, and chloroplasts (Ardini et al. 1998; Wilson and Nierhaus 2005). Moreover, concurrent with the appearance of the extracellular matrix in higher eukaryotes, S2 (SA) acquired an additional extraribosomal function of laminin-binding receptor during evolution (Ardini et al. 1998). At the same time, a functional role of S2 in ribosomal translational activity remains unclear. Recent observations suggest that in prokaryotes, S2 might be involved in protecting and stabilizing the Shine–Dalgarno (SD) helix docked in the “chamber” between the head and the platform of the 30S subunit (Kaminishi et al. 2007) as well as in binding the SD duplex at the post-initiation step (Yusupova et al. 2006). However, these data cannot explain an essential role of S2 in translational systems that follow the prokaryotic scenario but do not use the SD interaction during translation initiation, as is the case of the smallest γ-bacterial endosymbiont Carsonella ruddii (Nakabachi et al. 2006) or mammalian mitochondria (Koc et al. 2001), where the 3′-tail of a small ribosomal subunit RNA is naturally deleted. Thus, essential functions of a highly conserved protein S2 in protein synthesis in all cells are still waiting for a rational explanation.
According to the high-resolution structure of the small subunit from Thermus thermophilus, S2 within 30S has an elongated bidomain structure α2, α1β5α3, and contacts with two rather distant 16S rRNA regions involving helices 26 in the body and 35–37 in the head of the subunit (Wimberly et al. 2000; Brodersen et al. 2002). This suggests that, in spite of being one of the latest proteins in 30S assembly (Mizushima and Nomura 1970; Culver 2003), S2 is capable of 16S rRNA binding, but the binding surface of RNA must be preformed by the earlier r-proteins. Incorporation of S2 is absolutely necessary for binding r-protein S1 that accomplishes the assembly of the 30S subunit capable of recruiting mRNA in Escherichia coli and most likely in other S1-dependent translational systems (Bollen et al. 1979; Moll et al. 2002). Interestingly, a stoichiometric S1–S2 complex was copurified with RNA-polymerase and a global regulator Hfq from E. coli stationary-phase cultures (Sukhodolets and Garges 2003), implying that S1 and S2 are capable of interacting with each other even outside the ribosome. Other known protein–protein interactions of S2 in a cell include a chaperonin GroEL required for initial folding and conformational maintenance of S2 in vivo (Houry et al. 1999) and the ATP-dependent Lon protease that specifically degrades S2 in the presence of inorganic polyphosphate, providing an important source of amino acids for cell survival during starvation (Kuroda et al. 2001; Nishii et al. 2005).
In eubacteria, S2 is encoded in the rpsB-tsf operon that also codes for translation elongation factor Ts. It is logical to surmise that like in the case of other r-protein operons, production of these essential components of translational machinery might also be coordinated with the overall ribosome synthesis via the feedback mechanism. The S2 capability to modulate the rpsB-tsf expression has never been assayed, and the only argument in favor of the S2 autoregulation came from the observation that expression of extra copies of the plasmid-borne rpsB gene did not augment the S2 level in a cell (An et al. 1981; Bendiak and Friesen 1981). Furthermore, the promoter(s) responsible for the rpsB-tsf transcription in E. coli has so far remained unknown.
The main goal of this work was to study the regulatory mechanisms that govern rpsB-tsf expression in vivo. We determined the length of the rpsB 5′-untranslated (5′-UTR) region and located the rpsB promoter that appeared to belong to the extended −10 promoter class. Phylogenetic analysis revealed that both the promoter signature and the rpsB 5′-UTR structure are highly conserved in γ-proteobacteria. By using single-copy translational fusions with the chromosomal lacZ gene and Western-blot analysis, we show that S2 functions as a negative regulator of both its own and tsf expression in vivo, acting at a single target within the rpsB 5′-UTR. Surprisingly, the S2 repressor activity was found decreased in the rpsA∷IS10 mutant (ssyF29) producing a subnormal amount of truncated S1 (Boni et al. 2000), thus implying that S2 may cooperate with S1 in regulating the rpsB-tsf expression. Possible control mechanisms are discussed.
RESULTS
S2 negatively regulates its own gene expression in vivo
One of the approaches to examine autogenous regulation in vivo is monitoring expression of the corresponding translational fusion with the reporter (usually lacZ) gene under normal versus augmented or, on the contrary, reduced synthesis of a presumable regulator. This technique has been successfully used for dissecting the rpsA (Boni et al. 2000, 2001), rpsO (Mathy et al. 2004, and references therein), thrS (Sacerdot et al. 1998), rnc (Matsunaga et al. 1996), and L20 (Guillier et al. 2002, 2005) autoregulatory circuits. To maintain the dose of the reporter gene unaltered under different growth conditions, single-copy reporter constructs are preferable. Guided by this strategy, we assessed the capability of S2 to regulate its own gene expression.
We first constructed single-copy (chromosomal) rpsB′-′lacZ translational fusions and a plasmid expressing the rpsB gene as a source of S2 in trans (Fig. 1A). To create the plasmid, the E. coli chromosomal region encompassing the rpsB structural gene and its 5′- and 3′-flanks was amplified by PCR and then cloned into pACYC184. Taking into account that in E. coli translational starts of rpsB and of the preceding map gene (transcribed in opposite direction) are separated by 367 base pairs (bp), and corresponding promoters remained undetected, we arbitrarily chose the 208-bp region in front of the rpsB start codon for this construct. The rpsB and tsf genes in the operon are separated by a long (258 bp) intergenic region comprising a perfect inverted repeat followed by a run of T residues, a so-called attenuator/terminator (Merino and Yanofsky 2005) where about two-thirds of transcripts terminate, generating a monocistronic rpsB mRNA, while the remainder read through the attenuator until the major terminator downstream of tsf (An et al. 1981). This intergenic attenuator was included in our plasmid construct to terminate the rpsB transcription (Fig. 1A). The resulting plasmid, here referred to as pS2, complemented the temperature-sensitive rpsB1 allele (Bollen et al. 1979), indicating that it produces biologically active S2 (data not shown), and therefore the 208-bp region in front of the rpsB coding frame comprises the promoter. In a wild-type strain, pS2 slowed down the growth rate more than twofold. The harmful effect of S2 expressing plasmids was noted earlier (Bendiak and Friesen 1981) but still remains to be explained.
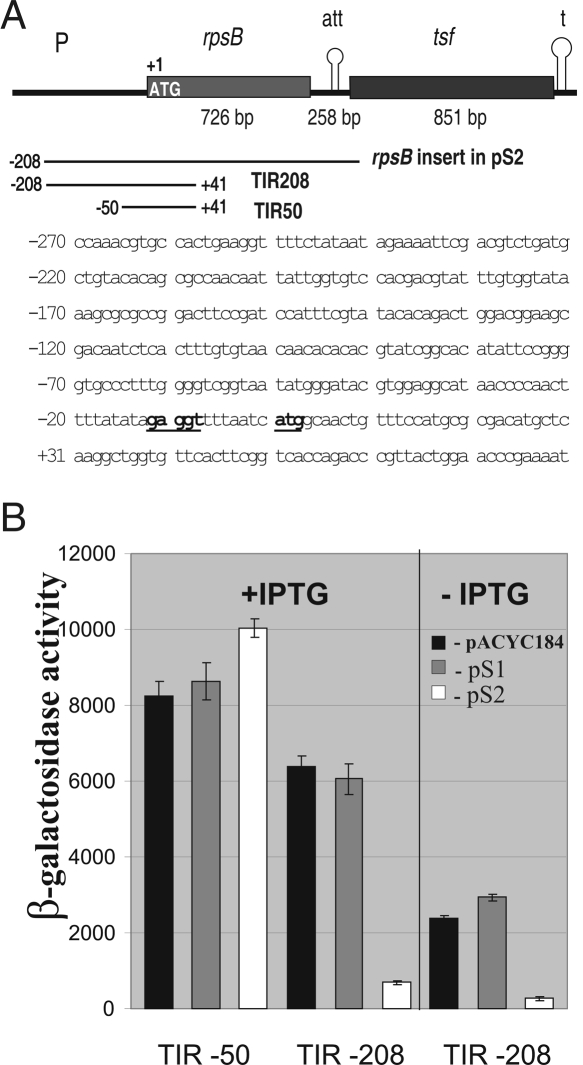
FIGURE 1.
S2 serves as a negative regulator of the rpsB expression. (A) Structure of the E. coli rpsB-tsf operon. (P) Promoter; (att) intergenic attenuator; (t) terminator. Regions amplified by PCR for constructing the plasmid pS2 and rpsB′-′lacZ fusions are shown by lines. (Bottom) The sequence of the 5′-UTR and beginning of the rpsB coding part; the Shine-Dalgarno element and the start codon are in bold underlined. (B) Effects of S2 in trans on rpsB TIR50 and TIR208 activities in the β-galactosidase assay. Average of three independent assays and standard deviations are shown. An empty vector pACYC184 and its derivative pS1 were used as specificity controls.
Next, we created the rpsB′-′lacZ chromosomal translational fusions, one of which comprised the same 208-nucleotide (nt) 5′-extension as in pS2 and the beginning of the rpsB coding part, while another had a much shorter 5′-UTR of 50 nt in length (Fig. 1A). The corresponding rpsB regions were amplified by PCR and inserted behind the lac-promoter/operator region of pEMBLΔ46 in-frame with the lacZ coding sequence (Dreyfus 1988), generating plasmids pES2TIR208 and pES2TIR50. The rpsB′-′lacZ fusions were next transferred onto the lac locus of the E. coli chromosome by homologous recombination as described earlier (Dreyfus 1988; Boni et al. 2000), generating strains LABrpsBTIR208∷lacZ and LABrpsBTIR50∷lacZ. In the first one, the β-galactosidase synthesis is under transcriptional control of both the lac promoter/operator and the rpsB promoter and governed by the full-length rpsB translation initiation region (TIR) at the translation level. The truncated TIR50 in the second construct comprises all conventional elements necessary for ribosome binding, but presumably not the rpsB promoter (Fig. 1A).
We then tested the rpsB′-′lacZ expression for the S2-mediated regulation by measuring the effect of pS2 on the β-galactosidase steady-state synthesis. An empty vector, pACYC184, its derivative pS1 expressing the rpsA gene coding for S1 (Skouv et al. 1990; Boni et al. 2000), and the isogenic strain bearing the rpsA′-′lacZ fusion autoregulated by S1 (Boni et al. 2000, 2001) were used to provide specificity controls. The drop in the β-galactosidase activity of the rpsBTIR208-lacZ construct in the presence of pS2 was about one order of magnitude compared with pACYC184 and pS1 controls, demonstrating the capability of S2 to function as a negative regulator of its own gene expression (Fig. 1B). The shorter TIR50 governed efficient expression unaffected by the presence of pS2. The latter fact, as well as the absence of the pS2-mediated inhibition of the noncognate rpsA′-′lacZ fusion (data not shown), provide evidence that the observed S2-mediated repression is specific and requires the region upstream of the position –50 relative to the start codon. Noteworthy, the TIR208 construct, but not TIR50, governed considerable and regulated β-galactosidase synthesis in the absence of lac-promoter induction (Fig. 1B), indicating the presence of the rpsB promoter(s) in the region between positions –50 and –208.
The single E. coli rpsB promoter belongs to the “extended −10” class
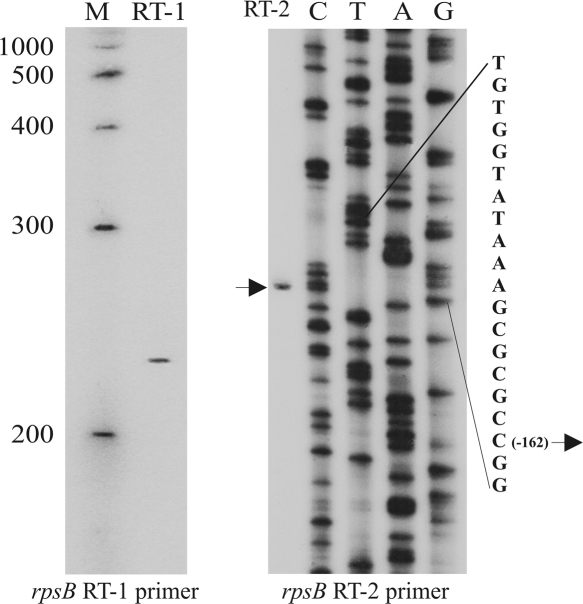
Most of the known E. coli promoters have been located by identifying transcriptional start sites (Hershberg et al. 2001). To locate the rpsB promoter(s), 5′-ends of the rpsB in vivo transcripts were mapped by primer extension, using total RNA isolated from exponential E. coli cultures and 5′-32P-labeled primers complementary to the beginning of the rpsB coding region. Only one extension product with the 5′-terminus corresponding to position –162 (relative to the A+1 in the initiator codon) was observed in sequencing gels, indicating the presence of a single promoter responsible for the rpsB expression (Fig. 2).
FIGURE 2.
Mapping the rpsB promoter by primer extension. The products of reverse transcription on total RNA with rpsB_RT1 primer were run along with 5′-32P-labeled ssDNA markers (M). To map the 5′-end of the single rpsB transcript, both primer extension on total RNA and pES2TIR208 sequencing were done with the rpsB_RT2 primer.
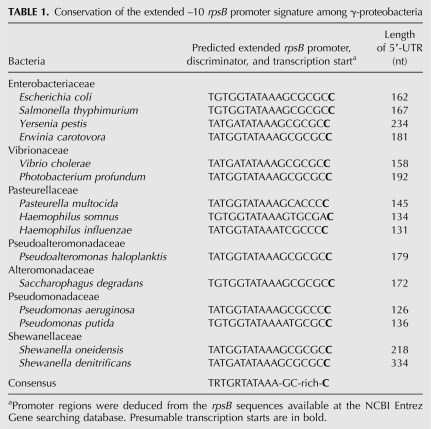
The identified 5′-position of the rpsB in vivo transcript represents most likely a real transcriptional start site (TSS). The TATAAA sequence situated at a proper distance upstream of this point almost perfectly matches the consensus –10 promoter element recognized by σ70 subunit of RNA polymerase, except for A at the position –7 relative the TSS in place of a highly conserved T. The 5′-TGTG(G)-extension of this –10 hexamer characterizes the E. coli rpsB promoter as a member of the minor class of extended –10 promoters (TRTGn-promoters, where R=purine), which exhibit decreased dependence on the –35 promoter region (Kumar et al. 1993; Barne et al. 1997; Burr et al. 2000). The TRTG extension has been shown to ensure a substantial promoter strength due to additional contacts with the region 2.5/3.0 of σ70 (Barne et al. 1997; Burr et al. 2000; Mitchell et al. 2003; Young et al. 2004), stabilizing the transcription initiation open complex (Voskuil and Chambliss 2002). The sequence GCGCGC between the extended rpsB promoter and the TSS identified is reminiscent of a GC-rich “discriminator” typical for promoters under growth-rate and stringent control (Pemberton et al. 2000; Haugen et al. 2006, and references therein). Remarkably, analysis of nucleotide sequences upstream of the rpsB start codon in fully sequenced bacterial genomes revealed that a combination of the extended −10 promoter with the GC-rich discriminator in front of rpsB is highly conserved in γ-proteobacteria, even in distant families (Table 1).
TABLE 1.
Conservation of the extended –10 rpsB promoter signature among γ-proteobacteria
Phylogenetic conservation of the rpsB regulatory elements
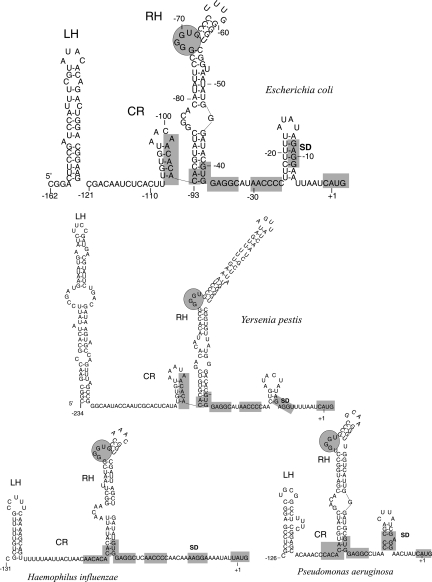
Conservation of the extended –10 promoter TRTGRTATAAA specific for the γ-proteobacterial rpsB-tsf operons (Table 1) enables comparing rpsB 5′-UTRs from diverse species. Although both the primary structure and the length of rpsB mRNA leaders appeared to vary significantly, computer modeling (Zuker 2003) revealed striking conservation of their fold (Fig. 3). The rpsB leader structure predicted by Mfold consists of two irregular double-stranded regions, left hand (LH) and right hand (RH), separated by a loosely structured (or unstructured, as in Haemophilus) central part (central region [CR]). As a rule, the helical regions LH and RH are interrupted by small bulges or internal loops, some of which are highly conserved even in distant species (Fig. 3, encircled). Several specific sequence elements are also remarkably conserved, indicating their potential importance for rpsB expression and/or regulation (Table 2). Thus, in most γ-proteobacterial families, the rpsB start codon (always AUG) is preceded by a 4–5-nt SD sequence embedded in a AU-rich context, and further upstream there resides a stretch of C residues and an SD-like GUGGAGG sequence, both universally conserved (Table 2). Due to these specific conserved features of the rpsB 5′-UTRs we noticed errors in annotation of the rpsB start codon in a number of sequenced genomes, e.g., in E. coli CFT073 and UTI89, Haemophilus influenzae 86-028NP and Rd KW20 (GenBank accession nos. AE014075, CP000243, CP000057, and L42023, respectively).
FIGURE 3.
Phylogenetic conservation of the rpsB 5′-UTR fold in γ-proteobacteria revealed by Mfold program. (LH, RH) Conserved stem–loop structures designated “left hand” and “right hand,” (CR) central loosely structured region. In E. coli, CR conventionally covers −121 to −93 positions relative to the AUG codon. Conserved elements within 5′-UTRs are shadowed, conserved GGGU-bulges in the top part of RH are encircled.
TABLE 2.
Conservation of primary structures of the γ-bacterial rpsB mRNAs immediately upstream of the start codon
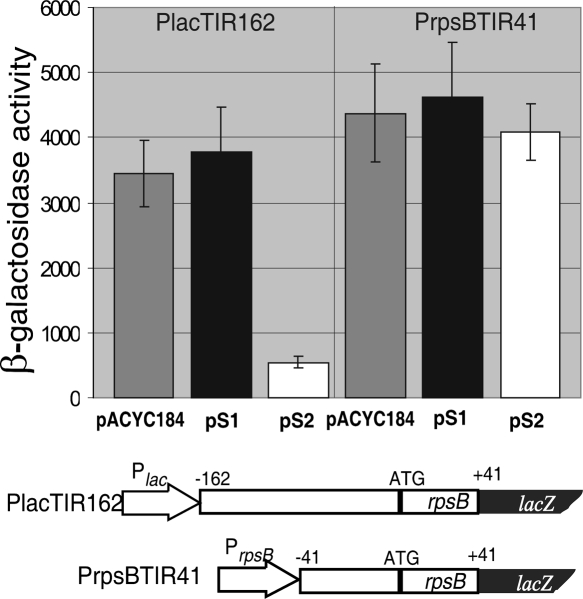
Thus, both the promoter signature and the 5′-UTR structure of rpsB appeared to be conserved in γ-proteobacteria. To find out which is essential for the S2-mediated control, we created two additional rpsB′-′lacZ constructs, PrpsBTIR41 and PlacTIR162 (Fig. 4). In the first one, the full-length rpsB 5′-UTR was truncated from –162 to –41 positions relative to the initiation codon, so that transcription from the rpsB extended promoter started with C–41. In the second one, transcription from a lac-promoter started with the authentic A (to keep the specificity of transcription starts for each promoter), immediately followed by the complete rpsB TIR (–162 to +41). Because the major lac-operator was deleted, the latter construct displayed constitutive expression without induction. The β-galactosidase assay unambiguously demonstrated that the mRNA region upstream of the conventional ribosome binding site, but not the extended promoter, was essential for the rpsB autoregulation (Fig. 4).
FIGURE 4.
The leader part of the rpsB mRNA, not the promoter design, is essential for the S2 autoregulation. The β-galactosidase activities in the presence of pACYC184, pS1, and pS2 are shown for the PrpsBTIR41 and PlacTIR162 constructs. PrpsBTIR41 is a derivative of the TIR208 with a deleted promoter-proximal region –162 to –41. In PlacTIR162, transcription from the lac-promoter starts from A, which is immediately followed by a complete rpsB TIR.
Deletion analysis of the rpsB-lacZ translational fusions
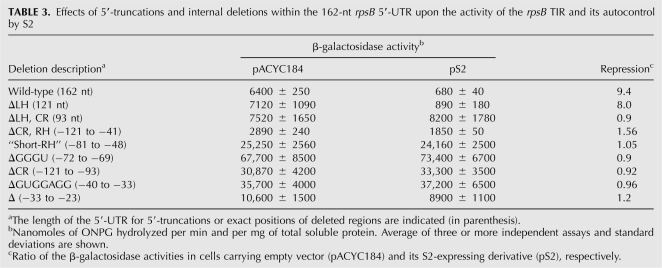
To identify sequence/structure cis-acting elements essential for the S2-mediated control, we introduced a series of 5′-truncations and internal deletions into the full-length rpsB 5′-UTR within pES2TIR208 (see above) and then created corresponding rpsB′-′lacZ chromosomal constructs to measure effects of the mutations on the expression level and inhibition in the presence of pS2. The results are summarized in Table 3. Analysis of β-galactosidase activities in 5′-truncated constructs revealed that the hairpin LH is dispensable, whereas both CR and RH regions are essential for the autocontrol. For internal deletions we chose the conserved features of the rpsB TIR and obtained prominent effects on both the expression level and the S2-mediated regulation (Table 3). In particular, deletions of a small G-rich bulge in the upper part of RH, an SD-like sequence upstream of the C-stretch, and the region including the C-stretch all augmented the translation activity of the rpsB TIR, simultaneously abrogating autogenous control (Table 3). This implies that within the rpsB TIR these universally conserved elements act as negative regulators implicated in autogenous control. A significant increase in the TIR activity and the loss of inhibition by pS2 was also obtained when the conserved G-rich bulge in RH was deleted from the rpsB leader of Pseudomonas aeruginosa, a γ-proteobacterium very distant from E. coli (data not shown). This indicates that the rpsB regulation displays higher extent of conservation within γ-proteobacteria than regulation of rpsA or S10 operons (Allen et al. 1999; Tchufistova et al. 2003), where the leader structures involved in autocontrol in E. coli and P. aeruginosa differ entirely.
TABLE 3.
Effects of 5′-truncations and internal deletions within the 162-nt rpsB 5′-UTR upon the activity of the rpsB TIR and its autocontrol by S2
S2 down-regulates the EF-Ts synthesis by acting at a single target within the rpsB 5′-UTR
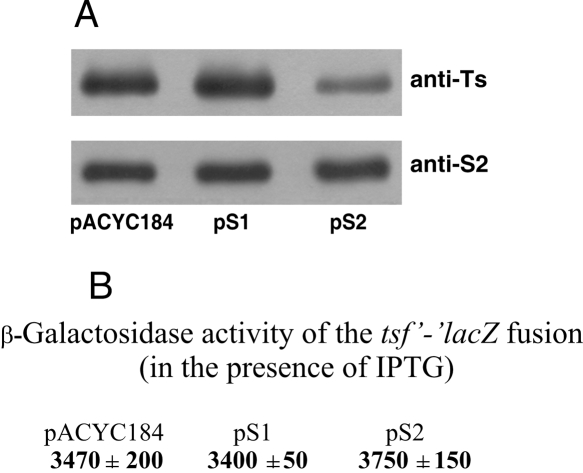
Earlier works (An et al. 1981; Bendiak and Friesen 1981) failed to identify an additional promoter in front of the E. coli tsf gene. Thus, EF-Ts is most likely synthesized from a bicistronic rpsB-tsf mRNA, and thereby its synthesis may be controlled by S2. The effect of S2 in trans on the EF-Ts steady-state synthesis was studied by Western blotting using polyclonal rabbit antibodies raised against purified Ts (anti-Ts). Significant reduction of the intracellular Ts amount in the presence of pS2, but not pS1, indicated that S2 had a specific negative impact on the Ts synthesis (Fig. 5A). In addition, consistent with the autoregulation circuitry, increase in the rpsB gene copies in the presence of pS2 did not proportionally augment the S2 level in a cell (Fig. 5A). Together with the above results obtained with the rpsB′-′lacZ fusions, this suggests that both rpsB and tsf expression are under S2-mediated negative control. Since Ts is a GDP/GTP exchange factor for EF-Tu, and hence an essential player in translation, inhibition of its synthesis is likely one of the major factors causing the growth defect in the presence of pS2.
FIGURE 5.
Effects of S2 in trans on the tsf expression as revealed by Western blotting (A) and the β-galactosidase assay of the tsf-lacZ translational fusion (B).
In many r-protein polycistronic mRNAs, the repressor binding inhibits translation of the downstream cistrons due to translational coupling (Zengel and Lindahl 1994). Since the rpsB and tsf cistrons are separated by a long 258-nt untranslated region comprising a number of strong stem–loop structures, the translation coupling between them seems improbable. Downstream from the tandem rpsB stop codons there resides a 127-nt enterobacterial repetitive intergenic consensus (ERIC) element of unknown function, which forms a long irregular hairpin due to the presence of inverted repeats (Hulton et al. 1991); further downstream, a set of stem–loops forms the terminator/antiterminator region (Merino and Yanofsky 2005). One cannot completely rule out that S2 might independently down-regulate the tsf expression by acting at some of these structural elements. To test this possibility, we constructed the tsf′-′lacZ translational fusion comprising the end of the rpsB cistron, the full-length intercistronic region, and the beginning of the tsf coding sequence. The β-galactosidase assay clearly showed that this construct is not regulated by S2 in trans (Fig. 5B). Furthermore, no detectable expression of tsf-lacZ was observed without lac-promoter induction, thus confirming the absence of a tsf-specific promoter within the rpsB-tsf intercistronic region. These results imply that regulation of both rpsB and tsf cistrons occurs from a single target within the rpsB 5′-UTR, and the S2-mediated repression of the rpsB translation most probably exerts a polar effect on expression of a downstream tsf via impairing transcription–translation coupling within rpsB.
Synthesis of S2 and Ts in rpsB mutants
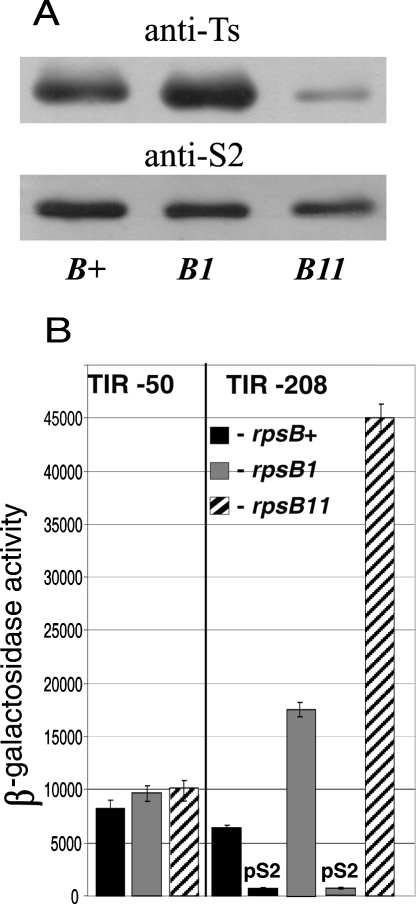
The negative control of the rpsB-tsf operon by S2 was then confirmed by evaluating effects of rpsB mutations on rpsB and tsf expression. Two previously characterized mutants, rpsB1(Ts) and rpsB11 (rpsB∷IS1), were used in these experiments. The rpsB1 mutation confers the temperature-sensitive phenotype, i.e., the mutant cannot grow above 40°C and grows much slower than wild-type cells at permissive 37°C (at which the mutant S2 is partially inactivated) because the denaturated S2 is unable to assist in 30S assembly (Bollen et al. 1979). The rpsB11 mutation is an insertion of a transposable element IS1 between the rpsB stop codons and ERIC, which destabilizes the rpsB mRNA, thereby decreasing the intracellular S2 level (Shean and Gottesman 1992). The rpsB11 cells form tiny colonies on agar plates and grow more than two times slower than rpsB+ in liquid medium (data not shown).
The two mutations were P1 transduced in strains bearing rpsB′-′lacZ fusions (see Materials and Methods). Western blot analysis revealed that in rpsB1 the level of EF-Ts is visibly higher than in wild-type (wt) cells, whereas in rpsB11 it is dramatically reduced (Fig. 6A). The increased Ts level in rpsB1 indicates the weakening of the S2-mediated repression, which was further confirmed by the β-galactosidase assay. The activity of the unregulated TIR50 construct did not alter in the presence of the mutations, whereas the activity of TIR208 increased about threefold in rpsB1 and more than sixfold in rpsB11 (Fig. 6B). These data indicate that normally (in rpsB+ background) the rpsB-tsf expression is partially repressed by S2 synthesized in excess over ribosomes, and in both mutants this intrinsic autogenous control is impaired. Analogous partial repression in wt cells and derepression caused by mutations in a repressor-encoding gene were previously shown for the rpsA (Boni et al. 2000), rpsO (Mathy et al. 2004), and rnc (Matsunaga et al. 1996) autoregulatory circuits. Most likely, in rpsB1 the mutant S2 lost, in part, its repressor activity at 37°C because of partial denaturation. Consistently, the presence of pS2 makes the TIR208 activity in rpsB1 indistinguishable from that in rpsB+/pS2 cells (Fig. 6B). The absence of a visible increase in the intracellular S2 level in rpsB1 (Fig. 6A) might be accounted for by proteolytic instability of the denatured S2.
FIGURE 6.
Effects of the mutations rpsB1(ts) and rpsB11 (rpsB∷IS1) on the rpsB-tsf expression. (A) Western blotting to evaluate the S2 and Ts level in the rpsB mutants. (B) β-Galactosidase activities of the rpsB′-′lacZ constructs were measured for cells exponentially grown in the presence of pACYC184 for TIR50, or pACYC184 and pS2 for TIR208. The rpsB11 cells cannot be transformed with pS2.
Destabilization of the rpsB mRNA in rpsB11 (Shean and Gottesman 1992), confirmed for our strains (data not shown), causes only a modest decrease in the S2 level (Fig. 6A). By destabilizing the rpsB mRNA, the rpsB11 mutation slows down accumulation of S2 in a cell, so that all synthesized S2 associates with 30S and no superfluous S2 to repress its own mRNA is generated, hence, an apparent increase in the rpsB TIR activity (Fig. 6B). The fact that a small amount of mRNA can supply a sufficient amount of the essential r-protein due to abolishing intrinsic autogenous repression emphasizes a vital importance of this control mechanism.
The dramatic reduction of the Ts level in rpsB11 (Fig. 6A) results from a polar effect of IS1 on the tsf expression. Transcriptional terminator within IS1 (Hubner et al. 1987) is supposed to impede synthesis of the bicistronic rpsB-tsf mRNA as a source of Ts, because unlike IS10 (Ciampi et al. 1982) IS1 does not possess its own outward promoter for transcription of downstream genes, it may only provide the −35 region for endogenous “−10”-like sequences (Prentki et al. 1986). Nevertheless, the rpsB11 cells are still able to synthesize Ts, though at a reduced level. Two possibilities can be considered: either polarity is not complete or IS1 is capable of activating a cryptic promoter in the intergenic sequence. In this regard, the fact that the rpsB11 cells cannot be transformed by pS2 gives a strong argument in favor of the first variant. Indeed, if a small amount of Ts in rpsB11 is produced from a bicistronic mRNA synthesized due to incompleteness of polarity, it should be further reduced in the presence of pS2 because of resuming S2-mediated repression (see Fig. 5A), making the cell nonviable. To prove this directly, we transformed the rpsB11 cells with two compatible plasmids, pS2 and pTrc99A-tsf (Karring et al. 2004), expressing the tsf gene under the control of the trc-promoter. The lethal effect of pS2 was suppressed by the second plasmid that supplied Ts necessary for survival, thus providing strong evidence for this interpretation.
Protein S1 may assist S2 in rpsB autoregulation
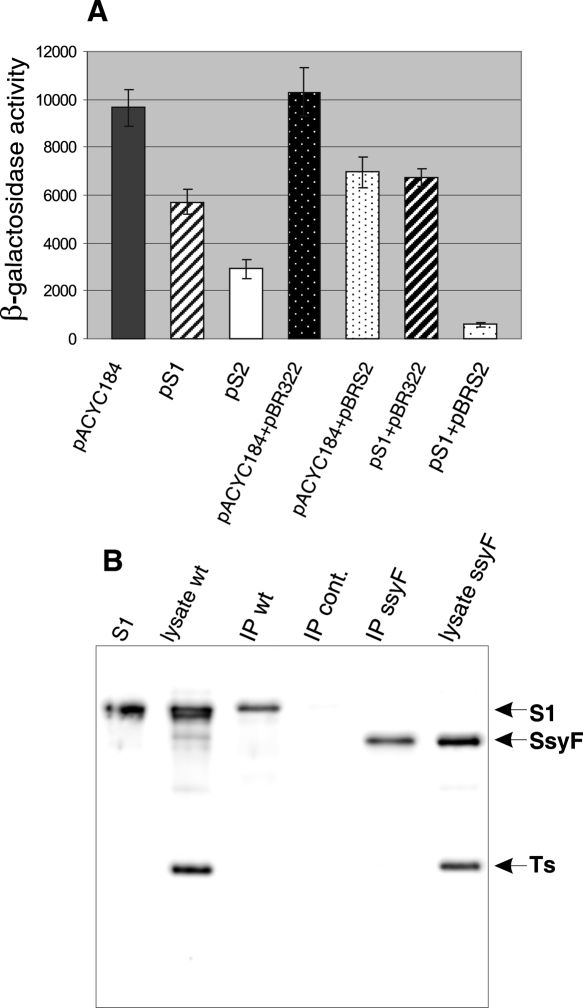
Surprisingly, the activity of the rpsB TIR in the TIR-208 construct appeared to be also augmented in the presence of the rpsA∷IS10 (ssyF29) mutation (Fig. 7A). This mutation causes production of the reduced amount of truncated protein S1 and confers slow-growth phenotype, because insertion of the IS10 mobile element within the 3′-terminal part of the rpsA gene destabilizes the rpsA mRNA (Boni et al. 2000). The observed increase in the TIR activity (by about 50%, compared with the TIR208 activity in wild-type cells) disappeared in the presence of pS1 (Fig. 7A), indicating that the enhancing effect is relevant to the S1 mutation. Moreover, S2 in trans did not repress the rpsB TIR208 activity in rpsA∷IS10 cells to the same extent as in rpsA + background, implying that the S2 autoregulation requires a normal supply of wild-type S1. To verify if this is the case, we cloned the rpsB gene into pBR322 (Ampr) compatible with pS1 (Cmr), generating a new construct expressing S2 and designated pBRS2. The β-galactosidase assay confirmed the decreased S2-mediated inhibition of the rpsBTIR208-lacZ expression in the presence of rpsA∷IS10 mutation and resuming autogenous regulation in the presence of both pBRS2 and pS1 in a cell (Fig. 7A). These results imply that S1 may assist S2 in regulating the rpsB expression, a so-far unknown activity for this multifunctional ribosomal protein. In this regard, several possibilities can be considered. Primary binding of S1 to the rpsB 5′-UTR due to its pronounced RNA-binding capacity (Subramanian 1983) could modify the RNA structure and facilitate further binding of S2 to its operator site to inhibit the rpsB expression. Alternatively, the preformed complex S1–S2 (both synthesized in excess over ribosomes) may act as a repressor. Interaction of S1 with S2 in the 30S subunit (Moll et al. 2002) and the described purification of a stoichiometric S1–S2 complex (Sukhodolets and Garges 2003) argue in support of the latter variant. Additional arguments were obtained using immunoprecipitation. A substantial amount of S1 was found in a protein fraction coprecipitated with S2 on the Protein A/G PLUS agarose in the presence of highly specific goat antibodies against S2 (Fig. 7B). Further studies (mainly in vitro experiments) are necessary to define the exact mode of cooperation of S2 and S1 in the rpsB autoregulation found here.
FIGURE 7.
The repressor activity of S2 depends on S1 concentration in a cell. (A) The reduced repressor activity of S2 in the ssyF29 (rpsA∷IS10) mutant producing a subnormal amount of truncated S1. The β-galactosidase activities were measured for TIR208 in the presence of pACYC184, pS1, and pS2, and in a biplasmid system where the high repression level can be achieved only when pS1 and pBRS2 (expressing wt S1 and S2) are simultaneously present. (B) Immunoprecipitation of S2–S1 complex from wild-type (IP wt) and ssyF29 cell lysates (IP ssyF) in the presence of polyclonal goat antibodies against S2. Western blotting of 12.5% PAAG revealed with anti-S1 and anti-Ts rabbit polyclonal antibodies (S1) −50 ng of purified S1; (lysate wt and lysate ssyF) 2 μg of total soluble cell proteins from rpsA+ and ssyF29 exponentially grown cells; (IP cont) a specificity control showing that no S1 is precipitated from wt cell lysate in the presence of goat polyclonal antibodies against ribosomal protein S15.
DISCUSSION
This work presents new data necessary for correct description of the essential E. coli rpsB-tsf operon, including its transcription organization and expression regulation. We report for the first time that transcription of the operon is governed by a single promoter TGTGGTATAAA belonging to the extended –10 promoter class, a rare case among ribosomal protein operons. A GC-rich spacer (“discriminator”) separating the rpsB promoter from the transcription start site suggests that the rpsB-tsf expression may be subjected to the growth rate control and stringent response (Pemberton et al. 2000; Haugen et al. 2006). The design of the rpsB promoter region appeared to be highly conserved in γ-proteobacteria. This conservation was supported experimentally. Corresponding DNA regions from Yersinia pestis, H. influenzae, and P. aeruginosa inserted in front of the E. coli chromosomal lacZ gene indeed possessed promoter activity (will be published elsewhere). It is currently difficult to appreciate the occurrence and conservation of this rare promoter type that combines the extended –10 motif TRTGRTATAAA, a GC-rich discriminator, and a rare C at the transcription start position (Table 1). Some advantages over predominant −35/−10 σ70 promoters may result from the increased tolerance to low temperatures (Phadtare and Severinov 2005, and references therein) and to anti-σ70 proteins similar to T4 AsiA that block interactions of σ70 with the –35 DNA region (Pineda et al. 2004). Further investigation is required to find the rationale for this peculiar design of the rpsB promoter and to characterize its regulatory features.
Not only the extended rpsB promoter (Table 1), but also a secondary structure of the rpsB 5′-UTR (Fig. 3) and several primary sequence elements upstream of the initiator AUG codon (Table 2) are highly conserved in γ subdivision of proteobacteria, thereby facilitating a correct annotation of rpsB genes in newly sequenced γ-bacterial genomes. It should be mentioned that a single rpsB transcription start experimentally determined here coincides with the annotated 5′-end of a so-called RNA T44, a small 136-nt-long RNA predicted on the basis of RNA structure conservation in the intergenic regions (Rivas et al. 2001; Hershberg et al. 2003). The corresponding gene t44 (tff) as residing in front of the E. coli rpsB gene was included in RegulonDB, the reference database of E. coli K-12 regulatory network and operon organization (http://regulondb.ccg.unam.mx/). Given that the hypothetical t44 gene appears to share the promoter with rpsB and, in addition, does not possess any reminiscent of transcription terminator, it seems very likely that the reported conservation actually reflects not the presence of a gene for sRNA with still-unknown functions, but the conservation of the rpsB 5′-UTR structure involved in the S2 autoregulation (the rpsB operator). Phylogenetic conservation among γ-proteobacteria of the hairpin structures implicated in autogenous control is typical for r-protein mRNAs (Allen et al. 1999, 2004; Boni et al. 2001; Tchufistova et al. 2003; Guillier et al. 2005), and the rpsB mRNA represents one more prominent example (Fig. 3).
Our findings demonstrate that r-protein S2, like some other r-proteins, has an extraribosomal function in E. coli and in related bacteria as a negative effector of expression of the essential rpsB-tsf operon. In all regulated r-protein operons, a repressor protein binds to the mRNA, preventing its translation (Zengel and Lindahl 1994). As we show here, the S2-mediated repression similarly operates at the RNA and not at the DNA level (Fig. 4). Both rpsB and tsf are negatively regulated by S2 from a single operator located within the rpsB 5′-UTR. Although the results obtained in vivo do not yet enable a detailed description of the operator site, deletion analysis shows that a large portion of the 5′-UTR is implicated in S2-mediated control, including several highly conserved cis-regulatory elements situated upstream of the conventional ribosome binding site (RBS). The fact that autogenous regulation requires a long structured region upstream of the start codon (Table 3) suggests that a simple direct competition between S2 and 30S for RBS binding is unlikely, rather more complex mechanisms (presumably S2-mediated structure rearrangements are inhibitory for 30S initiation complex formation) are involved.
S2 presents an unusual example of a regulatory r-protein, being a tertiary (one of the latest) and not a primary protein in ribosome assembly (Mizushima and Nomura 1970; Culver 2003), i.e., unlike most of the r-protein repressors, S2 is unable to recognize naked rRNA. Most of the primary r-proteins acting as autogenous regulators (e.g., S7, S8, S15, L1, L4, L20) recognize similar targets on free rRNA and on mRNA of its cognate operon (Zengel and Lindahl 1994; Guillier et al. 2002, 2005; Stelzl et al. 2003; Mathy et al. 2004), indicating that the autogenous control is based on molecular mimicry as was originally proposed by Nomura (Nomura et al. 1980). A detailed analysis of the refined crystal structure of the T. thermophilus 30S subunit at 3.05Å resolution revealed that S2 is located on the back of 30S and spans the head-body hinge region by forming contacts with 16S rRNA helices H26 in the body and H35–H37 in the head of the subunit (Brodersen et al. 2002). Direct contacts of S2 with a group of helices H35–H37 were also revealed by chemical probing (Powers et al. 1988). Although no visible resemblance exists between 16S RNA elements involved in S2 binding and conserved features within the rpsB 5′-UTR, it is too early to conclude that the S2-mediated inhibition is not based on molecular mimicry until the data on exact contacts between S2 as a repressor and its operator become available.
A noteworthy feature of the S2 autoregulation is that S2 alone seems to be unable to effectively repress the rpsB-lacZ expression. Indeed, while in wild-type cells the presence of the S2 expressing plasmid pS2 exerts a strong inhibitory effect (Fig. 1B), in the rpsA∷IS10 mutant (ssyF29, see Boni et al. 2000), which produces a subnormal amount of truncated S1, the repression is obviously reduced (Fig. 7A). Complete restoration of the repression can be obtained after simultaneous transformation of the ssyF29 cells with compatible plasmids pS1 and pBRS2 (Fig. 7A), suggesting that S2 probably cooperates with S1 in negative regulation of rpsB, a unique case for autogeneously controlled r-protein operons. The only case described so far when a complex of r-proteins serves as a translational repressor is the translational inhibition of the rplJL operon by a pentameric complex L10(L7/12)4 (Johnsen et al. 1982). However, in this case, both r-proteins are products of the same operon, their pentameric complex is very stable and forms an important functional domain of the 50S ribosomal subunit. As we revealed by immunoprecipitation, S1 and S2 are capable of forming the complex; moreover, S2 coprecipitates also with a truncated variant of S1 from the ssyF29 mutant (Fig. 7B). This suggests that the reason for the decreased S2 repressor capacity in ssyF29 is not the C-terminal S1 truncation, but most likely the scanty concentration of S1 in mutant cells. More details about cooperation of S1 and S2 to regulate the rpsB expression may be obtained in experiments in vitro that are currently in progress.
As we show here (Fig. 6A), autogenous inhibition of the rpsB expression exerts a polar effect on the downstream tsf gene. Two plausible reasons for this polarity may be considered. First, the S2 binding to the 5′-UTR promoted by S1 may provoke a rearrangement of the mRNA leader structure inhibitory for translation-initiation complex formation; in this case, translational repression of the rpsB mRNA may exert a polar effect on the tsf expression via breaking the transcription–translation coupling within rpsB. Alternatively, the S2–mRNA complex itself or in concert with other factors may directly stimulate preliminary transcription termination, as in the case of the L4-mediated repression of the S10 operon (Zengel and Lindahl 1994; Stelzl et al. 2003). Future experiments (mainly in vitro) will shed light upon the fine mechanism underlying the S2-mediated negative control of the rpsB-tsf expression.
MATERIALS AND METHODS
Bacterial strains and plasmids
Genetic constructs harboring single-copy rpsB′-′lacZ fusions were derivatives of a Lac−ENSO strain bearing a short chromosomal deletion encompassing the lac-promoter/operator region and the lacZ RBS (formerly HfrG6Δ12) (Dreyfus 1988). The rpsB fragments comprising 5′-UTR of variable length and the beginning of the coding sequence (Fig. 1) were amplified by PCR on the E. coli genomic DNA, using forward primers bearing BamHI site (italicized): −208for (5′-GTGGGATCCAACAATTATTGGTGTC) and −50for (5′-TATGGGATCCGTGGAGGCATAACC), and a common reverse primer S2TIRrev (5′-CCGAAGCTTACACCAGCCTTGAG) complementary to the rpsB coding region +28 to +41 and bearing HindIII site (italicized). The resulting PCR products were treated with BamHI and HindIII and then cloned in-frame with the lacZ sequence of pEMBLΔ46 (Dreyfus 1988), generating pES2TIRn plasmids, where n is the rpsB 5′-UTR length. Given the absence of the lacZ expression in pEMBLΔ46 because of the lacZ RBS deletion, proper in-frame clones can be readily identified by white-blue selection and confirmed by sequencing.
Deletion variants of pES2TIR208 were constructed by using PfuUltra II fusion HS DNA polymerase (Stratagene) in reverse PCR, with primers flanking the site to be deleted (to amplify the whole plasmid except for the region chosen for deleting). PCR products were purified on agarose gels, then ligated, treated with DpnI, and used to transform DH5α for selecting Lac+ clones.
All constructs were checked by sequencing and then used for transformation of ENSO. In transformed cells, the rpsB-lacZ fusions and the upstream lac-promoter/operator region were transferred onto the chromosome via homologous recombination (Dreyfus 1988; Boni et al. 2000), generating recombinant Lac+ clones designated LABrpsBTIRn∷lacZ. The rpsB1 (Bollen et al. 1979) and rpsB11 (Shean and Gottesman 1992) alleles were transferred into the strains by P1 transduction. The marker Tn10 from CAG18436 (Singer et al. 1989) was cotransduced with the rpsB mutations to provide selection of the tetracycline-resistant transductants. The rpsB1 allele in selected clones was further confirmed by its ts phenotype, and the rpsB11 clones differing from rpsB+ colonies by slow growth on agar plates were checked by PCR for the presence of IS1. The ssyF29 (rpsA∷IS10) allele was P1 transduced as described (Boni et al. 2000).
To create the tsf′-′lacZ fusion, a chromosomal region encompassing the end of rpsB, the intercistronic region, and the beginning of the tsf coding frame was amplified by PCR on the E. coli DNA with primers rpsBend-for 5′-CGCGGATCCGTAGAAGCTGAGTAATAAGGC (bearing BamHI site and overlapping the tandem of the rpsB stop codons) and tsfTIR-rev 5′-CATCAAGCTTGCGCCAGTACGCT (bearing the HindIII site and complementary to the tsf coding sequence +40 to +63 relative to the tsf start). The PCR fragment was cloned in pEMBLΔ46/BamHI, HindIII, with further manipulations to create the strain LABtsfTIR∷lacZ being exactly the same as described above for the rpsB-lacZ fusions.
To build plasmids for S2 expression, the rpsB structural gene flanked by the 208-nt 5′-UTR and the rpsB-tsf intergenic region including the attenuator was amplified from the E. coli genomic DNA using primers 208for (see above) and rpsBrev (5′-CAGGTCGACCTCGGAGATGTGATCTG) bearing SalI site (italicized). The PCR product treated with BamHI and SalI was ligated into respective sites within the tet-regions of pACYC184 or pBR322. The resulting plasmids able to complement the temperature-sensitive phenotypes of the rpsB1 mutant (Bollen et al. 1979) were designated pS2 (pACYC184 derivative) and pBRS2 (pBR322 derivative).
Cell growth and β-galactosidase assay
Cell cultures were grown at 37°C in LB medium supplemented, if necessary, with IPTG (0.2 mM) and appropriate antibiotics and harvested in exponential phase (OD600 ≅ 0.4–0.5). The β-galactosidase-specific activity was measured in clarified cell lysates essentially as described (Tchufistova et al. 2003) and expressed in nmol ONPG (o-nitrophenyl-β-D-galactopiranoside) hydrolyzed per minute per milligram of total soluble cell proteins. Protein concentrations in lysates were determined by Bradford assay (Bio-Rad).
Localization of the rpsB promoter by primer extension
E. coli cells were grown at 37°C in LB medium to OD600 = 0.5. Total RNA was isolated from 6 mL culture by using Aurum Total RNA Mini Kit (Bio-Rad) according to the manufacturer's protocol. Two 5′-32P-labeled primers complementary to the beginning of the rpsB coding part were used for primer extension by AMV reverse transcriptase (Promega) on 5−6 mkg of isolated total RNA: rpsB_RT1 5′-GTTCCAGTAACGGGTCTGGTGACC (+49 to +72) and rpsB_RT2 5′-GTCGCGCATGGAAACAGTTGCCATG (–1 to +24). RT products were run on 6% sequencing PAAG along with 5′-32P-labeled ssDNA markers (rpsB_RT1 extension on Fig. 2) or sequencing reactions for pES2TIR208 (Fig. 2, rpsB_RT2 extension).
Western blotting
Total soluble proteins prepared as for the β-galactosidase assay were analyzed in 10% Laemmli gel (5 μg per lane) and transferred onto nitrocellulose membrane (Bio-Rad). The blots were successively revealed with polyclonal rabbit antibodies raised against purified EF-Ts (prepared especially for this work) and with polyclonal goat anti-S2 antibodies (a gift of R. Brimacombe, Max-Planck-Institut, and O. Dontsova, Moscow State University). Secondary anti-rabbit (Bio-Rad) and anti-sheep/goat (Promega) horseradish peroxidase-labeled antibodies were used for visualization with the Immun-Star HRP Chemiluminiscent reagent (Bio-Rad) and Bio-Rad VersaDoc MP4000 image station.
Immunoprecipitation
Immunoprecipitation was carried out according to the protocol of Santa Cruz Biotechnology with minor modifications. Freshly prepared lysates of the exponentially grown E. coli cells (100 μg of total soluble proteins), were incubated with 3 μL (about 1 μg) of polyclonal goat anti-S2 antibodies and 10 μL 100 mM PMSF (phenylmethylsulfonyl fluoride) in 1 mL of PBS for 2 h at 4°C using a rotating device. In a control sample, the same amount of polyclonal goat anti-S15 antibodies was used. Then, 20 μL of Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) was added to each tube, and the resulting mix was further incubated under rotation at 4°C overnight. Agarose pellets were collected by centrifugation at 5000 rpm for 30 sec and supernatants were carefully discarded. The pellets were washed three to four times with PBS, each time repeating the centrifugation step above. After final supernatant discarding, pellets were resuspended in 40 μL of 2x Laemmli sample buffer, boiled for 3 min, and 10 μL of each probe was loaded on 12.5% Laemmli gel and run along with protein markers and 1−2 μg of the initial cell lysate. Immunoblotting was then performed using polyclonal rabbit antibodies against S1 and Ts (the latter to provide specificity control) as primary antibodies and goat-anti-rabbit horseradish peroxidase-labeled secondary antibodies. The Immun-Star HRP Chemiluminiscent reagent (Bio-Rad) and Bio-Rad VersaDoc MP4000 image station were used for visualization.
ACKNOWLEDGMENTS
We thank Max Gottesman for CS509 (rpsB11), Isabelle Iost for the rpsB1 strain, Vyacheslav Kolb for EF-Ts, Charlotte Knudsen for plasmids pTrc99A and pTrc99A-tsf, Elena Svirshevskaya for preparing rabbit polyclonal anti-Ts antibodies, Richard Brimacombe and Olga Dontsova for goat anti-S2 antibodies. We are indebted to Mathias Springer for helpful discussion. This work was supported by RFBR grant 06-04-48353 and partly by the Program of the Presidium of RAS for Basic Research in Molecular and Cellular Biology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1099108.
REFERENCES
- Allen T., Shen P., Samsel L., Liu R., Lindahl L., Zengel J.M. Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon. J. Bacteriol. 1999;181:6124–6132. doi: 10.1128/jb.181.19.6124-6132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.D., Watkins T., Lindahl L., Zengel J.M. Regulation of ribosomal protein synthesis in Vibrio cholerae . J. Bacteriol. 2004;186:5933–5937. doi: 10.1128/JB.186.17.5933-5937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Bendiak D.S., Mamelak L.A., Friesen J.D. Organization and nucleotide sequence of a new ribosomal operon in Escherichia coli containing the genes for ribosomal protein S2 and elongation factor Ts. Nucleic Acids Res. 1981;9:4163–4172. doi: 10.1093/nar/9.16.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E., Pesole G., Tagliabue E., Magnifico A., Castronovo V., Sobel M.E., Colnaghi M.I., Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol. Biol. Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- Barne K.A., Bown J.A., Busby S.J., Minchin S.D. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended –10” motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendiak D.S., Friesen J.D. Organization of genes in the four minute region of the Escherichia coli chromosome: Evidence that rpsB and tsf are co-transcribed. Mol. Gen. Genet. 1981;181:356–362. doi: 10.1007/BF00425611. [DOI] [PubMed] [Google Scholar]
- Bollen A., Lathe R., Herzog A., Denicourt D., Lecocq J.P., Desmarez L., Lavallé R. A conditionally lethal mutation of Escherichia coli affecting the gene coding for ribosomal protein S2 (rpsB) J. Mol. Biol. 1979;132:219–233. doi: 10.1016/0022-2836(79)90392-9. [DOI] [PubMed] [Google Scholar]
- Boni I.V., Artamonova V.S., Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in the autogenous control. J. Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni I.V., Artamonova V.S., Tzareva N.V., Dreyfus M. Non-canonical mechanism for translational control in bacteria: Synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. doi: 10.1093/emboj/20.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D.E., Clemons W.M., Jr, Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interaction with 16S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Burr T., Mitchell J., Kolb A., Minchin S., Busby S. DNA sequence elements located immediately upstream of the –10 hexamer in Escherichia coli promoters: A systematic study. Nucleic Acids Res. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M.S., Schmid M.B., Roth J.R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. 1982;79:5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M. Assembly of the 30S ribosomal subunit. Biopolymers. 2003;68:234–249. doi: 10.1002/bip.10221. [DOI] [PubMed] [Google Scholar]
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- Guillier M., Allemand F., Raibaud S., Dardel F., Springer M., Chiaruttini C. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: A possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA. 2002;8:878–889. doi: 10.1017/s1355838202029084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M., Allemand F., Dardel F., Royer C.A., Springer M., Chiaruttini C. Double molecular mimicry in Escherichia coli: Binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S RNA. Mol. Microbiol. 2005;56:1441–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- Haugen S.P., Berkmen M.B., Ross W., Gaal T., Ward C., Gourse R.L. rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Bejerano G., Santos-Zavaleta A., Margalit H. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001;29:277. doi: 10.1093/nar/29.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli . Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry W.A., Frishman D., Eckerskorn C., Lottspeich F., Harti F.U. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- Hubner P., Iida S., Arber W. A transcriptional terminator sequence in the prokaryotic transposable element IS1 . Mol. Gen. Genet. 1987;206:485–490. doi: 10.1007/BF00428889. [DOI] [PubMed] [Google Scholar]
- Hulton C.S., Higgins C.F., Sharp P.M. ERIC sequences: A novel family of repetitive elements in the genome of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Johnsen M., Christensen T., Dennis P.P., Fiil N.P. Autogenous control: Ribosomal protein L10-L12 complex binds to the leader sequence of its mRNA. EMBO J. 1982;1:999–1004. doi: 10.1002/j.1460-2075.1982.tb01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminishi T., Wilson D.N., Takemoto C., Harms J.M., Kawazoe M., Schluenzen F., Hanawa-Suetsugu K., Shirouzu M., Fucini P., Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine–Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Karring H., Mathu S.G., van Duin J., Clark B.F., Kraal B., Knudsen C.R. Qβ-phage resistance by deletion of the coiled-coil motif in elongation factor Ts. J. Biol. Chem. 2004;279:1878–1884. doi: 10.1074/jbc.M306605200. [DOI] [PubMed] [Google Scholar]
- Koc E.C., Burkhart W., Blackburn K., Moseley A., Spremulli L.L. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- Kumar A., Malloch R.A., Fujita N., Smillie D.A., Ishihama A., Hayward R.S. The minus 35-recognition region of Escherichia coli σ70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- Kuroda A., Nomura K., Ohtomo R., Kato J., Ikeda T., Takiguchi N., Ohtake H., Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli . Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Matsunaga J., Simons E.L., Simons R.W. RNase III autoregulation: Structure and function of rncO, the posttranscriptional “operator.”. RNA. 1996;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- Mathy N., Pellegrini O., Serganov A., Patel D.J., Ehresmann C., Portier C. Specific recognition of rpsO mRNA and 16S rRNA by Escherichia coli ribosomal protein S15 relies on both mimicry and site differentiation. Mol. Microbiol. 2004;52:661–675. doi: 10.1111/j.1365-2958.2004.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E., Yanofsky C. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Mitchell J.E., Zheng D., Busby S.J., Munchin S.D. Identification and analysis of “extended –10” promoters in Escherichia coli . Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., Nomura M. Assembly mapping of 30S ribosomal proteins in E. coli . Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- Moll I., Grill S., Gründling A., Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli . Mol. Microbiol. 2002;44:1387–1396. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella . Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nishii W., Suzuki T., Nakada M., Kim Y.T., Muramatsu T., Takahashi K. Cleavage mechanism of ATP-dependent Lon protease toward ribosomal S2 protein. FEBS Lett. 2005;579:6846–6850. doi: 10.1016/j.febslet.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Nomura M. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: Diversity and common principles. J. Bacteriol. 1999;181:6857–6864. doi: 10.1128/jb.181.22.6857-6864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Yates J.L., Dean D., Post L.E. Feedback regulation of ribosomal protein gene expression in Escherichia coli: Structural homology of ribosomal RNA and ribosomal protein mRNA. Proc. Natl. Acad. Sci. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Pemberton I.K., Muskhleshvili G., Travers A.A., Buckle M. The G+C-rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. J. Mol. Biol. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- Phadtare S., Severinov K. Extended –10 motif is critical for activity of the cspA promoter but does not contribute to low-temperature transcription. J. Bacteriol. 2005;187:6584–6589. doi: 10.1128/JB.187.18.6584-6589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda M., Gregory B.D., Szczypinski B., Baxter K.R., Hochschild A., Miller E.S., Hinton D.M. A family of anti-σ70 proteins in T4-type phages and bacteria that are similar to AsiA, a transcription inhibitor and co-activator of bacteriophage T4. J. Mol. Biol. 2004;344:1183–1197. doi: 10.1016/j.jmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Powers T., Stern S., Changchien L.-M., Noller H.F. Probing the assembly of the 3′- major domain of 16S RNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 1988;201:697–716. doi: 10.1016/0022-2836(88)90468-8. [DOI] [PubMed] [Google Scholar]
- Prentki P., Teter B., Chandler M., Galas D.J. Functional promoters created by insertion of the transposable element IS1 . J. Mol. Biol. 1986;191:383–393. doi: 10.1016/0022-2836(86)90134-8. [DOI] [PubMed] [Google Scholar]
- Rivas E., Klein R.J., Jones T.A., Eddy S.R. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Shean C.S., Gottesman M. Translation of the prophage λ c1 transcript. Cell. 1992;70:513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Caillet J., Graffe M., Eyermann F., Ehresmann B., Ehresmann S., Springer M., Romby P. The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol. Microbiol. 1998;29:1077–1090. doi: 10.1046/j.1365-2958.1998.00995.x. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker T.A., Schnitzler G., Deischel S.M., Goel M., Dove W., Jaaks K.J., Grossman A.D., Erickson J.W., Gross C.A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli . Microbiol. Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouv J., Schnier J., Rasmussen M.D., Subramanian A.R., Pedersen S. Ribosomal protein S1 of Escherichia coli is the effector for the regulation of its own synthesis. J. Biol. Chem. 1990;26:17044–17049. [PubMed] [Google Scholar]
- Stelzl U., Zengel J.M., Tovbina M., Walker M., Nierhaus K.H., Lindahl L., Patel D.J. RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J. Biol. Chem. 2003;278:28237–28245. doi: 10.1074/jbc.M302651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Tchufistova L.S., Komarova A.V., Boni I.V. A key role for the mRNA leader structure in translational control of ribosomal protein S1 synthesis in γ-proteobacteria. Nucleic Acids Res. 2003;31:6996–7002. doi: 10.1093/nar/gkg883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M.I., Chambliss G.H. The TRTGn motif stabilizes the transcription initiation open complex. J. Mol. Biol. 2002;322:521–532. doi: 10.1016/s0022-2836(02)00802-1. [DOI] [PubMed] [Google Scholar]
- Wilson D.N., Nierhaus K.H. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen D.E., Clemons W.M., Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Young B.A., Gruber T.M., Gross C.A. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- Yusupova G., Jenner L., Rees B., Moras D., Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zengel J.M., Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli . Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]