Abstract
DAZAP1 (Deleted in Azoospermia Associated Protein 1) is a ubiquitous hnRNP protein that is expressed most abundantly in the testis. Its ability to shuttle between the nucleus and the cytoplasm and its exclusion from the transcriptionally inactive XY body in pachytene spermatocytes implicate it in mRNA transcription and transport. We generated Dazap1 mutant alleles to study the role of DAZAP1 in mouse development. Most mice homozygous for the null allele as well as a hypomorphic Fn allele died soon after birth. The few Dazap1Fn/Fn mice that survived could nonetheless live for more than a year. They appeared and behaved normally but were much smaller in size compared to their wild-type and heterozygous littermates. Both male and female Dazap1Fn/Fn mice were sterile. Males had small testes, and the seminiferous tubules were atrophic with increased numbers of apoptotic cells. The tubules contained many germ cells, including pachytene spermatocytes with visible XY-bodies and diplotene spermatocytes, but no post-meiotic cells. FACS analyses confirmed the absence of haploid germ cells, indicating spermatogenesis arrested right before the meiotic division. Female Dazap1Fn/Fn mice had small ovaries that contained normal-appearing follicles, yet their pregnancy produced no progeny due to failure in embryonic development. The phenotypes of Dazap1 mutant mice indicate that DAZAP1 is not only essential for spermatogenesis, but also required for the normal growth and development of mice.
Keywords: DAZAP1, hnRNP protein, spermatogenesis, infertility, growth retardation
INTRODUCTION
Heterogeneous nuclear ribonuclear proteins (hnRNP proteins) are a group of nuclear proteins that bind to newly synthesized RNA transcripts and participate in their process and export (Dreyfuss et al. 1993). Several of them, including hnRNP A1 and hnRNP A2, have a general structure of 2xRBD-Gly, with two RNP-type RNA-binding domains (RBDs) near the N terminus and a glycine-rich domain near the C terminus. HnRNP A1, one of the most extensively studied, has been shown to shuttle between the nucleus and cytoplasm and play major roles in mRNA splicing, export, and translation (Mayeda and Krainer 1992; Mili et al. 2001; Bonnal et al. 2005). DAZ associated protein 1 (DAZAP1), also known as proline-rich RNA-binding protein (PRRP), is a more recently identified hnRNP protein (Tsui et al. 2000; Kurihara et al. 2004). It was initially isolated through its interaction with the germ-cell-specific RNA-binding protein Deleted in Azoospermia (DAZ) encoded by a region of the Y-chromosome that is frequently deleted in azoospermic men (Tsui et al. 2000). DAZAP1 also has two RBDs at its N-terminal portion. However, its C-terminal portion is rich in proline instead of glycine. It is expressed must abundantly in the testis and to a lesser degree in other tissues (Tsui et al. 2000; Dai et al. 2001; Pan et al. 2005). In somatic cells, DAZAP1 is present mainly in the nucleus, and its association with the hnRNP particles makes it a bona fide hnRNP protein (Lin and Yen 2006). In mouse testes, DAZAP1 shows a dynamic distribution during spermatogenesis. It appears abundantly in the nuclei of midpachytene spermatocytes, remains in the nuclei of round spermatids, and relocates to the cytoplasm during spermatid elongation (Vera et al. 2002).
The biological function of DAZAP1 remains unclear, although there are indications for its involvement in RNA transcription, splicing, transport, and possibly cell proliferation. A role of DAZAP1 in transcription is suggested by its specific exclusion from the transcriptionally inert XY body in the nuclei of pachytene spermatocytes, and the requirement of active transcription for its nuclear localization (Vera et al. 2002; Lin and Yen 2006). Its ability to shuttle between the nucleus and the cytoplasm supports its role in mRNA transport (Lin and Yen 2006). In addition, DAZAP1's ortholog in Xenopus, Prrp, has been shown to bind to a localization element in the 3′-untranslated region (3′-UTR) of the Vg1 mRNA that is essential for proper movement of the Vg1 mRNA to the vegetal cortex of mature oocytes (Zhao et al. 2001). However, DAZAP1 does not appear to participate directly in mRNA translation since, when in the cytoplasm, it is not associated with polyribosomes (Dai et al. 2001). DAZAP1 binds in vitro to RNA molecules containing two conserved elements, AAUAG and GU1–3AG, but its natural RNA substrates have not been identified (Hori et al. 2005). A recent study found that DAZAP1 bound to the T6 mutation of BRCA1 exon 18 mutant and inhibited its splicing (Goina et al. 2008). A possible role of DAZAP1 in cell proliferation is suggested by its involvement in a t(1;19)(q23;p13) translocation in a pre-B acute lymphoblastic leukemia (Yuki et al. 2004; Prima et al. 2005). The translocation fused DAZAP1 with the MEF2D oncogene and produced a MEF2D/DAZAP1 fusion protein that was a much more potent transcription activator than MEF2D itself and had the ability to promote the growth of HeLa cells when introduced into the cells.
Here we report the generation and characterization of Dazap1 mutant mice, with a focus on reproductive defects. We found that deficiency in DAZAP1 causes perinatal lethality, and the survivors had growth retardation in addition to reproductive defects, indicating a global role of DAZAP1 in mouse development.
RESULTS
Generation of Dazap1 mutant alleles
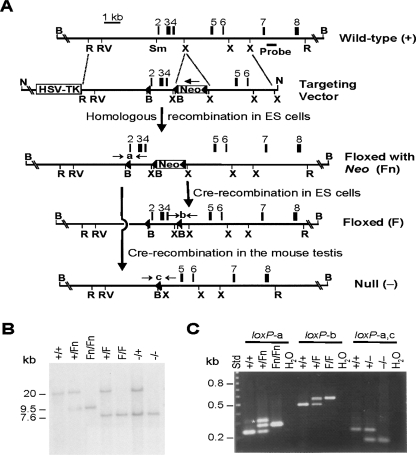
The mouse Dazap1 gene on chromosome 10 spans ∼23 kb and consists of 12 exons, with the start and stop codons residing in exons 1 and 12, respectively. We used homologous recombination in ES cells and subsequent breeding to generate three mutant alleles, targeting exons 2–4, which encode the first RBD (Fig. 1A). Deletion of exons 2–4 also causes frameshift of the coding region. A single embryonic stem (ES) cell line (#115) was identified that carried the floxed with Neo (Fn) allele containing three loxP sites and a neo gene inserted in intron 4 in the antisense direction. Subsequent expression of cre recombinase in #115 ES cells and in the testes of Dazap1+/Fn mice generated the floxed (F) allele that contain two loxP sites flanking exons 2–4, and the null (−) allele that lacks exons 2–4, respectively. The presence of the various alleles in ES cells and in mice was verified by Southern blotting (Fig. 1B) and by PCR amplification across the different loxP sites (Fig. 1C).
FIGURE 1.
Generation of Dazap1 mutant alleles. (A) Structure of the mouse Dazap1 gene and the scheme to generate the various mutant alleles. The restriction sites are (B) BamHI, (N) NotI, (R) EcoRI, (RV) EcoRV, (Sm) SmaI, and (X) XhoI. The loxP sites are depicted as close triangles. (B) Genotyping by Southern hybridization. Genomic DNAs from mice with the various genotypes were digested with BamHI and blotted with a probe from intron 7, indicated in A. The 20-, 9.5-, and 7.6-kb fragments are from the +, Fn, and F/null alleles, respectively. (C) PCR analyses of the Dazap1 alleles. Mouse tail DNA samples were assayed for the presence of loxP-a, loxP-b, and loxP-c using primers flanking the sites as indicated in A. (*) The heteroduplex in the PCR reaction of the loxP-a site.
Expression of an aberrant DAZAP1 protein from the Fn allele
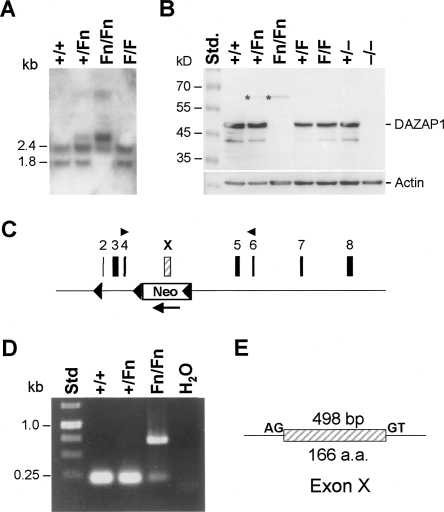
Dazap1 has two major transcripts, 2.4 and 1.8 kb, that encode a protein of 405 amino acid residues (Dai et al. 2001). Northern analyses of testis RNAs showed that while Dazap1F/F mice had the same transcripts as the wild-type mice, mice with the Fn allele expressed a longer transcript at 2.9 kb (Fig. 2A). Western blotting of testis extracts using an antibody that recognizes the C terminus of DAZAP1 detected a faint 64-kDa protein in mice carrying the Fn allele, the 45-kDa DAZAP1 in the Dazap1F/F and wild-type mice, and no DAZAP1 in the Dazap1 −/− mice (Fig. 2B). Further RT-PCR amplification of Dazap1Fn/Fn testis RNA using primers from exons 4 and 6 (Fig. 2C) produced a 700-base-pair (bp) fragment instead of the normal 210-bp fragment (Fig. 2D). Sequencing of the aberrant PCR fragment showed that it contained an extra 498-bp segment originating from the antisense strand of the neo gene that was flanked by the splicing consensus sequences (Fig. 2E). The inclusion of the extra exon (Exon X) maintained the reading frame of the transcript but added a 166-amino acid segment to the DAZAP1–Fn protein between the two RBDs. Thus, of the three Dazap1 mutant alleles we generated, the F allele expressed the wild-type DAZAP1, the null allele produced no DAZAP1, and the Fn allele produced an aberrant protein with a large insertion and likely significantly compromised function.
FIGURE 2.
Expression of an extra exon from the Dazap1 Fn allele. (A) Northern analyses of total testis RNAs using Dazap1 cDNA as a probe. (B) Western blots of testis extracts using an anti- DAZAP1 antibody. (C) The location of exon X within the Dazap1 gene. (D) RT-PCR analyses of testis RNAs using primers from exons 4 and 6 as indicated in C. (E) The structure of exon X. It is flanked by the splice donor and acceptor sequences.
Perinatal lethality and growth retardation of Dazap1Fn/Fn and Dazap1−/− mice
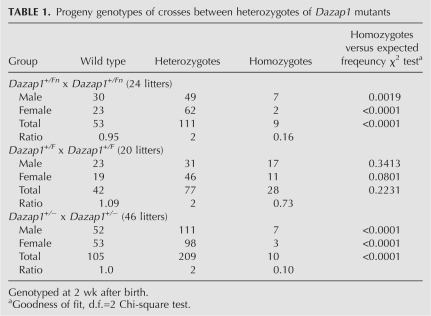
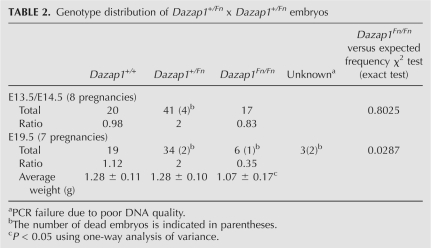
Mice heterozygous for all three Dazap1 mutant alleles had normal appearance and behavior, and were fertile. However, crosses between Dazap1 +/Fn or Dazap1 +/− heterozygotes, but not those between Dazap1 +/F heterozygotes, produced significantly fewer homozygous progeny than would be expected from Mendelian inheritance of the alleles when the mice were genotyped 2 wk after birth (Table 1). The ratios between wild-type, heterozygous, and homozygous mice were 0.95:2.0:0.16 and 1.0:2.0:0.10 for crosses between Dazap1 +/Fn heterozygotes and between Dazap1 +/− heterozygotes, respectively. Subsequent genotyping at earlier ages showed that most of the pups that died within the first few days of life were homozygotes. Further genotyping of embryonic day 19.5 (E19.5) embryos of crosses between Dazap1 +/Fn heterozygotes also showed an under-representation of the homozygotes (with a ratio of 1.12:2.0:0.35), although to a lesser degree (Table 2). The E19.5 Dazap1Fn/Fn embryos had normal appearance (data not shown) but weighed slightly less than their littermates (Table 2). The number of homozygotes at E13.5/E14.5 was not significantly different from that expected from Mendelian inheritance, suggesting that embryo lethality occurs mainly at later stages of fetal development. However, fibroblast cell lines established from E13.5/E14.5 Dazap1Fn/Fn embryos already showed poor growth in culture (Fig. 3A), indicating intrinsic defects in cell proliferation in the absence of normal DAZAP1.
TABLE 1.
Progeny genotypes of crosses between heterozygotes of Dazap1 mutants
TABLE 2.
Genotype distribution of Dazap1+/Fn x Dazap1+/Fn embryos
FIGURE 3.
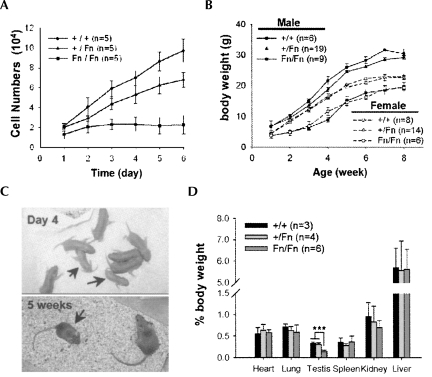
Growth retardation of Dazap1Fn/Fn mice. (A) Growth curves of mouse embryonic fibroblast cell lines derived from E13.5–E14.5 embryos. (B) Growth curves of Dazap1Fn/Fn mice and their littermates. (C) Dazap1Fn/Fn mice (arrows) compared to their littermates. (D) Percentage body weights of organs of male Dazap1Fn/Fn mice and their male littermates.
The Dazap1F/F homozygotes appeared and behaved normally. In contrast, many Dazap1Fn/Fn homozygotes died within the first 2 d after birth. By day 4, the surviving pups could be readily identified among their littermates by their small sizes (Fig. 3B,C). There was a complete concordance between small size and Dazap1Fn/Fn homozygotes that greatly facilitated the identification of the homozygotes in later experiments. The Dazap1Fn/Fn homozygotes remained small throughout their early development (Fig. 3B), appeared normal, and were active. Except for gonads, their organs had normal morphology and weights for their sizes (Fig. 3D). Male Dazap1Fn/Fn mice could live for >1 yr. However, female Dazap1Fn/Fn mice were much fewer in number, and none lived longer than 4 mo before dying suddenly of unknown causes.
The Dazap1 −/− mice had a more severe phenotype. Only about one-third of males and one-tenth of females survived the first week after birth. Similar to the Dazap1Fn/Fn mice, the Dazap1 −/− mice exhibited growth retardation and were much smaller than their littermates (data not shown). A few healthy-looking males lived to their adulthood, but all died suddenly of unknown causes before 2 mo of age if they were not already sacrificed for experiments. No females lived longer than a month. Because of the scarcity of Dazap1 −/− mice, subsequent characterization of the reproductive defects was carried out largely on Dazap1Fn/Fn mice.
Reproductive defects in male Dazap1 mutant mice
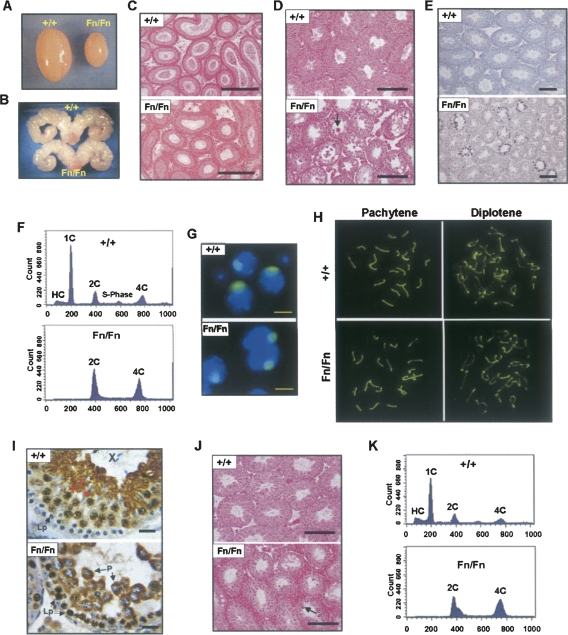
The Dazap1F/F homozygous mice were fertile, and the males had normal testis sizes and epididymis sperm counts (data not shown). In contrast, adult male Dazap1Fn/Fn mice were sterile. They had much smaller testes (40% of the normal for their body sizes) (Figs. 3D, 4A) but normal-size seminal vesicles (Fig. 4B), and their epididymides contained no sperm (Fig. 4C). Their serum testosterone levels were normal (2.43 ± 0.994 ng/mL; n = 8) compared to their littermate controls (2.44 ± 0.917 ng/mL; n = 7), indicating that their infertility was not caused by hormone imbalance. The Dazap1Fn/Fn seminiferous tubules were smaller in diameter and atrophic (Fig. 4D), and contained some multinucleated giant cells and a significantly increased number of apoptotic germ cells (Fig. 4E). There were many spermatogonia and spermatocytes but no meiotic or post-meiotic germ cells in the tubules (Fig. 4D). The absence of haploid cells in the Dazap1Fn/Fn testes was further confirmed by fluorescence activated cell sorting (FACS) analyses, which showed the presence of diploid cells (2C) and primary spermatocytes (4C) but no round (1C) or elongated (HC) spermatids (Fig. 4F). The XY bodies in Dazap1Fn/Fn spermatocytes appeared normal when stained with an anti-SUMO-1 antibody (Fig. 4G; Vigodner and Morris 2005). Staining of the synaptonemal complexes with an anti-SCP3 antibody (Lammers et al. 1994) showed normal synapsis of homologous chromosomes in pachytene spermatocytes and the presence of diplotene spermatocytes (Fig. 4H). However, there was an accumulation of post-pachynema spermatocytes in the mutant mice (72% of all spermatocytes compared to the wild-type, 38%). Our results thus indicate that in the Dazap1Fn/Fn mice, spermatogenesis is arrested right before the meiotic division. Additional immunostaining of testis sections showed that the mutant DAZAP1–Fn protein had an aberrant spatiotemporal expression pattern (Fig. 4I). In the wild-type mice, DAZAP1 is not expressed until the late pachytene stage, where it is present mainly in the nucleus (Vera et al. 2002). In the Dazap1Fn/Fn testes, DAZAP1–Fn appeared earlier in the nuclei of leptotene spermatocytes and was present in the cytoplasm instead of the nuclei of pachytene spermatocytes. The spermatogenetic defects in the Dazap1Fn/Fn mice did not worsen as the mice aged. Histological (Fig. 4J) and FACS (Fig. 4K) analyses showed the presence of many spermatogonia and spermatocytes in the testes of 1-yr-old homozygotes similar to those of the 6 wk olds.
FIGURE 4.
Spermatogenic defects in adult male Dazap1Fn/Fn mice. The animals were 5–7 wk old in A-I and 1 yr old in J and K. (A) The testes. (B) The seminal vesicles. (C) H&E staining of epididymis sections. (D,J) H&E staining of testis sections. The arrows point to multinucleated giant cells. (E) TUNEL staining of apoptotic cells on testis sections. (F,K) FACS profiles of testis cells sorted according to their DNA contents. Cell types in the various peaks are (HC) hypo-stained elongating and elongated spermatids; (1C) round spermatids; (2C) spermatogonia, secondary spermatocytes, and somatic cells; (S-phase) spermatogonia synthesizing DNA; and (4C) primary spermatocytes. (G) Immunofluorescence staining of the XY bodies in dispersed pachytene spermatocytes with an anti-SUMO-1 antibody. (H) Immunofluorescence staining of synaptonemal complexes (SC) in spermatocyte spreads with an anti-SCP3 antibody. Pachytene and diplotene spermatocytes were differentiated according to their SC structures. (I) Immunostaining of DAZAP1 on testis sections. The cell types are (P) pachytene and (Lp) leptotene spermatocytes. Scale bars, (C–E,J) 200 μm; (G) 10 μm; (I) 25 μm.
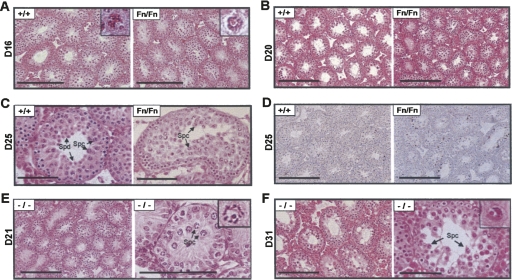
We next studied younger mice to determine the time when spermatogenic abnormality first became evident. During postnatal development of mouse testes, the seminiferous core normally acquires a lumen in many areas by day 16, and the first wave of meiotic division starts around day 18, when secondary spermatocytes and round spermatids first appear in the tubules (Bellve et al. 1977). The testes of 16-d-old Dazap1Fn/Fn males already showed some abnormality in that very few tubules contained a lumen (Fig. 5A). However, spermatogenesis appeared to have proceeded normally, and pachytene spermatocytes with visible XY-bodies could be readily identified (Fig. 5A, insets). The delay in lumen formation persisted in day 20 testes (Fig. 5B), yet there was no increase in apoptosis in the tubules (data not shown). At day 25, the most advanced germ cells in the mutant testes were pachytene/diplotene spermatocytes when the wild-type testis already contained many haploid round spermatids (Fig. 5C). In addition, there was an increase in apoptotic cells in the mutant testes (Fig. 5D). Thus the onset of spermatogenic defects in Dazap1Fn/Fn mice is consistent with the spermatogenic defects observed in adult mutant mice, with spermatogenesis arrested right before the meiosis division.
FIGURE 5.
Spermatogenic defects in premature male Dazap1 mutant mice. The ages (in days) of the animals are indicated at left. (A–C) H&E staining of testis sections of premature Dazap1Fn/Fn mice and their wild-type littermates. (Insets) Pachytene spermatocytes with visible XY-bodies. (D) TUNEL staining of apoptotic cells on testis sections. (E,F) Testis histology of Dazap1-null mutants shown in two different magnifications. (Insets) Pachytene spermatocytes with the XY-bodies. (Spc) Spermatocyte; (Spd) spermatid. Scale bars (A,B, left panels of E,F) 200 μm; (C, right panels of E,F) 100 μm; (D) 250 μm.
The testis histology of a limited number of Dazap1 −/− mice suggested a similar progression of spermatogenic defect. The seminiferous tubules of a day 21 mouse showed the presence of late pachytene spermatocytes and a delay in lumen formation (Fig. 5E), and those of a day 31 mouse were atrophic and contained many late pachytene spermatocytes but no post-meiotic haploid germ cells (Fig. 5F). Thus, in the Dazap1 −/− mice, spermatogenesis appears to arrest at the same stage as that of the Dazap1Fn/Fn mice.
Reproductive defects in female Dazap1 mutant mice
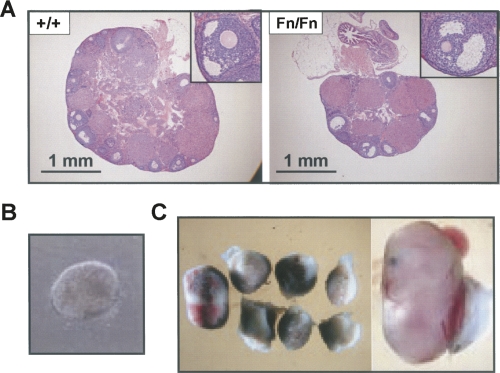
Very few female Dazap1Fn/Fn mice survived to sexual maturity. Adult homozygous females had smaller ovaries with fewer follicles (Fig. 6A). However, the development of the follicles appeared to be normal (Fig. 6A, insets). Mating between two 2-mo-old Dazap1Fn/Fn females with fertile Dazap1 +/Fn males over a 2-mo period failed to produce any progeny, whereas their littermates housed in the same cages all gave birth to pups. Dissection of two other females 3 wk after the detection of a vaginal plug but when they were showing no sign of pregnancy indicated that the animals were, in fact, pregnant. One had five decidua about the size of 8.5 dpc, but only one of them contained a poorly developed blastocyst-like embryo (Fig. 6B). The other female had eight decidua, but only one contained a viable fetus about the size of 14.5 dpc (Fig. 6C). The female Dazap1Fn/Fn mice therefore appeared to have normal oogenesis but were unable to sustain embryo development due to maternal effect.
FIGURE 6.
Reproductive defects in adult female Dazap1Fn/Fn mice. (A) Ovary histology. (Insets) Selected follicles at a higher magnification. (B) A blastocyst-like embryo in a Dazap1Fn/Fn uterus. (C) The decidua contents of another Dazap1Fn/Fn female.
DISCUSSION
Here we report the generation of three Dazap1 mutant mice from a single ES cell line. The presence of growth retardation and reproductive defects in Dazap1Fn/Fn and Dazap1 −/− mice, but not Dazap1F/F mice, indicates that the phenotype is, indeed, caused by the disruption of the Dazap1 gene. Our results indicate that DAZAP1's function is not restricted to testes, where it is most abundantly expressed. The association of perinatal lethality with DAZAP1 deficiency is not unexpected, considering that DAZAP1 is an hnRNP protein. Of the 30 or so genes encoding hnRNP proteins, so far only an insertion mutation of hnRNP C has been reported (Williamson et al. 2000). The homozygous mutant embryos failed to develop beyond E6.5, yet stem cell lines established from the blastocysts were able to grow and differentiate in vitro. Our Dazap1-null mutants have a less severe phenotype. Many embryos developed to term, and a few of the pups even survived to adulthood, although none of them lived longer than 2 mo. Compared to the Dazap1-null mutants, the Dazap1Fn/Fn mice had a milder phenotype and lived longer, indicating that the Fn allele is hypomorphic and the DAZAP1–Fn protein retains some biological functions. This is supported by our preliminary data showing that DAZAP1–Fn retains the ability to bind poly(G) and poly(U) in vitro. However, spermatogenesis appears to arrest at the same stage in the Dazap1Fn/Fn and the Dazap1 −/− mice, suggesting that the DAZAP1–Fn protein serves little function in male germ cell development. This could be due to the aberrant spatiotemporal expression of DAZAP1–Fn during spermatogenesis. Besides growth retardation and infertility, some of the surviving Dazap1Fn/Fn mice manifested other problems, such as obesity and loss of renal function, when growing older. In addition, most Dazap1Fn/Fn mice developed lesions around their eyes by 4 mo, suggesting defects in the immune system (data not shown). It should be pointed out that DAZAP1 is also expressed at a high level in the thymus, second only to the testis (http://symatlas.gnf.org/SymAtlas). Thus, the unexpected generation of the hypomorphic Fn allele uncovered additional functions of DAZAP1 that would not have been revealed by the null mutants due to their short lives. However, it will be difficult to study these additional defects in Dazap1Fn/Fn mice due to their short supply. Future investigation of possible roles of DAZAP1 in the immune as well as other systems will require the use of tissue-specific knockout approaches.
It is interesting that male and female Dazap1Fn/Fn mice are infertile for different reasons, and that DAZAP1 is required for spermatogenesis but not oogenesis. The spermatogenic defects in Dazap1 −/− and Dazap1Fn/Fn mutants resemble those of the Ccna1-null mutants (Liu et al. 1998; Nickerson et al. 2007). Ccna1 encodes cyclin A1, a member of the cyclin family that regulates cell cycle progression. Female Ccna1-null mutants are fertile. Male Ccna1 mutants have spermatogenic arrest at the late diplotene stage, and the first wave of spermatogenesis is arrested around day 22 with enhanced apoptosis but no overt degeneration (Salazar et al. 2005). In our Dazap1Fn/Fn mutant mice, spermatogenesis also proceeds to the diplotene stage and is arrested immediately before the meiotic division, and we observed increased apoptosis and atrophy in day 25, but not day 21 tubules. The possibility that cyclin A1 and DAZAP1 are involved in the same pathway remains to be investigated. In the Dazap1Fn/Fn female, oogenesis appears to proceed normally even though there were fewer follicles in the ovaries, and the eggs could be fertilized and implanted in the uterus. However, there appears to be a maternal effect on the development of the embryos regardless of their genotypes. The Dazap1Fn/Fn uterus seems to be incompetent in supporting embryo development, resulting in embryo death and infertility. The mechanisms underlying the spermatogenic arrest and the uterus defect are unknown, and the RNA targets of DAZAP1 remain elusive. Future identification of the downstream targets of DAZAP1 will provide new insights on the biological functions of DAZAP1 and its roles in the development of different cell lineages.
MATERIALS AND METHODS
Construction of the targeting vector and generation of Dazap1 mutant alleles
The targeting vector was constructed using pPNT-M2-loxP as the backbone (Chen et al. 2002), and the generation of Dazap1 mutant mice using TC1 ES cells (Yan et al. 1999) was carried out according to Figure 1A using standard protocols (Nagy et al. 2003). Clone #115 with the Fn allele was transfected with Cre plasmid pBS185 (GIBCO) to generate the F allele, and Dazap1 +/Fn female mice were mated with male protamine-cre transgenic mice (#58; The Jackson Laboratory) to generate the null allele. The mice were genotyped by PCR across the various loxP sites. The primers are loxP-a: F, 5′-atctctgtggcagcaaaggc and R, 5′-aagcccattcatggctggga; loxP-b: F, 5′-gagcgaacacggcccaag and R, 5′-gcagcctacagctaccatc. The loxP-c site was amplified using primers loxP-a-F and loxP-b-R.
All mice were housed in a specific pathogen-free animal facility, and the experiments were carried out under the approval of the Institutional Animal Care & Utilization Committee of Academia Sinica. The animals used in the study were from F 2 and F 3 intercrosses.
Generation of mouse embryo fibroblasts (MEFs) and growth curve analysis
MEFs were prepared from E13.5/E14.5 embryos using protocol 1 of Nagy et al. (2003) with some modifications. After mincing, the embryos were treated with trypsin for 5 min at 37°C and further broken up into single cells by repeated pipetting before being plated out. The growth curves of MEFs from the third passages were determined according to Mar and Hoodless (2006).
Other protocols
Western blotting of DAZAP1 in testis extracts was carried out according to Dai et al. (2001) using an antibody that recognizes the C terminus of the protein. Mouse testis dispersions were prepared according to Grabske et al. (1975), and the XY-bodies in pachytene spermatocytes were stained using a mouse anti-Sentrin-1 (SUMO-1) antibody (Zymed Laboratory). Synaptonemal complexes were stained using a rabbit anti-SCP3 antibody (Novus Biologicals) according to Uemura et al. (2000). TUNEL assays were performed on testicular sections using the In Situ Cell Death Detection Kit (Roche Diagnostics GmbH) according to the manufacturer's instructions. Serum testosterone levels were determined using the Testosterone ELISA Kit from IBL-Hamburg. Flow cytometric analyses of testis cell types were carried out according to Jeyaraj et al. (2002) using a BC FACSCalibur Flow Cytometer (BD Biosciences Clontech).
ACKNOWLEDGMENTS
We thank Chih-Cheng Chen for pPNT-M2-loxP; Yi-Tzu Lin, Kuang-Nan Hsiao, Li Ling, and Tiane Dai for technical assistance; Tateki Kikuchi for phenotype analyses; Chien-Hsiun Chen for statistical analyses of the data; Pei-Chun Tsai for blastocyst injection; and the IBMS Pathology Core for tissue section preparation. This work was supported by grants from the National Science Council (96-2311-B-001-026-MY3) and Academia Sinica in Taiwan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1152808.
REFERENCES
- Bellve A.R., Cavicchia J.C., Millette C.F., O'Brien D.A., Bhatnagar Y.M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A.C., Vagner S. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- Chen C.C., Zimmer A., Sun W.H., Hall J., Brownstein M.J. A role for ASIC3 in the modulation of high-intensity pain stimuli. Proc. Natl. Acad. Sci. 2002;99:8992–8997. doi: 10.1073/pnas.122245999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T., Vera Y., Salido E.C., Yen P.H. Characterization of the mouse Dazap1 gene encoding an RNA-binding protein that interacts with infertility factors DAZ and DAZL. BMC Genomics. 2001;2:6. doi: 10.1186/1471-2164-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Goina E., Skoko N., Pagani F. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.02253-07. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabske R.J., Lake S., Gledhill B.L., Meistrich M.L. Centrifugal elutriation: Separation of spermatogenic cells on the basis of sedimentation velocity. J. Cell. Physiol. 1975;86:177–189. doi: 10.1002/jcp.1040860119. [DOI] [PubMed] [Google Scholar]
- Hori T., Taguchi Y., Uesugi S., Kurihara Y. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 2005;33:190–200. doi: 10.1093/nar/gki153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj D.A., Grossman G., Weaver C., Petrusz P. Dynamics of testicular germ cell proliferation in normal mice and transgenic mice overexpressing rat androgen-binding protein: A flow cytometric evaluation. Biol. Reprod. 2002;66:877–885. doi: 10.1095/biolreprod66.4.877. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe H., Kawaguchi A., Hori T., Mishiro K., Ono M., Sawada H., Uesugi S. Dynamic changes in intranuclear and subcellular localizations of mouse Prrp/DAZAP1 during spermatogenesis: The necessity of the C-terminal proline-rich region for nuclear import and localization. Arch. Histol. Cytol. 2004;67:325–333. doi: 10.1679/aohc.67.325. [DOI] [PubMed] [Google Scholar]
- Lammers J.H., Offenberg H.H., van Aalderen M., Vink A.C., Dietrich A.J., Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.T., Yen P.H. A novel nucleocytoplasmic shuttling sequence of DAZAP1, a testis-abundant RNA-binding protein. RNA. 2006;12:1486–1493. doi: 10.1261/rna.42206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Matzuk M.M., Sung W.K., Guo Q., Wang P., Wolgemuth D.J. Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 1998;20:377–380. doi: 10.1038/3855. [DOI] [PubMed] [Google Scholar]
- Mar L., Hoodless P.A. Embryonic fibroblasts from mice lacking Tgif were defective in cell cycling. Mol. Cell. Biol. 2006;26:4302–4310. doi: 10.1128/MCB.02156-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mili S., Shu H.J., Zhao Y., Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Nickerson H.D., Joshi A., Wolgemuth D.J. Cyclin A1-deficient mice lack histone H3 serine 10 phosphorylation and exhibit altered aurora B dynamics in late prophase of male meiosis. Dev. Biol. 2007;306:725–735. doi: 10.1016/j.ydbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H.A., Lin Y.S., Lee K.H., Huang J.R., Lin Y.H., Kuo P.L. Expression patterns of the DAZ-associated protein DAZAP1 in rat and human ovaries. Fertil. Steril. 2005;84(Suppl 2):1089–1094. doi: 10.1016/j.fertnstert.2005.03.075. [DOI] [PubMed] [Google Scholar]
- Prima V., Gore L., Caires A., Boomer T., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23;p13.3) in acute lymphoblastic leukemia. Leukemia. 2005;19:806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- Salazar G., Joshi A., Liu D., Wei H., Persson J.L., Wolgemuth D.J. Induction of apoptosis involving multiple pathways is a primary response to cyclin A1-deficiency in male meiosis. Dev. Dyn. 2005;234:114–123. doi: 10.1002/dvdy.20533. [DOI] [PubMed] [Google Scholar]
- Tsui S., Dai T., Roettger S., Schempp W., Salido E.C., Yen P.H. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- Uemura T., Kubo E., Kanari Y., Ikemura T., Tatsumi K., Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct. Funct. 2000;25:149–159. doi: 10.1247/csf.25.149. [DOI] [PubMed] [Google Scholar]
- Vera Y., Dai T., Hikim A.P., Lue Y., Salido E.C., Swerdloff R.S., Yen P.H. Deleted in azoospermia associated protein 1 shuttles between nucleus and cytoplasm during normal germ cell maturation. J. Androl. 2002;23:622–628. [PubMed] [Google Scholar]
- Vigodner M., Morris P.L. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: Silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 2005;282:480–492. doi: 10.1016/j.ydbio.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Williamson D.J., Banik-Maiti S., DeGregori J., Ruley H.E. hnRNP C is required for postimplantation mouse development but Is dispensable for cell viability. Mol. Cell. Biol. 2000;20:4094–4105. doi: 10.1128/mcb.20.11.4094-4105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.T., Gritsman K., Ding J., Burdine R.D., Corrales J.D., Price S.M., Talbot W.S., Schier A.F., Shen M.M. Conserved requirement for EGF-CFC genes in vertebrate left–right axis formation. Genes & Dev. 1999;13:2527–2537. doi: 10.1101/gad.13.19.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki Y., Imoto I., Imaizumi M., Hibi S., Kaneko Y., Amagasa T., Inazawa J. Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13) Cancer Sci. 2004;95:503–507. doi: 10.1111/j.1349-7006.2004.tb03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Jiang C., Kroll T.T., Huber P.W. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 2001;20:2315–2325. doi: 10.1093/emboj/20.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]