Abstract
Tomographic neuroimaging techniques allow visualization of functionally and structurally specific signals in the mouse and rat brain. The interpretation of the image data relies on accurate determination of anatomical location, which is frequently obstructed by the lack of structural information in the data sets. Positron emission tomography (PET) generally yields images with low spatial resolution and little structural contrast, and many experimental magnetic resonance imaging (MRI) paradigms give specific signal enhancements but often limited anatomical information. Side-by-side comparison of image data with conventional atlas diagram is hampered by the 2-D format of the atlases, and by the lack of an analytical environment for accumulation of data and integrative analyses. We here present a method for reconstructing 3-D atlases from digital 2-D atlas diagrams, and exemplify 3-D atlas-based analysis of PET and MRI data. The reconstruction procedure is based on two seminal mouse and brain atlases, but is applicable to any stereotaxic atlas. Currently, 30 mouse brain structures and 60 rat brain structures have been reconstructed. To exploit the 3-D atlas models, we have developed a multi-platform atlas tool (available via The Rodent Workbench, http://rbwb.org) which allows combined visualization of experimental image data within the 3-D atlas space together with 3-D viewing and user-defined slicing of selected atlas structures. The tool presented facilitates assignment of location and comparative analysis of signal location in tomographic images with low structural contrast.
Keywords: 3-D reconstruction, atlas, brain, imaging, magnetic resonance imaging, positron emission tomography, stereotaxic, visualization
Introduction
Positron emission tomography (PET) and magnetic resonance imaging (MRI) techniques are increasingly used for investigations of the mouse and rat brain, in the context of numerous types of general brain mapping or disease model investigations. PET imaging reflects function and not primarily structure, and therefore, reveals few landmarks in the brain or skull. MRI imaging shows a variable degree of structural detail, depending on the specifications of the instrument and the imaging protocols. A common challenge is therefore to assign locations to the data collected with use of available brain atlases.
PET and MRI techniques produce three-dimensional (3-D) volume (tomographic) data, whereas standard brain atlases have until recently been available only in a two-dimensional (2-D) format. The established and most commonly used atlases of the mouse and rat brain (Paxinos and Franklin, 2001; Paxinos and Watson, 1998, 2005; Swanson, 2004) provide series of sections, cut at specified angles, with external surfaces and internal boundaries of areas and nuclei indicated, and names assigned to the delineated structures. Use of such atlases for neuroimaging studies are hampered by the 2-D format as well as a lack of an analytical environment for comparison and integration of atlas and image data. Recently, atlases with 3-D representations of major brain structures have been developed for the mouse (Boline et al., 2006; Chan et al., 2007; Gustafson et al., 2004; Lein et al., 2007; MacKenzie-Graham et al., 2004). Both the 3-D mouse brain atlas of the Biomedical Informatics Research Network, BIRN (Boline et al., 2006), and the 3-D surgical mouse atlas developed at the University of Toronto (Chan et al., 2007) support import of experimental data (e.g., TIFF and Analyze Image format), which can either be visualized as orthogonal slices in a separate window or in the 3-D viewer. Furthermore, the BIRN atlas facilitates alignment of the experimental data with the atlas, with a set of predefined transformation parameters (i.e., 180 degree rotation, swapping, and mirroring). As concerns 3-D rat brain atlases, several efforts are under development (see e.g., Gustafson et al., 2004, 2007), and a recently deployed neuroanatomical affiliation visualization-interface system demonstrates parts of the rat brain stem in 3-D (Palombi et al., 2006), but there is to our knowledge currently no comprehensive rat brain atlas available.
To further facilitate the task of establishing a spatial mapping between neuroimaging data and atlas space for the mouse and rat brain, we have established a pipeline for 3-D reconstruction of structures from two of the seminal high-resolution standard atlases (Paxinos and Franklin, 2001; Paxinos and Watson, 2005) including the stereotaxic space provided by the atlases. These particular rodent atlases are widely used and share a common nomenclature for both mouse and rat. The present version of the 3-D reconstructed atlases include 30 structures for the mouse brain and 60 structures for the rat brain. We have, furthermore, developed a multi-platform tool that allows sharing of the reconstructed atlas framework, surface modeling and rotation of selected structures, slicing at any chosen angle, and the possibility to add new structures to the framework according to the requirements of a given study. The tool allows import of PET and MRI data to facilitate comparison with the atlas and warping of experimental image data into atlas space. The approach taken uses linear registration, with the possibility to scale, rotate, and position the imported data, either globally, using the whole brain as template, or locally, using smaller parts of the brain as template. Here we demonstrate the use of this tool as a dynamic and analytical environment for assigning location to image data and discuss advantages of the approach taken as well as some of the remaining challenges.
Materials and Methods
Structures from two atlases, “The Rat Brain in Stereotaxic Coordinates” by Paxinos and Watson (2005) and “The Mouse Brain in Stereotaxic Coordinates” by Paxinos and Franklin (2001) were 3-D reconstructed. The diagrams in Paxinos and Watson (2005) were derived from coronal sections spaced at intervals of 120 μm, all collected from a single Wistar rat brain. The diagrams in Paxinos and Franklin (2001) were obtained from a sample of 26 adult C57BL/J6 mice. Both atlases use the same stereotaxic coordinate system (Paxinos and Watson, 1982), structure nomenclature and abbreviations. The reconstruction procedure is here exemplified with use of the Paxinos and Watson, 2005) atlas, but may be applied to any stereotaxic atlas providing digital drawings. The practical application of the reconstructed 3-D rat brain atlas is exemplified on basis of a set of MRI and PET data.
3-D atlas reconstruction and visualization
The coronal atlas of Paxinos and Watson (2005) provides digital diagrams (EPS format; Figure 1A) containing separate information layers (i.e., stereotaxic coordinate grid, structure names, and boundary lines). All diagrams were organized in separate layers and aligned to the stereotaxic grid of the atlas using Adobe Illustrator CS2 (Adobe Systems Inc., San Jose, CA). Regions of interest (ROIs, defined by closed boundary lines in the atlas) were segmented in relevant diagrams by assigning a specific color (Figure 1B). Occasionally, structures lacked unequivocally defined or closed boundaries, and contour lines were closed arbitrarily after following a set of predefined rules. Each segmented layer was exported separately as an image (TIFF) file to Amira 4.0 (Mercury Computer Systems Inc., Chelmsford, MA) for 3-D reconstruction. Since the export function in Illustrator is based on what is visible in the figure, all other layers and sub-layers were hidden. All exported images were enclosed by a standard invisible (white) rectangular frame, representing the stereotaxic X,Y space for each diagram, to ensure identical dimension and positioning of objects. Anterioposterior positions (Z-values) were defined by the bregma values given in the atlas diagrams. These values were entered upon loading stacks of image files to Amira, resulting in a 3-D atlas reconstruction aligned according to the stereotaxic framework of the atlas. Image colormaps (RGBA-tupels) were converted to intensity values represented as gray scale images, and voxels were assigned to the different structures using the “LabelVoxel” tool in Amira, employing thresholding algorithms (Figure 1C). Surfaces were rendered using the Amira SurfaceGen tool and exported as VRML (Virtual Reality Modeling Language) files. A custom Java3D visualization tool (m3d) was developed to import and visualize the VRML atlas structures (Figures 1D, and 2) as well as tomographic image data (PET or MRI) in NIfTI (Neuroimaging Informatics Technology Initiative, http://nifti.nimh.nih.gov) file format. The volumetric image data were visualized using texture mapping with transparency and thresholding applied to Texture3D Java3D class, and displayed in m3d together with the 3-D atlas structures. In addition, 2-D user-defined slices, prepared at any angle and orientation, of both the atlas (performed by ModelClip Java3D class extended by a cutting and contour-filling algorithm; Figures 2 and 3 and the image data were generated.
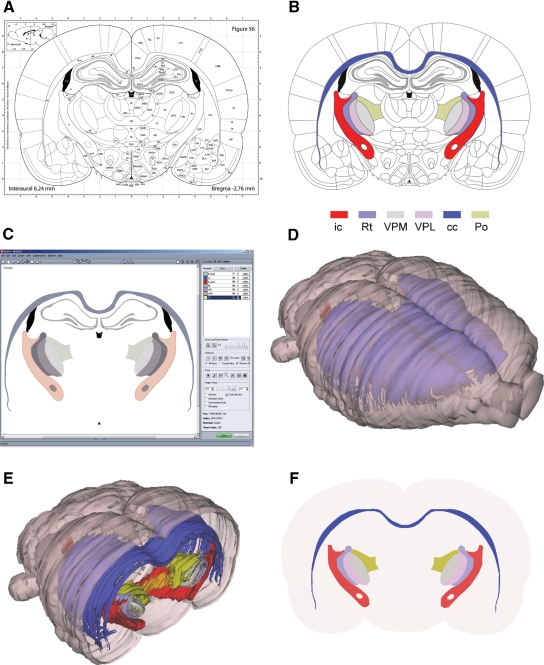
Figure 1.
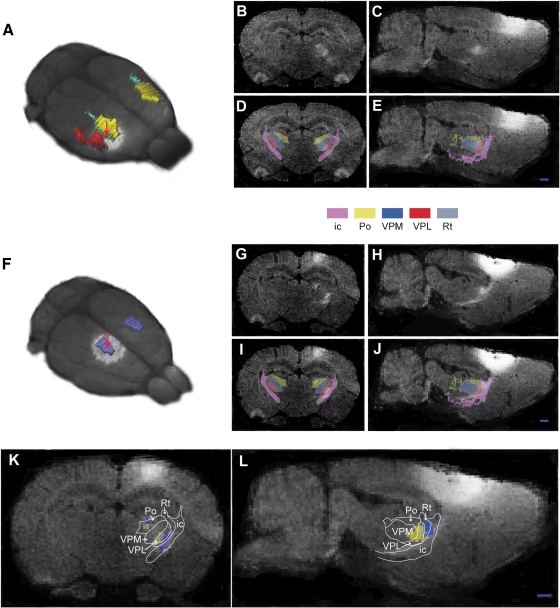
The stepwise 3-D atlas reconstruction procedure. (A) A coronal rat brain atlas diagram (reproduced from Paxinos and Watson, 2005 with permission) with the stereotaxic grid, section number, structure labeling and the distance from bregma indicated. (B) The same diagram imported to Illustrator, where regions of interest (ROIs)are selected and assigned a color code. (C) Image of the Amira GUI showing segmented ROIs. (D) Selected 3-D reconstructed structures visualized in the m3d tool. (E) The same reconstruction as shown in (C) coronally sub-divided using the m3d slicing module. (F) Customized atlas diagram, corresponding to (A), exported from m3d. A separate module in m3d is used to render the sub-divided surfaces as solid objects. ic, internal capsule; Rt, reticular thalamic nucleus; VPM, ventral posterolateral thalamic nucleus; VPL, ventral posteromedial thalamic nucleus; cc, corpus callosum; Po, posterior thalamic nucleus.
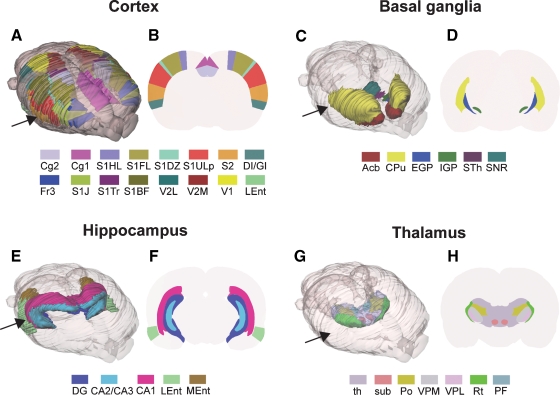
Figure 2.
Examples of rat brain regions included in the 3-D rat brain atlas. (A–H) Atlas representations of four major brain regions visualized as solid, color coded 3-D surfaces within the transparent outer surface of the brain, and as corresponding 2-D coronal slices, obtained from anteroposterior levels indicated by arrows. (A, B) 16 selected cerebrocortical brain regions. (C, D) Major regions of the basal ganglia. (E, F) Hippocampus and entorhinal cortex. (G, H) Selected sub-regions of the thalamus. In (G) thalamus (th)is made transparent in order to visualize the underlying sub-regions. Cg2, cingulate cortex area 2; Cg1, cingulate cortex area 1; S1, primary somatosensory cortex; S1HL, hindlimb region; S1FL, forelimb region; S1DZ, dysgranular zone; S1ULp, upper lip region; S2, secondary somatosensory cortex; DI/GI, insular cortex; Fr3, frontal cortex, area 3; S1J, jaw region; S1Tr, trunk region; S1BF, barrel field; V2L, secondary visual cortex, lateral area; V2M, secondary visual cortex, medial area; V1, primary visual cortex; Lent, lateral entorhinal cortex; Acb, accumbens nucleus; CPu, caudate putamen; EGP, external globus pallidus; IGP, internal globus pallidus; STh, subthalamic nucleus; SNR, substantia nigra; DG, dentate gyrus; CA2/CA3, field CA2 and CA3 of the hippocampus; CA1, field CA1 of the hippocampus; LEnt, lateral entorhinal cortex; MEnt, medial entorhinal cortex; th, thalamus, whole region; sub, submedius thalamic nucleus; Po, posterior thalamic nucleus; VPM, ventral posterolateral thalamic nucleus; VPL, ventral posteromedial thalamic nucleus; Rt, reticular thalamic nucleus; PF, parafasicular thalamic nucleus.
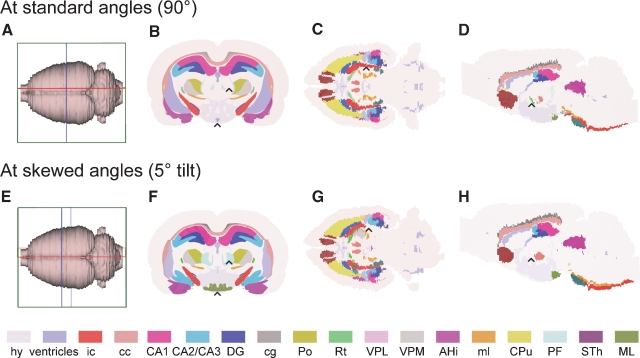
Figure 3.
Customized atlas diagrams. Series of customized, color coded coronal (B, F), horizontal (C, G), and sagittal (D, H) atlas diagrams, generated using the slicing and filling module in the atlas tool. Blue, red, and green bounding boxes in (A, E) indicate the levels from which the respective slices were obtained. (A–D) Standard orientation of atlas slices, oriented perpendicularly to axes of the stereotaxic coordinate system. (E–H) Example of atlas slices cut at non-standard angles, tilted 5 degrees from the planes used in (B–D). Black arrow heads indicate examples of structural differences between the corresponding diagram pairs, emphasizing the importance of having atlas diagrams with similar orientation as experimental slices. hy, hypothalamus, whole region; ic, internal capsule; cc, corpus callosum; CA1, field CA1 of the hippocampus; CA2/CA3, field CA2 and CA3 of the hippocampus; DG, dentate gyrus; cg, cingulum; Po, posterior thalamic nucleus; Rt, reticular thalamic nucleus; VPL, ventral posteromedial thalamic nucleus; VPM, ventral posterolateral thalamic nucleus; AHi, amygdalohippocampal area; ml, medial lemniscus; CPu, caudate putamen; PF, parafasicular thalamic nucleus; STh, subthalamic nucleus; ML, medial mammillary nucleus.
Image import and co-registration
Co-registration of experimental image data to atlas space can be achieved in several ways. The present version of the m3d program allows affine transformations that are implemented with use of a transformation tool interface for defining scaling, rotation, and translation. As exemplified below, co-registration is based on recognizable brain or skull landmarks, and different strategies may be employed depending on the nature of the image data. The precision of co-registration critically depends on the precision with which anatomical landmarks may be detected, which in turn also depends on voxel size and structural contrast (see also Discussion). Size and shape differences that may occur due to individual variation or by extraction of the brain from its enclosed position in the skull, can be adjusted by non-linear transformation (not included in the present software), or by limiting the co-registration to smaller ROIs.
Animal experiments
Animal procedures were approved by institutional animal welfare committees at the University of Oslo or Massachusetts General Hospital, and were in compliance with National Institutes of Health guidelines for the use and care of laboratory animals. During all experiments body temperature was kept constant by the means of feedback regulated heating pads. Eye ointment was applied in order to prevent dehydration of the cornea.
In vivo pathway tracing with Mn 2+-enhanced MRI
MR images were obtained at the Massachusetts General Hospital NMR-Center using a 3 Tesla Trio whole body human scanner (Siemens Medical Solutions, Iselin, NJ). Adult male Sprague–Dawley (SD) rats (Taconic farms, Germantown, NY) were anesthetized with urethane (Abbott, Chicago, IL; 1.2 g/kg i.p., with 10% supplements during surgery as needed). This provided stable anesthesia for the entire imaging session (see also Field et al., 1993). Animals were monitored by pulse oximetry, and body temperature was maintained at 37°C by a water-circulating heating pad (Gaymar T/Pump; Gaymar Industries, Orchard Park, NY). The T1 contrast agent manganese (Mn2+) was applied as outlined in Leergaard et al. (2003). Briefly, animal heads were immobilized in a stereotaxic frame, a small opening was made in the skull and dura overlying the SI cortex, and ∼10 nl of an aqueous solution of 2.0 M manganese chloride (MnCl2; ICN Biomedicals Inc., Aurora, OH) was slowly injected 1.0 mm below the pial surface through a glass pipette mounted on a 1.0 μl Hamilton syringe placed in a microinjection unit (Kopf 5001, Kopf Instruments, Tujunga, CA). Locations of injection sites were stereotaxically defined according to published topographical maps (Chapin and Lin, 1984). MR images were obtained using the (human) body RF coil for excitation, and a custom made loop surface receiver coil. T1 weighted volumetric images were obtained using a 3-D FLASH sequence with TR/TE/flipangle = 44 ms/13.37 ms/40 degrees. Images were obtained with 195 μm isotropic voxels, field of view = 50 × 50 × 50 mm3, matrix = 256 × 256 × 256, with a scan time of 48 minutes and 4 seconds. The animals were scanned continuously, starting within an hour after the MnCl2 injection, and continuing up to 8–9 hour after injection. At the end of the imaging experiments, animals were euthanized with an overdose of sodium pentobarbital (200 mg/kg).
To optimize visualization of the brain surface, the soft tissue surrounding the brain was stripped away from the raw MR images using PMOD software (PMOD Technologies Ltd., Zurich, Switzerland). The trimmed MR images were imported to the 3-D atlas tool (Figures 4D and 4G) and globally fitted to atlas space using affine transformations, based on measurements of multiple anatomical landmarks recognizable in the images (brain surface and midline, the genu and splenium of the corpus callosum, the decussation of the anterior commisure, the ventricular system, the boundary between the cortical gray matter and underlying white substance, the external boundaries of the hippocampus, the caudate putamen complex, the optic chiasm, the substantia nigra (intercalated between the cerebral peduncle and the medial lemnisc), and the rostral boundary of the pontine nuclei). Because the in vivo position of the cerebellum and of the flaccid brain stem caudal of the pons clearly differed from the atlas, these structures were disregarded in the alignment procedure. For this dataset, the spatial displacement of the different image landmarks with respect to corresponding atlas landmarks, was measured to 164 μm ± 23 SEM, (range of 11–446 μm), which is at the level of the resolution of the image data (voxel size = 195 μm). The size and location of the MnCl2 injections were analyzed by 3-D combined visualization of the MR based representation of the brain surface and the atlas representations of different cerebrocortical areas (Figures 4E and 4H). The position of Mn2+ signal enhancement within the thalamus was analyzed by slicing the combined image and atlas volumes in 200 μm thick slices using the m3d tool (Figures 5D, 5E, 5I, and 5J).
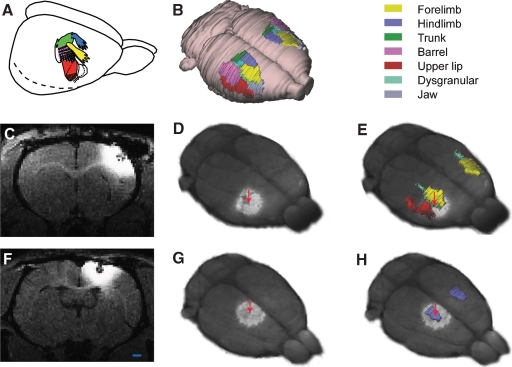
Figure 4.
MRI and 3-D atlas based localization of MnCl2 injection sites in the cerebral cortex. (A) Color coded drawing of major body representations in the primary somatosensory cortex (SI), modified from Welker (1971), with permission. (B) 3-D reconstructed atlas representation of the brain surface with SI regions indicated in corresponding colors. (C–E) and (F–H) show two different examples, in which MnCl2 was stereotaxically aimed at the forelimb or hindlimb representations of SI, respectively. (C, F) Raw T1-weighted MRI images acquired 9 hours after focal application of MnCl2. At the point of injection a central zone of low signal intensity is visible, surrounded by a halo of bright signal enhancement. (D, GD, G) 3-D reconstructed MRI volume registered to atlas space, showing Mn2+ signal enhancement surrounding the injection sites (arrows). (E, H) The same images combined with atlas reconstructions of relevant SI body representations. Comparison of injection site locations with the atlas surface reconstructions revealed that one injection is centered on the dysgranular zone lateral to the SI forelimb representation, while the other injection is centered on the SI hindlimb representation. Bar, 1 mm.
Figure 5.
Corticothalamic topographical organization revealed by MnCl2/MRI tracing and 3-D atlasing. Two cases are shown, case 1 (A–E), in which MnCl2 was injected in the dysgranular zone adjacent to the SI forelimb representation (A), and case 2 (F–J), in which MnCl2 was injected in the hindlimb representation of SI. (B, C and G, H) Raw coronal and sagittal T1-weighted MRI images obtained 9 hours after injection. (D, E and I, J) Corresponding images with overlay of selected reconstructed atlas structures. (K, L) Superimposed, pseudocolored images (case 1, yellow; case 2, blue)with an overlay of atlas derived boundary lines, demonstrating a somatotopical organization of the different signals within the Po and VPM. ic, internal capsule; Po, posterior thalamic nucleus; VPM, ventral posterolateral thalamic nucleus; VPL, ventral posteromedial thalamic nucleus; Rt, reticular thalamic nucleus. Bar, 1 mm.
PET experiments
PET data were acquired at the Small Animal Imaging Unit of the Center for Molecular Biology and Neuroscience, University of Oslo, Norway, with use of a small animal PET scanner (microPET Focus 120, Siemens Medical Solutions, Erlangen, Germany). Accompanying computer tomography (CT) data were collected with a small animal CT scanner (microCAT, Siemens Medical Solutions, Erlangen, Germany). The microPET scanner provides high sensitivity (6.5%) and high spatial resolution (<1.3 mm; Kim et al., 2007). A multi-modality animal bed was used to collect data with both the PET and CT scanner without re-positioning the animal.
Three different kinds of PET-data were imported to the 3-D atlas tool: individual dopamine D2/D3 receptor binding data acquired using [18F]-fallypride, individual brain and glioma tumor glucose metabolism data acquired using [18F]-fluorodeoxyglucose (FDG), and statistical parametric mapping (SPM) group data collected from a study of brain activity during nociceptive conditioning using FDG.
Individual PET receptor binding data
SD rats (National Lab Animal Center, Oslo, Norway) weighting 200–250 g, were anesthetized by inhalation of isoflurane 2% (Abbott Laboratories, Illinois, USA). 20 MBq [18F]-labeled fallypride (Department of Chemistry, University of Oslo, Norway) was injected intravenously, and the rats were scanned for 180 minutes. Data were collected in list mode and reconstructed using 3-D OSEM-MAP (Jinyi and Leahy, 2000; Jinyi et al., 1998) (2 OSEM iterations, 18 MAP iterations, β = 0.5, 128 × 128 × 95 matrix size, 0.87 × 0.87 × 0.80 mm3 voxel size). Two image volumes were extracted from the acquired scans: a sum image of the first 5 minutes of the scan reflecting tracer delivery (“perfusion”) and a late scan sum image (60–180 minutes) representing the D2/D3 receptor binding of [18F]-fallypride. Data were imported and globally aligned to the atlas space in m3d on basis of the “perfusion” image showing the external boundaries of the brain, as well as the receptor binding image, showing a negative image of the brain surface (Figures 6A–6D).
Figure 6.
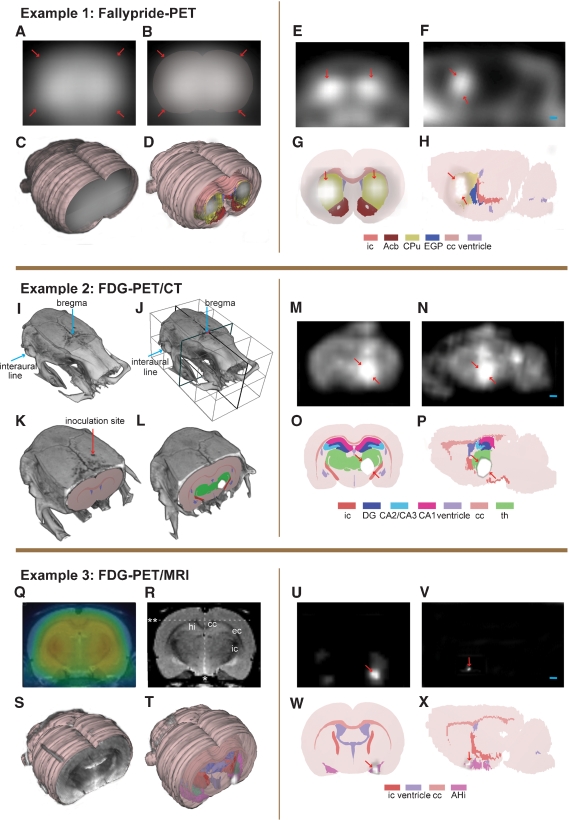
Alignment and localization of PET data. Example 1: (A–C) perfusion image of [18F]-fallypride image was used for alignment to 3-D atlas space. Arrows indicating surface boundaries. (D) Receptor binding image co-registered with the 3-D atlas model. (E, F) [18F]-fallypride-PET coronal and sagittal images showing high-uptake regions. (G, H) Coronal and sagittal atlas sections localizing the high-uptake PET signals to the basal ganglia. Example 2: (I, J) Bregma and the interaural line in the CT volume. (K) CT volume aligned with the 3-D atlas model. (L) FDG-PET signal aligned with the 3-D atlas on the basis of the same transformation matrix as for the CT volume. (M, N) Coronal and sagittal PET images showing high-uptake of FDG in the brain tumor. (O, P) Coronal and sagittal atlas diagrams locating the brain tumor in the right thalamic region. Example 3: (Q) co-registered FDG-PET image (colors)and MRI template. (R, S) MRI template aligned to the 3-D atlas space on the basis of defined landmarks. *, midline; **, upper boundary of cc. (T) SPM result volume aligned with atlas space using the transformation matrix obtained from the MRI alignment. (U, V) Coronal and sagittal SPM result images showing statistical significant voxels (p < 0.05). (W, X) Coronal and sagittal atlas diagrams locating the significant signal to the rostral part of the amygdala. ic, internal capsule; Acb, accumbens nucleus; CPu, caudate putamen; EGP, external globus pallidus; cc, corpus callosum; DG, dentate gyrus; CA2/CA3, field CA2 and CA3 of the hippocampus; CA1, field CA1 of the hippocampus; th, thalamus, whole region; hi, hippocampal region; ec, external capsule; AHi, amygdalohippocampal area. Arrow, indicate boundaries of regions with increased PET signal. Bar, 1 mm.
Individual FDG-PET and CT data
Inbred BDIX rats (body weight > 220 g; Harlan Olac Ltd. Bicester, UK) were used. The animals were anesthetized by subcutaneous injection (2 ml/kg) of a mixture of equal volumes of Hypnorm (fentanyl citrate 5 mg/mL, Janssen L.P., High Wycombe, UK) and Dormicum (midazolam 5 mg/mL, Hoffmann-La Roche Ltd., Basel, Switzerland). Fifteen days prior to the FDG-PET scan, 50.000 glioma cells (BT4Ce gliomasarcoma cell line) were inoculated. The inoculation was performed with the animal fixed in a stereotaxic frame and injected with a 30G blunt needle through a 1.0 mm burrhole. FDG (GE Healthcare AS, Oslo, Norway) was injected intravenously and the animals were scanned for 60 minutes. Attenuation correction was obtained by a 10 minute transmission scan with a 68Ge point source prior to injection. Data were collected in list mode and reconstructed using 3-D OSEM-MAP (2 OSEM iterations, 18 MAP iterations, β = 0.5, 128 × 128 × 95 matrix size, 0.87 × 0.87 × 0.80 mm3 voxel size). The animals were then transferred to the CT scanner utilizing the multi-modality bed ensuring the same position of the head. CT images were acquired using the same settings for all studies (80 kVp, 0.5 mA, 400 ms, 360 degrees rotation). Data were reconstructed using Feldkamp reconstruction (512 × 512 × 512 matrix size, 0.1 × 0.1 × 0.1 mm3 voxel size). The FDG-PET and CT data were co-registered in PMOD, re-slicing the resulting PET image to match the matrix and resolution of the CT volume. The CT data were imported into m3d and linearly fitted to atlas stereotaxic space on basis of the readily identifiable bregma and interaural line (Figures 6I and 6J). The same transformation parameters as for the CT data were used to fit the FDG-PET data set into exactly the same atlas space (Figure 6L).
SPM group results
Twenty female SD (National Lab Animal Center, Oslo, Norway) rats weighing 200–250 g, were adapted for minimum 10 days to the environment and given food and water ad libitum. For the experiments, the animals were anesthetized by inhalation of 2% isoflurane (Abbott Laboratories, Illinois, USA) and divided into two groups; nociceptive stimulation condition (n = 10) and sham (n = 10). In addition, all animals were scanned at baseline. FDG (20–25 MBq; GE Healthcare AS, Oslo, Norway) was injected through the tail vein and all animals were scanned for 60 minutes. Attenuation correction was obtained by a 10 minutes transmission scan with a 68Ge point source prior to FDG injection. Data were collected in list mode and dynamically reconstructed using 3-D OSEM-MAP (2 OSEM iterations, 18 MAP iterations, β = 0.1, 128 × 128 × 95 matrix size, 0.87 × 0.87 × 0.80 mm3 voxel size). Statistical comparisons (paired t-test stimulation-baseline) between the two groups were performed with SPM5 (Statistical Parametric Mapping, http://fil.ion.ucl.ac.uk/spm) after intra-subject co-registration and spatial normalization to a MRI template fitted to Paxinos and Watson atlas space (Schweinhardt et al., 2003; Figure 6Q). Significance was accepted at the 5% level. Localization of statistical significant activity was defined by the use of the 3-D atlas model. The MRI template was aligned with the atlas model in m3d (Figures 6R and 6S) by the means of identifiable anatomical landmarks (outer boundaries of the brain surface, gray and white matter contrast, genu and splenium of the corpus callosum, outer boundaries of the hippocampus and caudate putamen, and the ventricle system). Since spatial normalization between the PET data and the MRI template was already performed in SPM5, the same linear transformation obtained from the alignment could be applied to the PET data (Figure 6T).
Results
We have developed a procedure for 3-D reconstruction of a series of 2-D diagrams from conventional brain atlases. The 3-D reconstructed atlases are displayed and utilized in a multi-platform tool (m3d), prepared to include experimental data of different formats (i.e., JPEG, PNG, VRML, and NIfTI). The focus of the present report is to exemplify how a relatively straightforward atlas approach may be useful for the neuroanatomical analysis in the context of import of whole brain MRI and PET data.
The 3-D rat brain atlas currently includes 60 structures, while 30 structures have been incorporated in the 3-D mouse brain atlas. The structures included are the outer boundaries of the brain and areas, zones and nuclei of the cerebral cortex, hippocampus, basal ganglia, thalamus, amygdala, as well as major fiber tracts (Figures 2A, 2C, 2E, and 2G). The m3d atlas visualization tool allows interactive inspection of selected 3-D reconstructed structures as well as user-defined slicing to create customized 2-D atlas diagrams at any position and angle of orientation. This feature is particularly suitable for comparison with experimental data cut at angles different from the standard coronal, sagittal, or horizontal orientations used in printed brain atlases (Figures 3E–3H).
The current version of the m3d tool allows import of image data and subsequent registration of images to atlas space by interactive scaling, rotation, and translation on basis of recognizable landmarks. Depending on the modality of the experimental data, the alignment can be performed globally for the entire whole brain data set, using the outer boundaries of the brain as a template, and/or locally for parts of the whole brain data set, utilizing smaller readily identifiable landmarks in the brain. In addition, the quality of the registration can be enhanced by a step-by-step alignment of the atlas and image data with use of slicing at different locations and angles. In m3d, co-registered image and atlas representations can be co-visualized as 3-D objects and in 2-D custom defined slices.
In the following, we exemplify the use of the 3-D atlas for assigning location to MRI and PET image volumes.
MRI example
To illustrate the use of the 3-D atlas to assign location to discrete structural signals observed with MRI, we here describe two experiments in which high-resolution in vivo MnCl2/MRI pathway tracing (see e.g., Leergaard et al., 2003; Pautler et al., 1998; Saleem et al., 2002) was used to visualize specific corticothalamic connections, and 3-D atlasing was used to assign location to tracer injection sites and observed signal enhancement. To evaluate the position of injection sites and the corticothalamic signal enhancement, the two MR image volumes were registered to atlas space on basis of multiple recognizable anatomical landmarks and affine transformations.
In the two example experiments, MnCl2 was injected on basis of stereotaxic coordinates, respectively aimed at the forelimb or hindlimb representations of the primary somatosensory cortex (SI). When the animals were scanned ∼9 hours after the injection, injection sites were visible in the T1-weighted images as bright regions, surrounding a core with low signal intensity, presumably associated with high concentration of Mn2+ (Figures 4C and 4F; see also discussion in Leergaard et al., 2003). Mn2+ signal enhancement was observed at multiple cortical and sub-cortical locations, as reported in Leergaard et al. (2003). Discrete Mn2+ signal enhancement was observed in the internal capsule (ic) and at multiple sites within the thalamus (Figures 5B, 5C, 5G, and 5H).
Co-visualization in m3d of the co-registered MR image volumes and atlas reconstructions of SI body representations (Figures 4D–4H) demonstrated that the injection site center in one example was located slightly outside the targeted forelimb representation (Figure 4E), and in the other example that the injection site center was located centrally in the hindlimb representation (Figure 4F). Identical results were obtained with use of manual measurements of the mediolateral and anterioposterior distances from the injection site centers to the midline and the genu and splenium of the corpus callosum in the MR images. The manual measurements were converted to stereotaxic coordinates using the printed atlas of Paxinos and Watson (2005), and compared to known functional maps of SI (Chapin and Lin, 1984).
To evaluate the location of Mn2+ signal enhancement within the thalamus, series of contiguous 200 μm thick coronal and sagittal slices were obtained from the combined atlas and MRI volume. Overlay images of relevant atlas structures (thalamic sub-nuclei and the internal capsule) and MR images allowed atlas based anatomical segmentation of the MR images (Figure 5). Thus, aided by the atlas overlay, it was possible to distinguish Mn2+ signal enhancement located in the internal capsule and within the posterior complex thalamus (Po) and the ventral posterolateral nucleus thalamus (VPL) in both examples. Combined analysis of the two data sets further allowed assessment of topographical organization at a finer scale (Figures 5K and 5L). This analysis clearly demonstrated that the discrete clusters of signal enhancement where primarily distributed within the boundaries of Po and VPL (Figure 5K), with an inside-out relationship corresponding to the well known internal to external somatotopic distribution of hindlimb and forelimb representations in these thalamic sub-nuclei (Fabri and Burton, 1991). Signal enhancement was also seen in the reticular thalamic nucleus (Figure 5K), presumably representing pathways en route to the Po and VPL. Furthermore, in our first example (Figures 5B–5E) some signal enhancement was also observed in the ventral posteromedial thalamic nucleus (VPM), known to hold representations of the face (Fabri and Burton, 1991). This finding is presumably related to the location of the injection site center in the dysgranular zone between the forelimb and face representations in SI. We conclude that the present 3-D atlas approach provided an anatomical segmentation of the observed signal enhancement sufficiently accurate to resolve finer details of the well-known topographical organization found within sub-nuclei of the thalamus (see e.g., Deschenes et al., 1998; Fabri and Burton, 1991; Price, 1995).
PET examples
To illustrate the use of the 3-D atlas to assign location to PET data (Figure 6), we have used three different sets of experimental data.
Individual PET receptor binding data
The [18F]-fallypride data set represents the D2/D3-receptor binding in the brain. The receptor binding image itself contains little anatomical information and could not be aligned to atlas space (Figures 6E and 6F). “Perfusion” images of early frames representing the initial hematogeneous tracer distribution were used (Figures 6A–6C) to facilitate alignment, since these data show the external contours of the cerebrum and cerebellum, as well to a certain degree the outlining of the basal ganglia using different levels of voxel intensity scaling. The same transformation parameters were then used to fit the receptor binding image into the atlas space in m3d (Figure 6D). Visualization of the receptor binding showed the highest density of dopamine D2/D3 receptors to be present in the basal ganglia (Figures 6G and 6H). Only a course alignment of the experimental data with the atlas was possible due to limited structural information. A further judgment of details of distribution was not deemed possible with the present approach.
Individual FDG-PET and CT data
FDG is a glucose analog and FDG-PET images represent the rate of glucose metabolism of the cells. Cancer cells mostly utilize more glucose than healthy tissue and tumors can therefore be located as regions with high uptake of FDG in a PET scan (Figures 6M and 6N). To be able to decide the size, extent, and location of the brain tumor, the co-registered CT image was imported and aligned to the 3-D atlas space in m3d using the location of bregma and the interaural line which could be readily defined in the CT volume (Figures 6I and 6J). The inoculation site for the glioma cells is shown in Figure 6 K. The same transformation matrix was applied to the FDG-PET volume in order to fit the data to atlas space (Figure 6L). The increased FDG signal defining the brain tumor was located in the right thalamic region. The quality of the alignment procedure was in this case considerably enhanced by utilizing CT as a complementary imaging modality. The readily identifiable bregma and the interaural line provided concrete landmarks which could be aligned in 3-D atlas space. Inaccuracies can occur due to inter-subject variations in brain shape and size, and due to the fusion procedure of the PET and CT data prior to import into m3d.
SPM group results
SPM delivers a result volume (cluster of voxels) at a specified statistical significance value (Figures 6U and 6V). This parametric volume was already fitted to the space of a defined anatomic template. The MRI template was imported and aligned to the 3-D atlas space in m3d by the aim of well-defined landmarks, i.e., white matter and the contours of the brain (Figures 6R and 6S) The same parameters were applied to the significant parametric volume in order to fit the data to the atlas space (Figure 6T). The nociceptive conditioning is an aversive and stressful event, in which amygdala play a crucial role (Sah et al., 2003). After alignment, a significant effect of nociceptive conditioning could be localized to the rostral volume of the amygdala (Figures 6W and 6X). The alignment of the MRI template to the 3-D atlas space was performed using a best-fit approach based on large brain regions. Due to variations in the rat brain shape and size, a perfect fit is difficult to obtain. If focusing on specific areas within the brain, the transformation parameters might be altered using smaller local sub-regions as alignment landmarks. However, the resolution of microPET and averaging over several individuals has an inherent inaccuracy limiting the detailed localization and separation of smaller effects to specific, smaller sub-nuclei of any rat or even mouse brain structure. Once the MRI-template has been sufficiently aligned to a 3-D atlas, the same transformation routine and result reading can be established for all data processed with statistical parametric mapping. The 3-D atlas makes the data processing and interpretation less observer dependent. As shown in Figures 6T–6X, the 3-D atlas ensures a sufficiently precise allocation of a statistical significant result to a brain area in relation to neighboring structures.
Discussion
We present a multi-platform tool that provides a new dynamic and analytical environment for comparing experimental image data to a seminal atlas reconstructed in 3-D. Our 3-D reconstruction procedure was applied to two commonly used rodent brain atlases, but could be applied to other atlases as long as they provide diagrams from sections with limited distortions, mapped into a stereotaxic coordinate system. We further demonstrate how the atlas tool can be used to align 3-D data of different origin (PET, PET/CT, and MRI) to the same atlas space, facilitating analysis of localization in the experimental data.
In the example analyses provided, variable amounts of information was available to assist the user with the registration of the experimental data to the 3-D atlas space. The most limited structural information was present in the receptor binding PET data. The global registration performed for these data, based on the outer contours of the brain visible in the early frames recorded, was nevertheless sufficient to identify the caudate-putamen as the region containing the highest [18F]-fallypride receptor binding, in agreement with legacy data (Mukherjee et al., 1995). The acquisition of CT images in register with the experimental PET images facilitated the atlas registration considerably, since skull landmarks (readily detected with CT) could be directly mapped to the atlas coordinate space (which is based on the same skull landmarks). Use of MRI in combination with PET is an emerging new approach (Judenhofer et al., 2007). In our analyses, we demonstrate an indirect MRI-based co-registration procedure. With this procedure, the data sets were aligned to a common MRI template, which in turn was registered to the 3-D atlas. In this example, the location of the statistically significant increased activity was found in the expected location in the brain, in our case the amygdala following a nociceptive conditioning (Sah et al., 2003). Finally, direct import of experimental MRI data can be done with a high accuracy since a range of landmarks in the brain are visible in the MRI data. Our high-resolution MnCl2/MRI examples demonstrate the mapping of fine-scale topographical organization within thalamic sub-nuclei (see also Leergaard et al., 2003), corresponding with previous maps of the thalamus (Fabri and Burton, 1991).
The general validity of the linear co-registration approach used in the present study is indirectly demonstrated by the replication of known findings in the shown examples. For examples, in the FGD-PET/MRI example, the majority of the observed PET signal is largely confined to the rostral part of the amygdala, in accordance with Sah et al. (2003). In the MnCl2/MRI example, in which several anatomical landmarks were seen in the MR images, the mean spatial displacement between anatomical landmarks in atlas and image volume was found to be less than the size of one voxel. This correspondence is at a level sufficient to study topographical organization within sub-nuclei. The major displacement between atlas and images in this example, not corrected for in our approach, was found in the hindbrain. This displacement appears to be due to a dislocation of the hindbrain occurring at the time of extraction of the brain from the skull. Further analyses of the hindbrain would therefore require a second step of registration in which the hindbrain is treated in isolation from the remaining part of the brain. Adjustment for size variation alone will clearly facilitate comparison of brain stem data from different rat brains (Brevik et al., 2001; Leergaard et al., 2000). Thus, for tomographic material of the quality here exemplified, our linear procedures appear to provide sufficient accuracy to resolve most questions related to localization of data and comparison between animals. More optimized co-registration of multiple detailed anatomical landmarks derived from histological images (Lein et al., 2007; Toga et al., 1995) or high-resolution structural MRI (Ali et al., 2005; Badea et al., 2007; Benveniste et al., 2000; Ma et al., 2005) would require the use of non-linear methods.
An important feature of the 3-D atlas tool provided is the ability to co-register image data to an atlas framework. The m3d tool supports the import of voxel data in NIfTI file format, which allows import of volume data such as PET, CT, and MRI. Future developments will include other data formats. As discussed above, several approaches can be employed for aligning image data to atlas space. In m3d, image data can be linearly transformed (i.e., transformed with rotation, scaling, and translation) in order to fit the 3-D atlas space. The transformation matrix is calculated based on identifiable landmarks in the image data. A PET image volume typically lacks well-defined structural information which may complicate the alignment. The quality of the co-registration may be enhanced when the researcher has access to other modalities such as MRI and/or CT. The alignment of an MRI volume is based on anatomical landmarks such as the outer boundaries of the brain and white matter. In a CT volume, the definition of bregma and the interaural line provides a rigid aid in calculating the transformation matrix. Since the present approach is based on linear transformations only, local alignment errors may occur due to inter-subject variations in rodent brain shape and size. High-resolution MRI based probabilistic atlasing of the mouse brain indicate that this variation is relatively low (average structure volume variation coefficient in the order of 7%; Badea et al., 2007; Ma et al., 2005). Given that such variations may be partly compensated for by linear transformations, the need to adjust for remaining variability must be viewed in relation to the resolution employed in the tomographic image volumes to be analyzed.
The present approach, with a common analytical environment for experimental data and 3-D atlas reconstructions, not only facilitates direct assignment of anatomical location, but may also be useful in the context of generating probabilistic representations of the brain. Probabilistic maps are typically based on manually segmented cases, but may also incorporate information from reconstructed atlases. Co-registration of manually segmented cases into a common atlas space would thus enable the construction of a probabilistic atlas, containing a priori probabilities of various anatomical structures at each location in atlas space. Such a probabilistic atlas, in combination with the image contrast properties of different structures, can in turn be used to automate the process of anatomical segmentation (Ali et al., 2005; Fischl et al., 2002).
A major advantage of reconstructing standard stereotaxic atlases (rather than constructing new customized atlases) is the wide recognition and use, and the high-resolution and comprehensive nomenclature, of these atlases. Similarly, for human brain imaging data, the frequently used reference atlas space (Talairach and Tournoux, 1988) has been made available in many different formats in order to have a common reference space in the scientific community. Digital versions of the human brain atlas exist in three orthogonal planes with a high-speed database server for querying and retrieving data about human brain structures over the internet (ric.uthscsa.edu/projects/talairachdaemon.html). The present application of the 3-D atlases of the rat and the mouse brain makes it simpler and more reliable to use these atlases in combination with 3-D imaging modalities, and to bring data from different modalities into the same environment. A further challenge would be to bring different tomographic data modalities together with primarily 2-D histological or other section-based data, such as optical imaging data and autoradiography. At present, the skull-based stereotaxic coordinate system of Paxinos and Watson (1982) would seem to be a suitable common reference space.
Tools availability
Information on the availability of tools and atlas data can be found at The Rodent Brain Workbench (http://rbwb.org).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Acknowledgments
Funded by grants from The Research Council of Norway to J. G. B., T. B. L., and F. W., and grants from the NIH (R01-EB00790, U24-RR021382) to A. M. D. We thank Bent Schoultz and Gjermund Henriksen, Department of Chemistry, University of Oslo, for synthesis of [18F]-fallypride, Dag R. Sorensen and Hong Qu for expert technical assistance related to PET/CT scanning, Anna Devor and Andre van der Kouwe for expert technical assistance related to MnCl2/MRI tracing, and Johan F. Storm for valuable discussions and facilitation of the present project.
References
- Ali A. A., Dale A. M., Badea A., Johnson G. A. (2005). Automated segmentation of neuroanatomical structures in multispectral MR microscopy of the mouse brain. Neuroimage 27, 425–435 [DOI] [PubMed] [Google Scholar]
- Badea A., li-Sharief A. A., Johnson G. A. (2007). Morphometric analysis of the C57BL/6J mouse brain. Neuroimage 37, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste H., Kim K., Zhang L., Johnson G. A. (2000). Magnetic resonance microscopy of the C57BL mouse brain. Neuroimage 11, 601–611 [DOI] [PubMed] [Google Scholar]
- Boline J. K., MacKenzie-Graham A. A., Shattuck D. W., Yuan H., Anderson S. P., Sforza D. M., Wang J., Williams R. W., Wong W., Martone M. E., Zaslavsky I., Toga A. W.A Digital Atlas and Neuroinformatics Framework for Query and Display of Disparate Data. (2006). Society for Neuroscience 2006. Abstract and itinerary viewer. Program No. 100. 12 [Google Scholar]
- Brevik A., Leergaard T. B., Svanevik. M., Bjaalie J. G. (2001). Three dimensional computerised atlas of the rat brain stem precerebellar system: approaches for mapping, visualization, and comparison of spatial distribution data. Anat. Embryol. (Berl.) 204, 319–332 [DOI] [PubMed] [Google Scholar]
- Chan E., Kovacevic N., Ho S. K. Y., Henkelman R. M., Henderson J. T. (2007). Development of a high resolution three-dimensional surgical atlas of the murine head for strains 129S1/SvImJ and C57Bl/6J using magnetic resonance imaging and micro-computed tomography. Neuroscience 144, 604–615 [DOI] [PubMed] [Google Scholar]
- Chapin J. K., Lin C. S. (1984). Mapping the body representation in the SI cortex of anesthetized and awake rats. J. Comp. Neurol. 229, 199–213 [DOI] [PubMed] [Google Scholar]
- Deschenes M., Veinante P., Zhang Z. W. (1998). The organization of corticothalamic projections: reciprocity versus parity. Brain Res. Rev. 28, 286–308 [DOI] [PubMed] [Google Scholar]
- Fabri M., Burton H. (1991). Topography of connections between primary somatosensory cortex and posterior complex in rat: a multiple fluorescent tracer study. Brain Res. 538, 351–357 [DOI] [PubMed] [Google Scholar]
- Field K. J., White W. J., Lang C. M. (1993). Anaesthetic effects of chloral hydrate, pentobarbitone and urethane in adult male rats. Lab. Anim. 27, 258–269 [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D. H., Busa E., Albert M., Dieterich M., Haselgrove C., van der K. A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A. M. (2002). Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 [DOI] [PubMed] [Google Scholar]
- Gustafson C., Tretiak O., Bertrand L., Nissanov J. (2004). Design and implementation of software for assembly and browsing of 3D brain atlases. Comput. Methods Programs Biomed. 74, 53–61 [DOI] [PubMed] [Google Scholar]
- Gustafson C., Bug W. L., Nissanov J. (2007). Neuroterrain—a client-server system for browsing 3D biomedical image data sets. BMC Bioinformatics 8, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinyi Q., Leahy R. M. (2000). Resolution and noise properties of MAP reconstruction for fully 3-D PET. Med. Imaging, IEEE Trans. 19, 493–506 [DOI] [PubMed] [Google Scholar]
- Jinyi Q., Leahy R. M., Chinghan H., Farquhar T. H., Cherry S. R. (1998). Fully 3D Bayesian image reconstruction for the ECAT EXACT HR+. Nucl. Sci., IEEE Trans. 45, 1096–1103 [Google Scholar]
- Judenhofer M. S., Catana C., Swann B. K., Siegel S. B., Jung W. I., Nutt R. E., Cherry S. R., Claussen C. D., Pichler B. J. (2007). PET/MR images acquired with a compact MR-compatible PET detector in a 7-T Magnet. Radiology 244, 807–814 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lee J. S., Im K. C., Kim S. J., Kim S. Y., Lee D. S., Moon D. H. (2007). Performance measurement of the microPET Focus 120 Scanner. J. Nucl. Med., in press. [DOI] [PubMed] [Google Scholar]
- Leergaard T. B., Lyngstad K. A., Thompson J. H., Taeymans S., Vos B. P., De Schutter E., Bower J. M., Bjaalie J. G. (2000). Rat somatosensory cerebropontocerebellar pathways: spatial relationships of the somatotopic map of the primary somatosensory cortex are preserved in a three-dimensional clustered pontine map. J. Comp. Neurol. 422, 246–266 [DOI] [PubMed] [Google Scholar]
- Leergaard T. B., Bjaalie J. G., Devor A., Wald L. L., Dale A. M. (2003). In vivo tracing of major rat brain pathways using manganese-enhanced magnetic resonance imaging and three-dimensional digital atlasing. Neuroimage 20, 1591–1600 [DOI] [PubMed] [Google Scholar]
- Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chi Chin M., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H. W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K. R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Feng Yuan X., Zhang B., Zwingman T. A., Jones A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- Ma Y., Hof P. R., Grant S. C., Blackband S. J., Bennett R., Slatest L., Mcguigan S. J., Benveniste H. (2005). A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience 135, 1203–1215 [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A., Lee E. F., Dinov I. D., Bota M., Shattuck D. W., Ruffins S., Yuan H., Konstantinidis F., Pitiot A., Ding Y., Hu G., Jacobs R. E., Toga A. W. (2004). A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 204, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J., Yang Z. Y., Das M. K., Brown T. (1995). Fluorinated benzamide neuroleptics–III. Development of (S)-N-[(1-allyl-2-pyrrolidinyl)methyl]-5-(3-[18F]fluoropropyl)-2, 3-dimethoxybenzamide as an improved dopamine D-2 receptor tracer. Nucl. Med. Biol. 22, 283–296 [DOI] [PubMed] [Google Scholar]
- Palombi O., Shin J., Watson C., Paxinos G. (2006). Neuroanatomical affiliation visualization-interface system. Neuroinformatics 4, 299–318 [DOI] [PubMed] [Google Scholar]
- Pautler R. G., Silva A. C., Koretsky A. P. (1998). In vivo neuronal tract tracing using manganese-enhanced magnetic resonance imaging. Magn. Reson. Med. 40, 740–748 [DOI] [PubMed] [Google Scholar]
- Paxinos G., Franklin K. B. (2001). The mouse brain in stereotaxic coordinates (San Diego, Elsevier Academic Press; ). [Google Scholar]
- Paxinos G., Watson C. (1982). The rat brain in stereotaxic coordinates (New York, Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Paxinos G., Watson C. (1998). The rat brain in stereotaxic coordinates (San Diego, Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Paxinos G., Watson C. (2005). The rat brain in stereotaxic coordinates (San Diego, Elsevier Academic Press; ). [Google Scholar]
- Price J. L. (1995). Thalamus. In The Rat Nervous System, Paxinos G., ed. (San Diego, Academic Press; ), pp. 629–648 [Google Scholar]
- Sah P., Faber E. S., De Lopez A. M., Power J. (2003). The amygdaloid complex: anatomy and physiology. Physiol. Rev. 83, 803–834 [DOI] [PubMed] [Google Scholar]
- Saleem K. S., Pauls J. M., Augath M., Trinath T., Prause B. A., Hashikawa T., Logothetis N. K. (2002). Magnetic resonance imaging of neuronal connections in the macaque monkey. Neuron 34, 685–700 [DOI] [PubMed] [Google Scholar]
- Schweinhardt P., Fransson P., Olson L., Spenger C., Andersson J. L. R. (2003). A template for spatial normalisation of MR images of the rat brain. J. Neurosci. Methods 129, 105–113 [DOI] [PubMed] [Google Scholar]
- Swanson L. W. (2004). Brain maps: Structure of the rat brain (San Diego, Elsevier Academic Press; ). [Google Scholar]
- Talairach J., Tournoux P. (1988). Co-planar stereotaxic atlas of the human brain (New York, Thieme Medical Publishers, Inc.). [Google Scholar]
- Toga A. W., Santori E. M., Hazani R., Ambach K. (1995). A 3D digital map of rat brain. Brain Res. Bull. 38, 77–85 [DOI] [PubMed] [Google Scholar]
- Welker C. (1971). Microelectrode delineation of fine grain somatotopic organization of (SmI) cerebral neocortex in albino rat. Brain Res. 26, 259–275 [PubMed] [Google Scholar]