Abstract
The complexity of the nervous system requires high-resolution microscopy to resolve the detailed 3D structure of nerve cells and supracellular domains. The analysis of such imaging data to extract cellular surfaces and cell components often requires the combination of expert human knowledge with carefully engineered software tools. In an effort to make better tools to assist humans in this endeavor, create a more accessible and permanent record of their data, and to aid the process of constructing complex and detailed computational models, we have created a core of formalized knowledge about the structure of the nervous system and have integrated that core into several software applications. In this paper, we describe the structure and content of a formal ontology whose scope is the subcellular anatomy of the nervous system (SAO), covering nerve cells, their parts, and interactions between these parts. Many applications of this ontology to image annotation, content-based retrieval of structural data, and integration of shared data across scales and researchers are also described.
Keywords: neuroanatomy, neuronal cell types, electron microscopy, subcellular anatomy, data integration
Introduction
In neuroscience, scientifically relevant complexity occurs at every spatial and temporal scale that is currently open to examination. Unfortunately, our current complement of experimental and analytical techniques generally locks an investigation into a very limited dimensional range, leading to a fragmented and incomplete view of nervous systems across scales. This fundamental “multiscale problem” of neuroscience is, at its core, a problem of information integration. One indication of the extreme difficulty of information integration in the neurosciences is the conspicuous lack of any widely practiced automated methods for integrating information among major classes of neuroscientific data: structural, functional, and behavioral. Many tools have been developed to provide infrastructure to organize and analyze brain data, resulting in large part from the Human Brain Project, funded through the US National Institutes of Health (Huerta et al., 1993; Koslow and Huerta, 1997). Such tools have included databases for storing primary data (e.g., CCDB; Martone et al., 2003, WebQTL; Wang et al., 2003, etc.), knowledge bases for derived information (e.g., BAMS; Bota et al., 2005 and CoCoMac; Stephan et al., 2001), tools for performing novel analyses of brain data and mining the literature (e.g., Textpresso; Muller et al., 2004). However, the integration of diverse types of information still occurs largely through the efforts of individuals who examine the data and construct the necessary bridges between different data based on their knowledge of neuroscience.
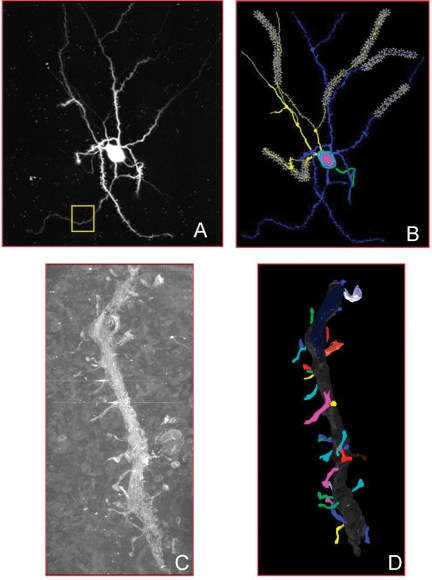
The grand challenge of neuroinformatics is the creation of systems that seamlessly integrate data across spatial and temporal scales such that information, for example, about white matter bundles derived from diffusion tensor imaging can be analyzed in context with electrophysiological data recorded from the neurons whose axons make up the bundles. The difficulties in performing this type of integration from data alone is illustrated in Figure 1, which shows an intracellularly injected medium spiny neuron from the mouse nucleus accumbens, imaged using correlated light and electron microscopy. At each level, different types of visualization and analytical tools are applied to extract meaningful content, for example, the branching structure of the dendritic tree, the surface area of dendritic spines. The knowledge required to richly inter-relate these different data representations and analytical results, however, largely resides in the domain scientists with specific, detailed understanding of the links between the various data types and the biological objects from which they derive.
Figure 1.
Multiple representations of the same medium spiny neuron taken from the CCDB. In (A), a light-level fill of the neuron. The yellow box shows the portion of the dendritic branch shown in (C). In (B), the Neurolucida segmentation of that neuron. In (C), the EM image of the portion of the dendrite featured in (A). In (D), the 3D reconstruction of the dendrite from (C) after segmentation.
In this paper, we describe specific steps toward creating generic information bridges by constructing a formal ontology designed to provide the knowledge necessary to integrate data acquired across multiple scales in structural neuroscience. An ontology is a formal representation of knowledge in a domain (Gruber, 1993). It defines the inter-related set of concepts representing a knowledge area and the common terms used to describe them, for example, “neuron is a cell” and “cell has part plasma membrane.” A critical aspect of modern ontologies is the encoding of these entities and relationships in a standard form where the semantics of the domain are machine interpretable using open source tools and software libraries. Ontologies are used by people, databases, and applications to share information in a semantically precise way within and across particular domains (Gruber, 1993).
The ontology for subcellular anatomy (SAO) focuses on the spatial scale that has come to be known as the “mesoscale,” roughly defined as the dimensional range encompassing macromolecular complexes, subcellular structures up to the level of cells and cellular networks. The SAO describes neurons, glia, their parts, and how these parts come together to create the dense feltwork of processes that characterizes the nervous system. The SAO was constructed through the Cell Centered Database (CCDB) project (Martone et al., 2002, 2003, 2007), an on-line resource for disseminating data derived from light and electron microscopic imaging. The CCDB project, as its name implies, takes the view that the cell should provide the rallying point for information integration in biological tissues. Thus, the SAO starts with the cell and models how cell parts, including molecules, fit into coarser levels of anatomy. This view contrasts with the approaches of many ontologies that start at the level of gross anatomy and traverse down to the level of the cell, for example, the Foundational Model of Anatomy (FMA) (Rosse and Mejino, 2003) and BAMS (Bota et al., 2005).
The SAO was built as a reference ontology with the ultimate goal of describing data, principally derived from light and electron microscopy, through the use of multiple annotation applications. It is built using the Web Ontology Language (OWL; http://www.w3.org/TR/owl-features) a W3C open standard for ontologies. Version 1.0 of the SAO was presented in Fong et al. (2007), which concentrated on the use of OWL and the associated tools for its construction. In this paper, we present an updated version (1.2) of the SAO, provide considerably greater detail on the design principles from a neuroscience point of view, describe new examples of reasoning, and describe new examples of data that are marked up using the SAO. We also briefly illustrate how it is being used as the semantic “glue” that binds together an environment of tools capable of annotating disparate types of structural data from imaging studies of the nervous system.
Materials and Methods
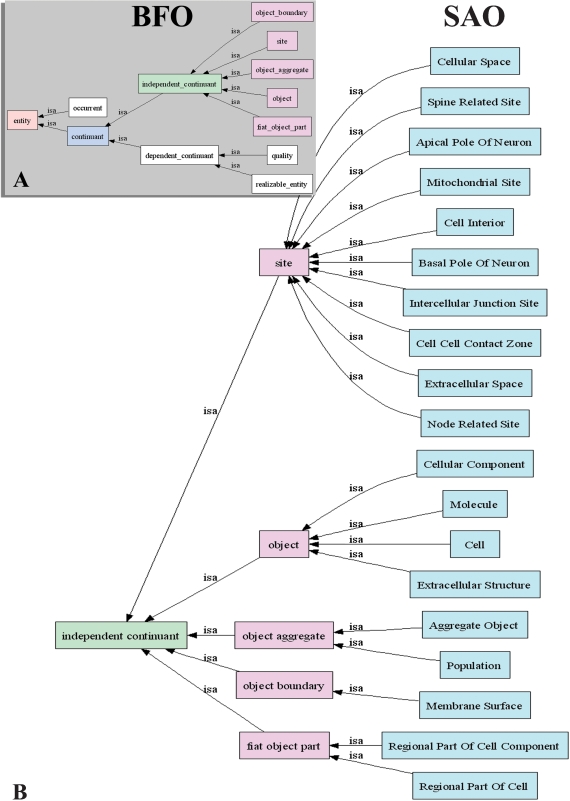
The primary source for subcellular anatomy used for the construction of the SAO was Peters et al. (1991) The Fine Structure of the Nervous System Ed 2, the standard reference for neuronal ultrastructure. Additions and modifications to this framework were also made from more recent literature. The source of each entity in the ontology is indicated as an annotation to the concept. As a way to keep epistemological distinctions clear, we adopted as an organizing framework the Basic Formal Ontology version 1.0 (BFO 1.0; Grenon, 2003) (Figure 2). The structure/function dichotomy is expressed in the BFO through the division of all possible entities into continuants (objects, qualities, sites, etc.) and occurrents (dynamic processes, temporal intervals). A continuant is an entity in the world that endures through time (Grenon et al., 2004). Examples of continuants are basic cell structures such as mitochondria and nuclei, as well as lumens and membranes. On the other hand, an occurrent refers to a process, event, activity, or change. Examples include the cell cycle phases, cell secretion, and motility. The BFO further divides continuants into dependent and independent continuants. An independent continuant is an entity that exists irrespective of its relationship to anything else, for example, cell, organism. A dependent continuant is an entity that inheres in an independent continuant, for example, color, age.
Figure 2.
High level class structure of the SAO. The BFO entities are shown in (A) and in the green and pink boxes in (B). Spatial regions, subclasses of occurrents, and subclasses of realizable entity have been omitted because the SAO does not currently use them. SAO classes that are under the BFO hierarchy are shown in blue in (B).
The SAO is available for download and browsing at (http://ccdb.ucsd.edu/SAO) and has been incorporated into the BioPortal1, a resource maintained by the National Center for Biomedical Ontologies (http://www.bioontology.org/ncbo/faces/index.xhtml). The SAO is expressed in OWL DL. OWL is a vocabulary extension of the Resource Description Framework (RDF) and is derived from the DAML + OIL OWL. Together with RDF and other components, these tools make up the growing semantic web community (Neumann and Prusak, 2007). One of the goals of the semantic web is to create tools for achieving highly interoperable data resources. The SAO was composed using Protege version 3, an open source authoring tool for OWL ontologies (Noy et al., 2001). The OWL standard is designed as a kind of description logic, which means that an application domain described in OWL is automatically described using formal logic-based semantics. One benefit of this is that tools like Protege and additional reasoning tools such as Pellet (Evren et al., 2005) and Swoop (Kalyanpur et al., 2005) can identify statements that are logically inconsistent. It also supports machine-based inferencing to generate new knowledge and to provide classification. The other major benefit is the machine-readability of OWL, which can be expressed as an XML document. This means that arbitrary software applications can take advantage of the knowledge and data that is encoded in an ontology as their underlying data model. It also means that ontologies written in OWL can be automatically imported and cross-linked by other ontologies.
An OWL ontology contains a series of classes, properties, and annotations. The classes are simply the entities that are organized in a top-down hierarchical graph structure (Figure 2A). Classes contain subclasses, for example, neuron and glia are subclasses of nerve cell. Subclasses are related to superclasses through the is a relationship, for example, neuron is a nerve cell. Properties are parts or attributes a class, for example, nucleus is a part of a cell; age is an attribute of organism. Properties are typically related to a set of classes through some form of “has a” relationship, for example, cell has part nucleus. Properties may be related to other properties through inverse, symmetric or transitive relationship, for example, is part of is the inverse of has part. Annotations are used to record metadata about the entity, for example, definitions, abbreviations, synonyms, sources of data, comments, and references. OWL allows for the placing of “restrictions” on classes, defining necessary and sufficient conditions for classification, and providing constraints on what properties need to be filled in for a given class, for example, (Neuron has regional part some Regional Part of Neuron) is a restriction that requires that a Neuron be related with the property has regional part to the class Regional Part of Neuron.
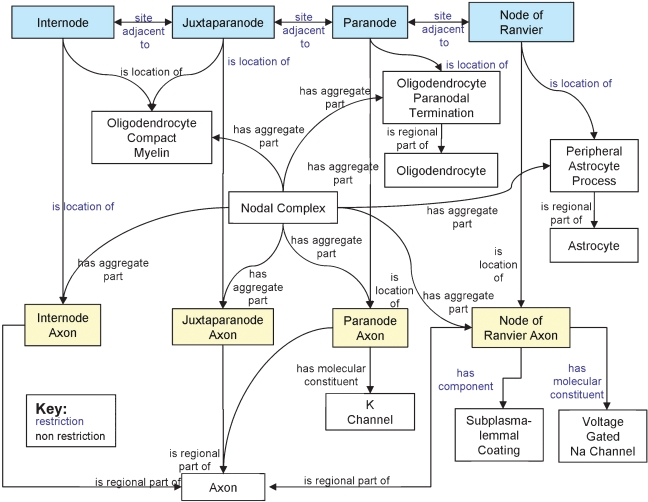
In the OWL language, all properties are first-class entities, meaning they exist independently of classes they are used to describe. Consequently, whether using properties as attributes, or as relations, the same underlying logical mechanism is invoked. Therefore, OWL properties do not have the facility to distinguish between structural properties (i.e., attributes) and relationships between classes (i.e., relations). Instead, structural properties are defined through the use of OWL restrictions, which we have used throughout the SAO. These can be seen in Figure 3, where arrows with blue text describe relationships enforced by restrictions, where arrows with black text describe relationships defined only for this particular instance.
Figure 3.
Diagram of a Node of Ranvier instance description in the SAO. The boxes indicate instances of classes that are related to one another as a description of a particular instance of a Node of Ranvier. The blue text indicates relationships that are enforced between classes through the use of OWL restrictions, while the black text indicates relationships defined for this instance alone.
In constructing the SAO, we have tried to adhere to best practices recommended by the OBO Foundry project (Smith et al., 2005). These practices include unique identifiers for each concept, re-use of existing ontologies where possible, provision of human-readable definitions that are consistent with the machine interpretable definitions encoded within the ontology. The SAO follows the principle of single inheritance as recommended by Smith et al. (2005). Single inheritance results in a is a hierarchy that is a simple tree, where children have only one parent. Through the assignation of the part of relationships, we utilize some of the features of OWL to cross-cut the is a hierarchy such that new hierarchies can be generated. Examples of this concept will be illustrated in the Results section.
For the SAO, we incorporated several existing ontologies using the owl:imports mechanism of OWL within Protégé 3. In this way, we do not reinvent content that is already substantially covered in other ontologies. The import mechanism allows wholesale incorporation of existing ontologies into the SAO while maintaining the integrity and source of the original ontology. In addition to the BFO, we imported an extensive set of annotation properties from the BIRNLex (www.nbirn.net/birnlex). Entities may be added to a merged resource, but entities may not be deleted or modified nor the class structure changed. Additional resources of relevance, for example, the cell component hierarchy from Gene Ontology, that were not encoded in OWL, were imported manually and cross referenced to the appropriate identifiers.
Results
Structure of the SAO
Classes
The high level structure of the SAO is illustrated in Figure 2B. The main classes of biological independent continuants within SAO are Cell, Regional Part of Cell, Cell Component, Extracellular Structure, and Molecule. The current version primarily covers structural entities that would be observed within the adult mammalian nervous system. Each class is assigned a unique identifier. We utilize the class identifier as the class name, but also assign a commonly used human understandable label to each class, for example, sao1224657022 corresponds to the label ‘Nerve Cell’.
Cell
We have included a set of cell types found in the nervous system (Figure S1) that include neurons and glial cells, as well as other classes of cells that one would encounter in structural studies of the nervous system, for example, vascular cells, endothelial cells, muscle cells, and macrophages. The class “Nerve Cell” contains neurons and glia, that is, cells that are derived from the neuroepithelium. We also include neuronal stem cell under this category. The SAO lists neurons (Figure S2) according to common names reflecting a mixture of classification criteria, for example, morphology (“pyramidal neuron”), proper names (“Purkinje neuron”). The SAO utilizes these names merely as labels that were assigned to cells and does not further classify cell types into subtrees based on these names, except in instances where the hierarchy is fairly straightforward, for example, layer 3 cortical pyramidal neuron is a cortical pyramidal neuron. The name chosen is meant to have meaning to a neuroscientist and not express the importance of a particular criterion for classification. In other words, we chose the label “layer 3 cortical pyramidal neuron” because we believe that there is a class of cell defined by a set of properties, not because we think its location in layer 3 is its defining characteristic. We deliberately chose to keep the cell classification flat because the SAO can be used to classify neurons along multiple dimensions according to their specific properties (see Subsection User-Defined Reclassification and Query). Rather, we have focused on providing a comprehensive model of subcellular parts and how these parts relate to the parent cell. As we discuss in a later section, we utilize the relationships between cell parts and features to infer hierarchies as they are required. The SAO organizes glial cell types (Figure S3) from a morphological perspective rather than from a strict lineage perspective. Macroglial cells include astrocytes, ependymoglial cells, oligodendrocytes, and NG2 cells, according to classifications outlined in recent literature, for example, Reichenbach and Wolberg (2005). The reference from which a particular entity was drawn is included as an annotation property for that entity.
The SAO does not aim to provide a comprehensive list of nerve cells as this domain is covered in other resources, for example, BAMS (Bota et al., 2005) and the Cell Type Ontology (Bard et al., 2005). Because the SAO is meant to be applied to data, we anticipate that users will add cell types from these resources to the SAO as they are encountered.
Part of cell
The SAO comprises two main classes of cell parts, following the structure of the FMA: regional part and component part. Regional part of cell is elaborated under the BFO concept Fiat Object Part. A fiat object part is a part of an object that possesses at least one boundary where there is no obvious physical discontinuity or landmark structure. For example, the transition between a dendrite and the cell soma has no clear boundary. Regional parts of neurons include processes, such as dendrites and axons, the cell soma and protrusions such as dendritic spines. Regional part of glia include the cell soma and glial processes such as astrocytic endfeet and myelinating processes. Each of these regional parts may in turn be further subdivided into finer parcellations. For example, dendrites are divided into trunk, that is, the primary dendrite emanating from the cell somata, branches, and terminal specializations. Component parts are considered to be independent objects and represent the building blocks common to all cells, for example, plasma membrane, mitochondrion. Components are largely drawn from the Gene Ontology cell component hierarchy (Gene Ontology Consortium, 2002), with additional neuron-specific parts such as post-synaptic density added when necessary.
Molecules
Macromolecules are also elaborated within SAO under the independent continuant class. Just as with cell types, the SAO does not contain an exhaustive list of macromolecules, because we anticipate that these entities are covered in other resources. As molecules are encountered in biological data, they may be added to the SAO. Because the SAO is designed for annotation of data, we include separate entities for the RNA, DNA, and protein forms of a molecular entity. In this way, users can capture the target of a labeling study according to the molecular species localized and assign the species to the correct subcellular compartment.
Properties
We have devised three major groups of properties in the SAO: part of, morphological and spatial relationships, again largely following the model of the FMA. Regional parts are assigned to each cell class using restrictions, for example, neurons may only have neuronal regional parts. The geometrical relationships among cell parts are specified by relationships such as continuous with, for example, dendrites are continuous with the cell somata; dendritic spines are continuous with dendrites. Thus, each regional part is assumed to belong to a parent cell. Although some properties are assigned at the level of cell class, for example, morphological type, most are assigned at the level of cell part. In this way, cell components and macromolecules are assigned to the particular part of the nerve cell in which they are found. Similarly, because nerve cells are large and may span many brain regions, the property has anatomical location, designed to situate the cell within a regional part of the nervous system, is assigned separately to each part of the cell. The SAO thus differs from most anatomical ontologies, for example, BAMS (Bota et al., 2005) where anatomical location is assigned at the level of cell class.
We have employed ‘restrictions’ within OWL to associate regional parts with the appropriate cell class. Thus, a neuron may only have regional parts of a neuron; an astrocyte may only have regional parts of an astrocyte. In contrast, component parts may be found in any cell. Although certain neuronal classes are distinguished by features such as a characteristic number of dendrites, the presence of spines or a myelinated axon, we have largely avoided creating many restrictions along these lines. Unlike gross anatomy, we usually have very few examples of a given class from which to infer these types of rules and there tends to be considerable variation within and across species of these parameters. We therefore have chosen to create a fairly generic model of a neuron in the SAO which can be used to describe individual instances of neuronal cell classes in a standard way.
The SAO places molecules within their cellular contexts through the has molecular constituent property and its inverse is molecular constituent of. This property is defined as a special type of has part. Most of these molecules will be localized using techniques such as immunocytochemistry and in situ hybridization. Molecules may be assigned to any aspect of the cell, both regional and component parts, and at whatever level of granularity can be determined from the technique. An exception to this rule is the assignment of neurotransmitter. Because neurotransmitter has traditionally been one of the defining properties of a neuron to most neuroscientists, we included the property has neurotransmitter as a special type of has molecular constituent and assigned it at the level of cell class. In theory, we should be able to derive the neurotransmitter from a consideration of the types of molecules located within the synaptic region, but because techniques such as immunocytochemistry often determine neurotransmitter indirectly, for example, through the localization of a synthetic or degradative enzyme for a neurotransmitter, and because determination of a neurotransmitter usually involves additional physiological or pharmacological criteria, we decided to assign this as a simple property for now.
Through the properties has anatomical location, the SAO situates cells and parts of cells into higher order brain regions. The SAO divides anatomical localization into three categories: has general anatomical location; has specific anatomical location; has atlas location. General anatomical location is assigned to the level of the cell class and is meant to encode the generally known location of a cell class. This property again was included for expediency, because neuroscientists are so used to naming individual cells as parts of anatomical regions, even though only the cell soma may be located there. The level of specification may be fairly coarse in this case, for example, Purkinje cell has general anatomical location cerebellar cortex. Specific anatomical location is meant to be assigned at the instance level and is intended to be assigned at as fine a level of granularity as possible, for example, my Purkinje cell dendrite has specific anatomical location outer third of cerebellar molecular layer. If known, anatomical location can be recorded as a set of atlas coordinates through the has atlas anatomical location property. This property type contains the atlas referenced, the coordinates, and the reference point from which the coordinates are derived, for example, bregma. Currently, the SAO assigns anatomical location in the form of free text. We are in the process of changing the anatomical location to an object property that is drawn from the BIRNLex anatomical ontology, which in turn draws its anatomical entities largely from the Neuronames hierarchy (Dubach and Bowden, 2002).
Supracellular structures
One of the biggest challenges in constructing the SAO was to provide the specification of supracellular entities like the Node of Ranvier and the synapse. Although these entities are treated by other ontologies (e.g., Zhang et al., 2007) as if they are independent entities, in fact neither of these objects exist independently within complex tissue. Rather, they represent sites where certain configurations of subcellular objects are found (e.g., neuropil, synapses, glomeruli, and the Node of Ranvier) and where certain functions are presumed to occur. Thus, although in preliminary versions of the SAO, we classified synapses and Nodes as objects, starting in v1.0 we utilized the structure of the BFO to classify supracellular domains through the object aggregate and site classes.
An object aggregate in BFO 1.0 is defined as “an independent continuant that is a mereological sum of separate objects and possesses non-connected boundaries. Examples: a heap of stones, a group of commuters on the subway, a collection of random bacteria, a flock of geese, the patients in a hospital.” A site is defined as “an independent continuant consisting of a characteristic spatial shape in relation to some arrangement of other continuants and of the medium which is enclosed in whole or in part by this characteristic spatial shape. Sites are entities that can be occupied by other continuants.” The BFO further clarifies sites in this way: “In BFO, ‘site’ allows for a so-called relational view of space which is different from the view corresponding to the class ‘spatial region.’ Space and ‘spatial region’ entities are entities in their own rights which exist independently of any entities which can be located at them. This view of space is sometimes called ‘absolutist’ or ‘the container view.’ In BFO, the class ‘site’ allows for a so-called relational view of space, that is to say, a view according to which spatiality is a matter of relative location between entities and not a matter of being tied to space. The bridge between these two views is secured through the fact that while instances of ‘site’ are not ‘spatial region’ entities, they are nevertheless spatial entities.” (BFO 1.1; http://www.ifomis.org/bfo/1.1).
We considered supracellular domains as object aggregates because they represent a somewhat ad hoc grouping of cell parts into a higher order structures. However, many of these ad hoc groupings are given special designations because they are believed to be the locations at which a particular function occurs. For example, the Node of Ranvier is the site of action potential propagation down the axon; the synapse is the site at which neurotransmission occurs. The location of that function is inferred because of the presence of one of more molecules or cell components that have been demonstrated to be involved in the expression of these dynamic processes. Figure 3 shows the SAO structure for describing the Node of Ranvier from the central nervous system. We define the Node of Ranvier as a site on the axon in the gap between two segments of myelin. Neuroscientists have identified different compartments of the node based on the locations of certain structural configurations and molecules such as ion channels. We thus constructed a set of entities, grouped under “Node Related Sites,” utilizing the parcellation described in Sosinsky et al. (2005) to describe the different sites, the cellular objects located at each site and the spatial relationships among them. Note the difference between “Internode” (transitively a subclass of “Site”) and “Internode Axon” (transitively a subclass of “FiatObjectPart”). “Internode” is not the parent class of “Internode Axon,” because they refer to distinct entities in the axon. The distinction between the two reflects the difference between material and location. If we were to ask “what is the material located at the Internode site?” the answer would be not only “the Internode axon,” but would also include compact myelin, protein channels and other macromolecules. Conversely, if we were to ask “where is the Internode Axon?” in the sense of asking where the material substance of this regional part of an axon is located, the answer would be, “at the site called the Internode.” Similarly, asking “where is there both compact myelin and a regional part of an axon?” would also give the answer, “at the site called the Internode.” In this way, the SAO can provide a very precise specification of the different macromolecules and provides a formal basis for creating rules by which a structure can be recognized.
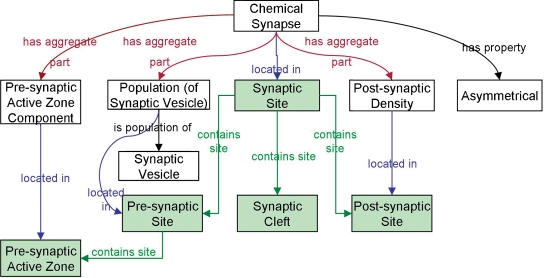
The synapse is modeled using the object aggregate and site classes (Figure 4). We created an aggregate object consisting of a pre-synaptic part, a post-synaptic part, and a junctional part, similar to the Synapse Ontology of Zhang et al. (2007) and then localize them to the synaptic site. Each of these parts have cell components, for example, synaptic vesicles, located within them that define the extents of these parts, that is, the pre-synaptic part is the part of the presynaptic structure (axon terminal, dendrite, or soma) containing synaptic vesicles. In our earlier versions of the SAO, which classified the synapse as a single material entity rather than a site, we encountered the problem that our designation of cellular structures as pre- or post-synaptic provided no way to distinguish the part that participated in the synaptic contact from the whole structure. When we say that the neuron soma is the post-synaptic structure, we are usually saying is that there is a contact on a part of the cell body. Through the relationships encoded in the SAO, we can restrict the definition of the synapse to that part of the cellular structure where certain structures, for example, synaptic vesicles, or molecules are localized.
Figure 4.
Diagram of a chemical synapse instance description in SAO. Sites are indicated by green backgrounds. The boxes indicate Instances of classes that are related to one another as a description of a particular instance of a chemical synapse.
Anatomical qualities
Version 1.2 of the SAO has included a more extensive list of morphological qualities under the dependent continuant class that are used to modify objects within the SAO (Figure S4). Generic morphological qualifiers such as “round” or “spherical” are imported into SAO through the Phenotype and Trait Ontology (Gkoutos et al., 2005). However, we included a set of qualities that were specific for subcellular anatomy, for example, spine shapes (mushroom, thin, stubby), nuclear shape (round, lobulated, indented), and cell soma shape (pyramidal, fusiform). We elected in most cases not to precoordinate these terms with the independent continuants they describe, because these qualities can be assigned at the time of annotation. By pre coordination, we mean the creation of a set of independent continuants which incorporate the qualifier, for example, mushroom-shaped spine; lobulated nucleus. Precoordination was used for morphological classes that required unique identification like spine classes, where the designation of mushroom shape confers a set of unique properties to that class. We chose not to precoordinate when the qualifier was considered descriptive of an instance and not necessarily indicative of a member of a distinct class. In these cases, we apply the qualifier to the instance, for example, instance of nucleus with morphological quality “indented” at the time of annotation. In this way, we do not have to generate large numbers of classes that differ on what might be a superficial detail. Additional qualities that are assigned to each object are morphometric quantities such as length, surface area, etc., orientation, and polarity.
Annotation properties
Annotation properties contain information about the ontology entities. We imported the annotation properties from the BIRNLex, a lexicon developed for the Biomedical Informatics Research Network (BIRN) project (www.nbirn.net/birnlex). These properties cover lexical entities such as definitions, synonyms, alternative spellings, and the curation status of each entity. The label assigned to the class name is also an annotation property. The BIRNLex, in turn, imported many entities from the Simple Knowledge Organization System (SKOS; http://www.w3.org/2004/02/skos/), a set of RDF properties and classes for describing the entities in a knowledge resource.
The definition property provides a human-readable definition for each entity in the SAO. We believe that such definitions are critical for human annotators to reference when using ontology class terms to describe data, because the equivalence between the descriptions of objects observed in an investigation and the ontology elements provides the ontology with its semantic power. Thus, a human must clearly understand the way the term is defined in the ontology in order to apply it. Because of the somewhat artificial and complicated structure imposed on some entities (see Figures 3 and 4), the definition cannot be easily extrapolated by a human from the structure of the ontology itself. Thus, following the recommendations of the OBO Foundry, we provide a human-readable definition in the form of A is a type of B which exhibits C. A is a B provides the location of the entity within the class hierarchy, for example, A protoplasmic astrocyte is a type of astrocyte, translates easily into “protoplasmic astrocyte” is a “astrocyte” in the SAO. “Which exhibits C” provides the extensional property or properties differentiating the entity from others in a class, for example, a protoplasmic astrocyte is a type of astrocyte which is characterized by many fine processes and relatively few intermediate filaments. From this definition, the property has regional part process and has component intermediate filament may be inferred. The goal is to provide a human-readable definition that is consistent with the machine-processable definition encoded in the ontology.
User-defined reclassification and query
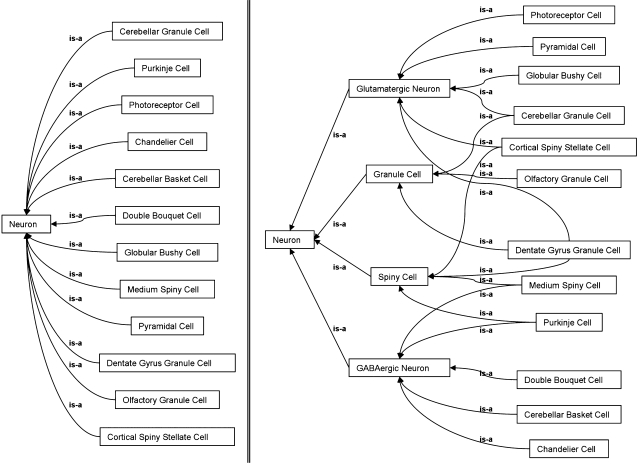
To illustrate how properties in OWL can be used to infer additional hierarchies from the SAO, we constructed some OWL classes which reclassify the neuron cell types based on their properties assigned by the SAO. We classified neurons based on neurotransmitter, morphological type, or the presence of spines simply by defining using OWL and Protégé that these categories ought to include any cell which had the main property of that category (e.g., that the neuron was known to use glutamate or GABA as a neurotransmitter, etc). After defining these categories, we used the open source ontology reasoner Pellet (Sirin et al., 2007) to transform the flat version of the SAO neuron type hierarchy in Figure 5A into the inferred hierarchy in Figure 5B. The inferred hierarchy demonstrates that a cell like the a Medium Spiny cell is both spiny and GABAergic while a Dentate Gyrus granule cell can be classified as spiny, glutamatergic, and granule at the same time. Any arbitrary reclassification may be performed using the combinations of properties that suits the purpose of the user. Since the parent-child (is-a) relationships of the inferred hierarchy are not written back to the ontology, this allows us to maintain a hierarchy with single parents in the authored version of the ontology. However, the classes of the inferred hierarchy, Spiny Cell, Glutamatergic Neuron, Granule Cell, and GABAergic Neuron are implicitly embedded in the authored ontology as children of the class Neuron. These classes use OWL restrictions to define the kinds of children that it must logically have, and thus implicitly allows cells to exist in multiple inferred categories.
Figure 5.
Inferred hierarchies using OWL. On the left, a subset of the hierarchy under the Neuron class prior to inference. On the right, the automatic reclassification of that subset under four user-defined groupings, GlutamatergicNeuron, GABAergicNeuron, SpinyCell, GranuleCell, based on the properties of the cells alone.
SAO as semantic “glue”
In order to use the standard names of the SAO to annotate images in different data formats, the SAO is itself used as a data exchange format between three image annotation software applications. To apply the ontology to actual data, we have incorporated annotation with the SAO into our routine segmentation tools for light and electron microscopy. We have created a programmatic interface to the OWL ontology that may be called by Jinx, our 3D segmentation tool for electron tomography data. Through Jinx, users describe the objects contained in electron microscopic volumes of neural tissue as instances of the SAO, rather than as a set of user-defined objects with no relationship among them. The application of SAO captures each object and allows the definition of related objects. Instances of the SAO are then stored in a large instance store, which we call the Cellular Knowledge Base (Fong et al., 2007), where they can be queried (Chen et al., 2006). The data files used to generate the instances are stored in the CCDB which tracks their experimental and data provenance. We are in the process of incorporating SAO into additional analysis tools for analyzing neuronal branching patterns and for annotation of spatially varying signals using our GIS-based brain atlas, the SMART Atlas (Martone et al., 2007b).
The SAO and Cellular Knowledge Base architecture enable us to integrate these different data types through the shared semantic representation of biologically significant elements. For example, the image of a dendritic tree generated with two-photon fluorescent microscopy (Figure 1A), is annotated as an instance of sao:DendriticTree, which is part of medium spiny neuron, and has part Dendrite. The instance of dendrite has regional part Dendritic segment. This same instance of dendritic segment is visible in the correlated electron microscopic volume of the same medium spiny neuron (Figure 1C), where we can further assign has regional part Dendritic Spine to this dendrite. An algorithm with access to the SAO infer that the dendritic spine is part of the dendritic tree, and apply properties derived from the electron tomography study to that acquired from the light microscopic imaging. Without this common interlingua and the codified knowledge explicitly declaring the shared semantic context, programmatic combination and cross query of these images and data types is much more difficult and requires customized algorithms to encode the semantic information.
By structuring the SAO in OWL, we have made its encoded knowledge available to OWL reasoners and RDF query engines. Consequently, we use instances stored in the Cellular Knowledge Base and the knowledge encoded in the ontology to determine what molecular constituents are found in the Node of Ranvier, and which sites on the Node are they respectively found in. We can also query about the glial cell types associated with the Node, and how the parts of the glial cells relate to the different parts of the Node.
Discussion
We created an OWL ontology representing the subcellular anatomy of the nervous system to provide the necessary scaffold for integrating molecular and anatomical data through accurate description of mesoscale anatomy. By codifying it in OWL, we have enabled algorithmic query and analysis of that knowledge. Moreover, we have enabled the use of formalized knowledge as a standard for making connections between data formats, making connections between other ontologies, and as a data exchange format for image annotation tools. This scaffold is amenable both to tool development and to semantically driven information exchange across the field. It also provides individual researchers a means to perform reasoner-based quality control and inferential analysis of annotated neuroimages. Applying formal semantic representation techniques to neuroanatomical structure has been preliminarily addressed in the macroscopic domain (Martin et al., 2001; Mechouche et al., 2006); little exists in the mesoscopic neuroanatomical domain as yet. A Synapse Ontology was recently constructed (Zhang et al., 2007), but it does not situate synapses in their cellular and tissue contexts, nor is it built on top of community-shared foundational ontologies. Our motivation for creating the SAO was to provide the necessary tools for describing the types of subcellular and supracellular entities located in the dimensional range now termed the mesoscale. The SAO is designed as a reference ontology, defined by Brinkley et al. (2006) in the following way: “Unlike application ontologies, reference ontologies are not designed for any specific application, but are intended to be re-used in multiple application contexts […] Reference ontologies are designed according to strict ontological principles, whereas application ontologies are designed according to the viewpoint of an end-user in a particular domain.” We elected to tackle the more difficult task of creating a reference ontology with formal semantics, because we believe that such resources are needed to build models of mesoscale structures that combine information from multiple domains and to be able to utilize information obtained at the mesoscale at coarser and finer scales of granularity. Through application of the ontology, researchers can work in a narrow dimensional range, but their observations are immediately linked across scales. For example, a researcher segmenting a reconstruction derived from electron tomography may make the observation that an endoplasmic reticulum of a dendritic spine from a Purkinje cells expresses the IP3 receptor. Through the SAO, the following inferences can be made: There exists a Purkinje cell dendrite that expresses the IP3 receptor; the cell class Purkinje cell expresses the IP3 receptor; the cerebellar cortex expresses the IP3 receptor; and the cerebellum expresses the IP3 receptor.
The SAO is meant to describe structure, not function nor dynamic processes, following the parcellation of biomedical reality established by the BFO. However, although we try to adhere as much as possible to this distinction within the formal class structure of the ontology, as can be seen by the labels assigned to SAO classes, many labels that are applied to our SAO entities have a functional flavor to them, for example, ‘chemical synapse’. Where possible, we tried to remove entities that mixed a structure with a function, for example, myelinating oligodendrocyte or with a physiological state, for example, activated microglia. However, we also felt in some cases that it was important to assign the labels that are commonly employed by the community. Although these labels appear in the figures and text provided in this paper, SAO classes are actually identified using semantically neutral numeric labels (e.g., SAO class sao1507566336 has the preferred label Post-synaptic Component). The human-readable preferred label is assigned as an annotation property, as are a variety of lexical term variants, such as alternate labels, abbreviations, synonyms, acronyms, and so on. This practice is standard in the ontology community, and although it makes working with the ontology at times cumbersome for humans because of the need to associate the label with the class, we find it philosophically appealing. The entity is the same entity regardless of what we call it, that is, “a rose by any other name would smell as sweet.” So the fact that our neuron labels reflect mixtures of classification schemes does not impact the class structure of the SAO; rather, the class of neuron to which the label is applied is defined by the set of properties assigned to it.
Ultimately, the goal of anatomy is to provide the structural substrate for mapping function and understanding the structural constraints on dynamic processes. Anatomy is a mature discipline with a rich history. Many structures have been described, and continue to be described, particularly in electron microscopy, for which no functional property is known. The classic view of structure-function relationships assumes that structural differences reflect functional differences as well. However, mapping function onto structure is a complex issue that is currently beyond the domain of the SAO. We chose to adhere to a strict structural approach to keep the SAO scope tractable. We also, however, believe that by not mixing structural and functional classes together, it will be easier in the future to utilize the SAO within a functional ontology. As an example, the term synapse, as is recounted in all introductory textbooks, was a functional concept introduced by Sherrington to describe the transmission of information between cells. The morphological correlate of the synapse was described by Palay and colleagues using electron microscopy in the 1950s, and is also familiar to beginning students of neuroscience. SAO currently provides a formal description of the set of entities to describe the morphological correlates of what are assumed to be the sites and machinery for synaptic transmission in the nervous system. Although the labels employed, pre-synaptic and post-synaptic compartment, do have functional significance, the precise mapping of the functional aspects onto the morphological correlate is not straightforward. Though these familiar functional labels date back to work on the cellular physiological correlate of Sherrington's synapse first described by Katz and colleagues in the 1940s, as a recent paper indicating evidence for “ectopic release” from the chick ciliary ganglion synapse illustrates (Coggan et al., 2005), our understanding of neural signaling at the cellular level continues to evolve. If release of neurotransmitter can occur at sites other than the active zone visualized in electron micrographs, then the functions associated with a synapse cannot be restricted to this domain. However, by modeling a synapse as a site where objects, and eventually dynamic processes, are located, the definition of a synapse can expand as our functional understanding of synaptic transmission expands. We believe that mapping of function onto structure will be one of the greatest challenges faced by those who are creating ontologies for biomedical science.
Reasoning and inference with OWL
Biological objects are complex entities that do not fit neatly into single hierarchies. We have chosen to follow the recommended practice of single inheritance for all SAO classes, even when that means providing a very flat hierarchy with minimal utility for classification purposes. However, the power of OWL as an ontology formalism is that it not only enables us to explicitly express the complex qualities and inter-relatedness of entities, the standard tools built around the OWL formalism allows us to automatically infer multiple valid hierarchies for an entity, depending on what is required. For complex entities such as neuronal classes, we can use the OWL inference engine to infer hierarchies based on neurotransmitter, morphological properties, anatomical location, or circuit type (Figure 5). The same can be done for other classes of subcellular structures, for example, dendritic spines. This approach provides maximal flexibility to the end user and allows us to begin to cluster and define neurons based on a set of properties rather than along a single dimension (Migliore and Shepherd, 2005).
We have only begun to experiment with the power of OWL to infer new knowledge about objects that is not explicitly encoded in the ontology that allows information to be inferred across scales. In Larson and Martone (2007), we provide an example of this cross scale reasoning using OWL and rules about how cell parts relate to cells and brain regions. In this example, we showed how annotation of a synapse between a terminal of a thalamocortical axon and the dendritic spine of a cortical neuron observed through axonal tracing and electron microscopy could be used to infer knowledge about regional brain connectivity. Through relationships encoded in SAO, we inferred from the presence of a labeled axon terminal that there must be a neuron in the thalamus that has an axon projecting to the cortex. From the presence of a spine, we inferred that there existed a neuron to which the spine belonged in cortex. From the local observation that an axon terminal synapsed on a dendritic spine, we could infer that thalamic cells synapse with cortical cells, and that thalamus projects to cortex. While the reasoning itself does not provide new insight about brain function, we show here that a computational algorithm was able to infer the same logical cross-scale consequences of the subcellular arrangement of cell parts as would a neuroscientist without our having to write custom code to embed that knowledge in the program.
Application of the ontology
In construction of the SAO, we have attempted to provide a formal structure for describing data, balancing the needs for a “top-down” versus a “bottom-up” approach. By top-down, we mean that the biological theory governing a domain is used to classify data products; by bottom-up, we mean that we do not impose prior knowledge constraints on interpreting data but let the data speak for themselves (Murphy, 2005). OWL classes are essentially descriptive templates that constrain the possible properties and relationships which instances may have. As such, we only encode knowledge into the class level when we are sure that it ought to constrain all further instances that may be seen. This criterion enforces a certain amount of rigor when describing the properties of biological entities. What are those things that must always be true of a biological entity? Unlike the case of gross anatomy, where we can be reasonably certain of the canonical form taken by the human body, for example, we do not believe that we are at the stage with subcellular anatomy where we can comfortably define such canonical forms. Thus, although we sacrifice some of the reasoning power of OWL through the minimal placement of restrictions on the classes, we designed version 1.2 of the SAO to serve as the basis by which such rules can be derived from the instances.
When describing data, we apply the ontology only down to the level of granularity of which we are reasonably certain. For example, if we know the type of neuron we are describing, we can assign instances of properties to that specific class; if we do not, we can assign the observed properties to the class “neuron.” Using the reasoning power of OWL, it may turn out that the properties of this unidentified neuron are equivalent to a known class, but that can be inferred from the actual instance. In this way, the structure of the OWL standard forces the SAO to make careful and conservative descriptions about subcellular anatomy while still allowing a place for uncertainty.
Instances within the SAO also serve another important function by allowing us to annotate the biological description of a piece of data with the data and experimental properties from which it was derived. Entities within SAO are not directly observable by humans but must be imaged through a device such as a microscope and recorded in some form on a particular medium. Biologists are well aware that how a specimen was prepared, imaged, and analyzed will impact the types of observations that are made. In many cases, subcellular structures that are observed under certain conditions, for example, chemical fixation, are determined to be artifactual when recorded under different conditions. Most experimentalists are uncomfortable with knowledge management systems that attempt to divorce the biological reality from the methods used for acquisition, visualization, and analysis, because these methods largely determine the form that the reality will take. We must recognize, however, that the entities that we are attempting to describe in the SAO are assumed to transcend any technique. That is, we are assuming that there is such as thing as a dendrite, even though its properties can only be described in a specific experimental context. So, although the SAO itself does not assign technique or data type to the biological entity, for each instance of the entity, we provide a link to the experimental evidentiary context and the data type from which it was derived (e.g., this “instance” of dendrite was stained with a Golgi stain and imaged in a light microscope).
Through the construction of the SAO, we have made progress toward the goals of building information bridges in neuroscience in three broad areas: formalization, externalization, and standardization. By formalization, we mean the process of describing concepts in a fully explicit manner in order to clarify and sharpen the meanings of the terms being used. The lengths that we have gone to either find or impose structure on implicit concepts in subcellular anatomy reflect the absence of prior efforts to bring them into a single cohesive framework. Such a framework is important for the growing community interested in producing detailed computational models of structure and function in the nervous system. It is vitally important that experimental neuroscientists be able to communicate with this community and provide increased levels of explanation of their experimental systems. Providing a formal way of communicating, these explanations make it much easier to begin the modeling process. Ontologies in general, and the SAO in particular, is crucial “connective tissue” to help place these goals within reach for neuroscience.
In order for formalized information to be used by software applications, the information must be capable of externalization. By externalization, we mean to draw attention to the ability to transform the information into “code,” as opposed to the translation of abstract concepts into a human-only readable explicit representation. Once knowledge has been formalized and subsequently codified into a computer-readable form, that knowledge becomes externalized as an entity that is capable to programmatically interact with other knowledge. This makes information much more flexible than if it resided on the printed page, and it allows algorithms to answer questions for us, saving time and effort. The process of constructing an OWL ontology formalizes the knowledge it contains, but encoding it in OWL and saving it on a computer in its underlying RDF/XML format externalizes the information for other systems to digest and manipulate via standard open source code frameworks.
Through externalization, we are able to remix knowledge into other forms. It allows us to generate diagrams, to view it in different software interfaces (e.g., Jinx), to reclassify hierarchies on demand, and to run rule-based reasoning or other automatic inferencing mechanisms. The benefits of this are obvious in the context of the goals of data sharing and model construction. Externalization is also needed in order to construct algorithms that are capable of assisting neuroscientists do their own work, such as to guide them in a literature search or to suggest the name of a structure they are segmenting.
Once an information bridge has been formalized, and also externalized, it can be used for the final important purpose of standardization. In this context, the aspect of standardization that we focus on is the ability for OWL ontologies to serve as semantic “glue” which allow disparate data, ontologies, and applications to interoperate. The strategy we have employed in our knowledge environment is to leverage the externalized knowledge in the SAO by embedding it in tools that have first contact with primary data. By embedding the SAO in these tools, we enable the user not only easy access to SAO terms to use in annotating their data, but also we make the tools more intelligent to minimize the amount of implied knowledge that a user must contribute.
Future directions
We are continuing to develop the SAO, apply it to the type of biological data contained within the CCDB and to refine the structure of the ontology. Current development is focused on the development of a set of entities to describe cellular inclusions observed in neurodegenerative disease, and entities from subcellular anatomy in domains outside of neuroscience. We welcome any feedback or contributions to the ontology from the biological community, and are working on a web-based interface through the NCBO BioPortal (http://www.bioontology.org/ncbo/faces/index.xhtml) that will facilitate this process. The process of ontology construction is laborious and contains many fits and starts that leave legacy errors within the ontology. Besides the complicated nature of the domain, we face additional challenges in developing the SAO using emerging community standards, (e.g., the BFO), that are themselves still developing. Consequently, we periodically have to refactor the ontology as new versions of the constituent come on-line. However, we believe that it is important for neuroscience ontologies to align themselves as much as possible with the broader life sciences community, because ultimately we hope to be able to integrate neuroscience with the broader domains. The act of formalizing knowledge is to make explicit what was once implicit, and in so doing clarifying the boundaries of definitions. Giving something a name gives power over it (Winston, 1992). Once we have assigned appropriate labels, the creation of a system of axioms that interrelate the labeled entities gives us additional power to describe the interactions between the entities. This practice has been at the heart of scientific understanding since the beginning of history. The poster child of formalization is mathematics itself, which is a system where the entities are variables, and the system of axioms consists of mathematical operations. The impact of mathematics, a precise and consistent means of communicating ideas, was to provide extraordinary leverage to thinkers throughout history to build truths upon truths in the service of understanding. A key example of this was the expression in calculus of the fundamental relationships between electric fields, magnetic fields, electric charge, and electric current by Maxwell's equations. It required the formal language of calculus to clarify and distill the knowledge of those physical concepts. As such, we see our attempt to formalize the concepts of the structure and function of the brain with ontologies, whose underpinnings are first-order logic, to be part of a broader pattern in the history of science. The issues we have explored through our formalization efforts might be considered to be part of a larger movement underway to develop formal means to describe biological entities.
Supplemental Data
The Supplemental Data for this article can be found online at http://ccdb.ucsd.edu/SAO/Larson2007/.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants NIDA DA016602 (CCDB), NINDS RO1NS058296, NCRR RR04050, and RR08605. The Bioinformatics Research Network is supported by NIH grants RR08605-08S1 (BIRN-CC) and RR021760 (Mouse BIRN). The Protégé resource is supported by grant LM007885 from the United States National Library of Medicine. The authors wish to thank Eric A. Bushong for his help with glial cell types and Sarah M. Maynard for her help with biological entities relevant to neurodegenerative diseases.
Footnotes
References
- Bard J., Rhee S. Y., Ashburner M. (2005). An ontology for cell types. Genome Biol. 6, R21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M., Dong H. W., Swanson L. W. (2005). Brain architecture management system. Neuroinformatics 3, 15–48 [DOI] [PubMed] [Google Scholar]
- Bowden D. M., Dubach M. F. (2003). NeuroNames 2002. Neuroinformatics 1, 43–59 [DOI] [PubMed] [Google Scholar]
- Brinkley J. F., Suciu D., Detwiler L. T., Gennari J. H., Rosse C. (2006). A framework for using reference ontologies as a foundation for the semantic web. AMIA Annu. Symp. Proc. 96–100 [PMC free article] [PubMed]
- Chen L., Martone M. E., Gupta A., Fong L., Wong-Barnum M. (2006). OntoQuest: exploring ontological data made easy. Abstract, Very Large Databases Conference1183–1186 [Google Scholar]
- Coggan J. S., Bartol T. M., Esquenazi E., Stiles J. R., Lamont S., Martone M. E., Berg D. K., Ellisman M. H., Sejnowski T. J. (2005). Evidence for ectopic neurotransmission at a neuronal synapse. Science 309, 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evren S., Parsia B., Grau B. C., Kalyanpur A., Katz Y. (2005). Pellet: a practical OWL-DL reasoner. UMIACS Technical Report, 2005–2068
- Fong L. L., Larson S. D., Gupta A., Condit C., Bug W. J., Chen L., West R., Lamont S., Terada M., Martone M. E. (2007). An ontology-driven knowledge environment for subcellular neuroanatomy. CEUR Workshop Proceedings 258, ISNN
- Gene Ontology Consortium. (2002). Creating the gene ontology resource: design and implementation. Genome Res. 11, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos G. V., Green E. C., Mallon A. M., Hancock J. M., Davidson D. (2005). Using ontologies to describe mouse phenotypes. Genome Biol. 6, R8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon P. (2003). BFO in a nutshell: a bi-categorial axiomatization of BFO and comparison with DOLCE. IFOMIS, (ISSN ).
- Grenon P., Smith B., Goldberg L. (2004). Biodynamic ontology: applying BFO in the biomedical domain. Stud. Health Technol. Inform. 102, 20–38 [PubMed] [Google Scholar]
- Gruber T. R. (1993). Toward principles for the design of ontologies used for knowledge sharing. Int. J. Hum. Comput. Stud. 43, 907–928 [Google Scholar]
- Huerta M., Koslow S. H., Leshner A. I. (1993). The Human Brain Project: an international resource. TINS 16, 436–438 [DOI] [PubMed] [Google Scholar]
- Kalyanpur A., Parsia B., Sirin E., Cuenca-Grau B., Hendler J. (2006). Swoop: a web ontology editing browser. J. Web Semant. 4(2), 144–153 [Google Scholar]
- Koslow S. H., Huerta M. F. (1997). Neuroinformatics: an overview of the human brain project (Mahwah, New Jersey, Lawrence Erlbaum Associates; ). [Google Scholar]
- Larson S. D., Martone M. E. (2007). Rule-based reasoning with a multi-scale neuroanatomical ontology. CEUR Workshop Proceedings 258, ISSN
- Martin R. F., Mejino J. L., Bowden D. M., Brinkley J. F., Rosse C. (2001). Foundational model of neuroanatomy: implications for the Human Brain Project. Proc. AMIA Symp. 438–442 [PMC free article] [PubMed] [Google Scholar]
- Martone M. E., Gupta A., Wong M., Qian X., Sosinsky G., Ludaescher B., Ellisman M. H. (2002). A cell centered database for electron tomographic data. J. Struct. Biol. 138, 145–155 [DOI] [PubMed] [Google Scholar]
- Martone M. E., Sargis J., Tran J., Wong W. W., Jiles H., Mangir C. (2007a). Database resources for cellular electron microscopy. Methods Cell Biol. 79, 799–822 [DOI] [PubMed] [Google Scholar]
- Martone M. E., Zhang S., Gupta A., Qian X., He H., Price D. L., Wong M., Santini S., Ellisman M. H. (2003). The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics 1, 379–395 [DOI] [PubMed] [Google Scholar]
- Martone M. E., Zaslavsky I., Gupta A., Memon A., Tran J., Wong W., Fong L., Larson S. D., Ellisman M. H. (2007b). The smart atlas: spatial and semantic strategies for multiscale integration of brain data. In Anatomy Ontologies for Bioinformatics: Principles and Practice, Burger A., et al., eds. (London, Springer-Verlag; ), in press. [Google Scholar]
- Mechouche A., Golbreich C., Gibaud B. (2006). Towards an hybrid system for annotating brain MRI images. Workshop on OWL Experiences and Directions 216
- Migliore M., Shepherd G. M. (2005). Opinion: an integrated approach to classifying neuronal phenotypes. Nat. Rev. Neurosci. 6, 810–818 [DOI] [PubMed] [Google Scholar]
- Muller H. M., Kenny E. E., Sternberg P. W. (2004). Textpresso: an ontology-based information retrieval and extraction system for biological literatures. PLOS Biol. 2, e309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. F. (2005). Cytomics and location proteomics: automated interpretation of subcellular patterns in fluorescence microscope images. Cytometry A 67, 1–3 [DOI] [PubMed] [Google Scholar]
- Noy N. F., Sintek M., Decker M., Crubezy , Fergerson R. W., Musen M. A. (2001). Creating semantic web contents with Protege-2000. IEEE Intell. Syst. 16, 60–71 [Google Scholar]
- Neumann E., Prusak L. (2007). Knowledge networks in the age of the semantic web. Brief. Bioinform. 8, 141–149 [DOI] [PubMed] [Google Scholar]
- Peters A. L., Palay S., Webster H. D. F. (1991). The fine structure of the nervous system (New York, Harper and Row; ). [Google Scholar]
- Reichenbach A., Wolberg H. (2005). Astrocytes and ependymal glia. In Neuroglia H., Kettenmann, Ransom B. R., eds. (New York, Oxford University Press; ), Chapter 2. [Google Scholar]
- Rosse C., Kumar A., Mejino J. L. V., Jr., Cook D. L., Detwiler L. T., Smith B. (2005). A strategy for improving and integrating biomedical ontologies. AMIA 2005 Symp. Proc. 639–643 [PMC free article] [PubMed]
- Rosse C., Mejino J. L., Jr. (2003). A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 36, 478–500 [DOI] [PubMed] [Google Scholar]
- Sirin E., Parsia B., Grau B. C., Kalyanpur A., Katz Y. (2007). Pellet: a practical OWL-DL reasoner. J. Web Semant. 5 [Google Scholar]
- Smith B., Rosse C. (2004). The role of foundational relations in the alignment of biomedical ontologies. Medinfo 11, 444–448 [PubMed] [Google Scholar]
- Smith B., Ceusters W., Klagges B., Kohler J., Kumar A., Lomax J., Mungall C., Neuhaus F., Rector A. L., Rosse C. (2005). Relations in biomedical ontologies. Genome Biol. 6, R46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky G. E., Deerinck T. J., Greco R., Buitenhuys C. H., Bartol T. M., Ellisman M. H. (2005). Development of a model for microphysiological stimulations: small Nodes of Ranvier from peripheral nerves of mice reconstructed by electron tomography. Neuroinformatics 3(2), 133–162 [DOI] [PubMed] [Google Scholar]
- Stephan K. E., Kamper L., Bozkurt A., Burns G. A., Young M. P., Kotter R. (2001). Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1159–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston P. H. (1992). Artificial intelligence (New York, Addison-Wesley; ). [Google Scholar]
- Wang J., Williams R. W., Manly K. F. (2003). WebQTL: web-based complex trait analysis. Neuroinformatics 1(10), 299–308 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang Y., Zheng H., Zhang C., Xiong W., Olyarchuk J. G., Walker M., Xu W., Zhao M., Zhao S., Zhou Z., Wei L. (2007). SynDB: a synapse protein database based on synapse ontology. Nucleic Acids. Res. 35, D737–D741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.