Abstract
Newly formed centrioles can spring forth from clouds of pericentriolar material, violating the precise regulation of centriole counting. These observations challenge the long-standing view that centriole number is determined by the periodic activation of an assembly template thought to reside on pre-existing centrioles.
Armed only with the light microscope and keen skills of observation, cell biologists of the early part of the twentieth century first recognized that the centrosome is fundamental for organizing the cytoplasm and controlling chromosome dynamics during cell division. In these early investigations, centrioles, which reside at the very centre of the centrosome, gained attention because of their enigmatic autonomy — centriole number doubles from two to four in a process that is strictly coordinated with DNA replication, and then their number is reduced again as pairs of centrioles partition into daughter cells during cell division (Fig. 1a). Fascination with centrioles grew when electron microscopy first revealed their elegant microtubule-based, ninefold symmetry and the remarkable orthogonal relationship between older and younger members of a centriole pair.
Figure 1.
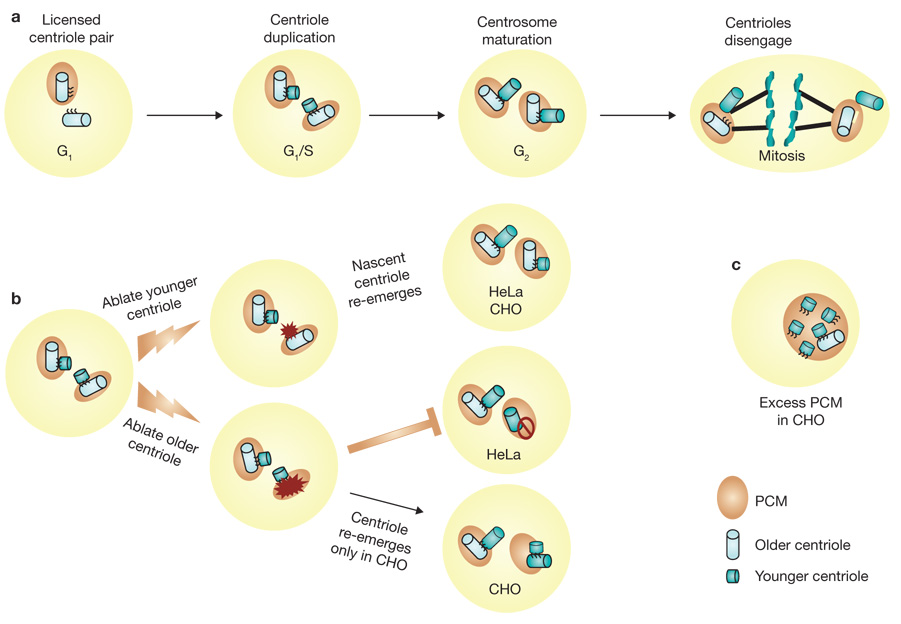
Alternative routes for centriole duplication. (a) Centrosomes of metazoan cells, including those of humans, consist of a pair of centrioles and a surrounding protein lattice called pericentriolar material (PCM), which serves to secure γ-tubulin-containing microtubule nucleation sites. As cells enter S phase, new centrioles begin to form near each of the two pre-existing centrioles and establish an orthogonal arrangement for each centriole pair with the nascent centriole extending from the proximal wall of the older centriole. During late S and G2 phases of the cell cycle, the newly formed centrioles elongate to near mature length. As cells prepare for mitosis, the centrosome matures by acquiring additional PCM and γ-tubulin, and then the two pairs of centrioles and their associated PCM separate to function as two mitotic spindle poles of the dividing cell. As cells pass through anaphase, the individual centrioles of each pair separate a short distance from one another in a process called centriole disengagement. Finally, with the completion of cell division, each G1 daughter cell inherits one spindle pole (centrosome) that contains a disoriented pair of centrioles. (b) Laser ablation studies show that when the younger centriole of a pair is destroyed, a new centriole emerges within several hours in both HeLa and CHO cells. However, when the older centriole of a pair is destroyed, a new centriole re-emerges only in CHO cells. (c) CHO cells expressing excess pericentrin form multiple nascent centrioles.
These features and other considerations led to the prevailing notion that formation of new centrioles is patterned on a structural template that is closely associated with each pre-existing centriole and is under cell-cycle control. In this manner, it is thought that centriole duplication is limited to yielding only two centriole pairs in each cell cycle. However, in a study published on page 322 of this issue, Alexey Khodjakov and colleagues1 have reached a strikingly different conclusion. Using the surgical precision of laser ablation to destroy centrioles, and correlative optical and electron microscopy methods, they present innovative studies that suggest that the size of the cloud of surrounding pericentriolar material (PCM), and not the centrioles themselves, limits the number of new centrioles that can form under certain conditions.
To mark centrioles in living cells, Khodjakov and colleagues used recombinant centrin–GFP (green fluorescent protein) expressed in HeLa and Chinese hamster ovary (CHO) cells. Centrin is one of the firs proteins to localize at sites of newly forming centrioles and is required for structural integrity of pre-existing centrioles2,3. Importantly, the older centriole of a pair labels more intensely with centrin–GFP, and so it can be distinguished from the younger centriole. Using precisely focused laser pulses, Khodjakov and colleagues were able to destroy either the older or younger centriole of a pair in living cells and then follow the re-emergence of centrioles labelled with centrin–GFP. In HeLa cells, when the younger centriole of a pair was ablated in this manner, a nascent centriole re-emerged in its place within a few hours. Examination of the same cells by electron microscopy revealed that the new centriole was as oriented orthogonal to the older centriole. Nascent centrioles re-emerged even after repeated destruction of the younger centriole. In contrast, a new centriole failed to form when the older centriole was ablated. Furthermore, they report that, in approximately one third of the trials, when the older centriole was destroyed, the remaining (younger) centriole ultimately deteriorated after losing its connection to the PCM. These experiments suggest that in HeLa cells, the older centriole exerts control over the time and place that a younger centriole forms, and seem consistent with the traditional ‘pre-existing centriole template’ model for stringent control of centriole duplication.
In contrast, the CHO cell line shows aberrant regulation of centriole duplication. CHO cells ‘re-duplicate’ centrioles inappropriately when DNA synthesis is blocked using drugs such as hydroxyurea or aphidicolin4. A similar feature is also seen in some cancer cells where an excess of centrioles and PCM result in centrosome ‘amplification’, a condition thought to be responsible for at least one form of chromosomal in instability5. In CHO cells, a nascent centriole formed when the younger centriole of a pair was destroyed by laser ablation. However, in contrast to HeLa cells, formation of a nascent centriole was also seen when the laser destroyed the older centriole of a centriole pair. A small percentage of CHO cells arrested in S phase also showed groups of three centrioles (‘triplosome’) consisting of one older centriole and two younger centrioles that showed coordinated movement within the cytoplasm, suggesting that they are physically linked. When laser pulses were used to destroy both of the younger centrioles, a single nascent centriole re-emerged. Surprisingly, when only a single younger centriole of a triplosome was destroyed, leaving an older and younger centriole pair, no new centriole was formed. In approximately one third of these cells, 15–25 h later, the remaining centrioles no longer showed coordinated movement, suggesting that they had lost their physical connection to one another, and the older centriole then underwent re-duplication. These observations suggest that, even in CHO cells, the older centriole maintains some control over centriole duplication and re-duplication after laser ablation, albeit with imperfect counting. Yet when the older centriole is destroyed in CHO cells, jurisdiction over centriole formation and number seems to be transferred to a proxy control mechanism.
Khodjakov and colleagues then addressed the question of where such an alternative mechanism may reside by examining the role of pericentrin, a coiled-coil protein that itself is not required for centriole formation, but that causes excessive accumulation of PCM when overexpressed in cells6. Remarkably, they found that centriole re-duplication could be exacerbated in S-phase-arrested CHO cells that also over-expressed pericentrin. This was driven by an initial accumulation of centrin–GFP aggregates that coalesced near pre-existing centrioles, and marked the sites in the cloud of PCM where multiple new centrioles emerged. Similar results in S-phase-arrested CHO cells without overexpression of pericentrin have been reported in an earlier study7. Bona fide centriole formation was confirmed by correlative electron microscopy, which also showed the new centrioles to be scattered about and not orthogonally related to pre-existing centrioles. From these studies, Khodjakov and colleagues concluded that formation of new centrioles is initiated in the PCM cloud and that the role of the older centriole is not to provide a template for the assembly of new centrioles, but rather to organize and restrict the size of the PCM, thereby limiting new centriole formation.
In the search for a mechanistic explanation for how centriole as assembly is controlled, it seems premature to cast aside the tenets of a century of centriole biology. Minimally, caution is prudent when overexpression of centrin–GFP is used both to mark the target for destruction and to monitor centriole re-emergence, especially when prevailing evidence implicates centrin in the control of the very process under examination7. These considerations and other aside side, any theory for the control of centriole duplication must integrate information from diverse systems and remain faithful to the unifying doctrine of evolutionary biology: as centrioles were an early achievement of eukaryotic cell evolution, it is likely that fundamental mechanisms controlling centriole duplication are ancient and largely remain common to all extant organisms that possess centrioles.
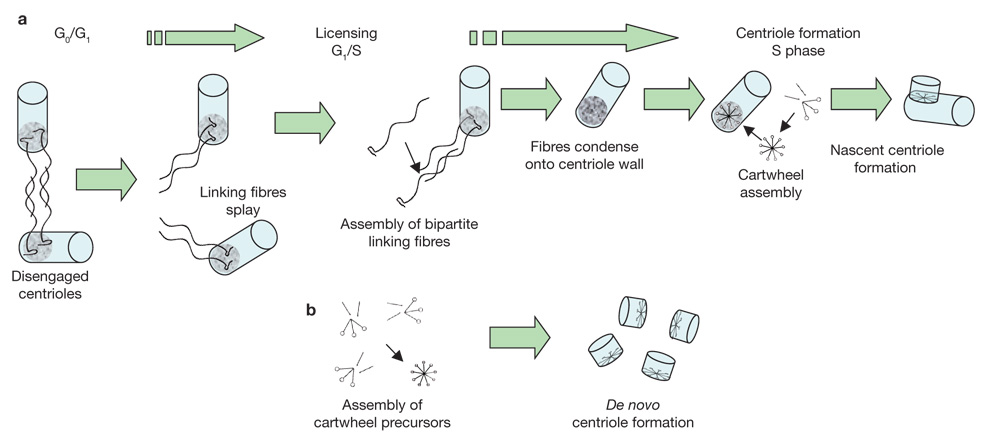
At least two structurally defined processes operate in centriole formation: most cycling cells show semi-conservative centriole duplication, in which a pre-existing centriole provides a ‘template’ or ‘seed’ that directs the assembly of a new centriole and therefore provides an un ambiguous mechanism for precise control of centriole number. In other cell types, centrioles can also arise through de novo centriole formation (for example, during sperm formation in the water fern Marsilea vestita, at the time of ciliogenesis in ciliated epithelia and in exceptional experimental circumstances such as those in the studies by Khodjakov), which does not exercise strict control over centriole number, where new centrioles form in the cytoplasm without any obvious contribution of a pre-existing centriole. A simple model that uses first principles of self-assembly can reconcile both these processes (Fig. 2).
Figure 2.
A unifying model for control of centriole number. (a) Semi-conservative centriole duplication: cells are produced in G0/G1 with a pair of disengaged centrioles. Licensing centriole duplication involves the dissociation (splaying) of linking fibre arrays, half of which remain attached to each pre-existing centriole. This is followed by formation of a complete ‘unit of duplication’ through the assembly of new bipartite fibre arrays. The fibres then contract or condense on the proximal wall of the centriole to which they are attached and form the preferred site for assembly of the cartwheel structure. The cartwheel then becomes a template for formation of a nascent centriole. In this manner, strict control over centriole number is maintained by the older centriole. (b) De novo centriole formation: in some cells, and under exceptional experimental conditions, new centrioles can form when the concentration of precursor molecules becomes high enough to cause cartwheel formation despite the lack of a favoured assembly site. De novo centriole formation does not exercise strict control over centriole number.
First, consider the concept of ‘licensing’ centriole formation whereby a two-step process involves a triggering or licensing step. This occurs just once in the cell cycle, thus providing a preferred site where a centriole precursor can form when proteins reach critical levels that support self-assembly8. Prevailing evidence from diverse systems suggests that a ‘cartwheel’ structure is the earliest patterned as assembly seen at sites where centriole formation occurs2,3. Microtubules elongate from the cartwheel, first as nine single microtubules, then as doublets and ultimately triplets, which elongate to form the nine blades of the nascent centriole. The cartwheel is common to both the semi-conservative and de novo pathways for centriole genesis. Also, consider that when centrioles occur in pairs they are linked near their proximal ends by a system of fine fibres that show contractile or elastic properties9. When these fibres are taut, the centrioles nestle against one another in the classic orthogonal arrangement, and when the fibres relax or extend, centriole pairs disengage a short distance from one another yet remain connected10. Importantly, the proximal end of centrioles is where the linking fibres are attached and where the cartwheel is first located early in centriole formation. Drawing clues from the behaviour of the yeast spindle pole body (SPB, the yeast centrosome), the fundamental unit of duplication can be defined as a bipartite array of centrin/Sfi1p fibres known as the bridge that links SPB pairs11. It is reasonable to conclude that fibres linking centrioles and those linking SPB are homologous structures, both functionally and in composition12. Now, if the fibres extending between the proximal ends of centriole pairs consist of a bipartite array similar to that of the yeast SPB, and the array splays at the time of the licensing step, their newly freed ends would provide a site near each existing centriole that is predisposed to act as an assembly nucleus for the formation of a cartwheel and ultimately, a nascent centriole. Assembly of a new centriole would be favoured at the centriole-tethered site when the local concentration of cartwheel precursor- molecules reached a critical level. In the case of the de novo pathway, or if the older centriole is eliminated, assembly of centriole precursors would nevertheless proceed, though only when precursors reached a higher local concentration that bypassed the requirement of a tethered assembly nucleus. In this case, centriole precursors would accumulate in a region of the centrosome initially through a dynein microtubule-based mechanism. In this manner, centriole duplication and strict control of centriole number remains within the purview of a pre-existing centriole, and when one is not present, centriole formation can proceed essentially in accordance with the same fundamental assembly mechanism.
References
- 1.Loncarek L, Hergert P, Magidson V, Khodjakov A. Nature Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beisson J, Wright M. Curr. Opin. Cell Biol. 2003;15:1–9. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 3.Bettencourt-Dias M, Glover DM. Nature Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 4.Balczon R, et al. J. Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukasawa K. Nature Rev. Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 6.Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey S. J. Mol. Biol. Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuriyama R, Terada Y, Lee KS, Wang CL. J. Cell Sci. 2007;120:2444–2453. doi: 10.1242/jcs.008938. [DOI] [PubMed] [Google Scholar]
- 8.Sluder G, Nordberg JJ. Curr. Opin. Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Paintrand M, Moudjou M, Delacroix H, Bornens M. J. Struct. Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 10.Tsou MF, Stearns T. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 11.Li S, et al. J. Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salisbury JL. J. Cell Physiol. 2007;213:420–428. doi: 10.1002/jcp.21226. [DOI] [PubMed] [Google Scholar]