Abstract
The human SWI/SNF (hSWI/SNF) ATP-dependent chromatin remodeling complex is a tumor suppressor and essential transcriptional coregulator. SWI/SNF complexes have been shown to alter nucleosome positions, and this activity is likely to be important for their functions. However, previous studies have largely been unable to determine the extent to which DNA sequence might control nucleosome repositioning by SWI/SNF complexes. Here, we employ a minicircle remodeling approach to provide the first evidence that hSWI/SNF moves nucleosomes in a sequence dependent manner, away from nucleosome positioning sequences favored during nucleosome assembly. This repositioning is unaffected by the presence of DNA nicks, and can occur on closed-circular DNAs in the absence of topoisomerases. We observed directed nucleosome movement on minicircles derived from the human SWI/SNF-regulated c-myc promoter, which may contribute to the previously-observed “disruption” of two promoter nucleosomes during c-myc activation in vivo. Our results suggest a model wherein hSWI/SNF-directed nucleosome movement away from default positioning sequences results in sequence-specific regulatory effects.
The precise localization of nucleosomes is one of the most important factors influencing transcription factor binding and gene regulation. ATP-dependent chromatin remodeling complexes are expected to act as transcriptional coregulators at least in part by altering nucleosome positions (1, 2). However, very little is known about the influence of DNA sequence on nucleosome repositioning by these complexes. Human SWI/SNF (hSWI/SNF 1) is an evolutionarily-conserved multi-protein chromatin remodeling complex containing a core catalytic ATPase domain of either BRG1 or hBRM. It functions as a transcriptional coactivator or corepressor when recruited to promoters by over two dozen different activator and repressor proteins, including p53 and Rb as well as virtually all of the nuclear hormone receptors (3, 4). Mammalian SWI/SNF function is necessary for embryonic development and for the differentiation of muscle, adipose and blood cells (3, 5, 6). Mutations or decreased expression of several hSWI/SNF subunits have also been observed in many types of human cancers, and BRG1 mutant heterozygous mice are highly predisposed to cancer formation (3, 7).
Human SWI/SNF activity is important for the transcriptional activation of many genes, including the proto-oncogene c-myc, which is transcriptionally upregulated in many forms of human cancer (8, 9). Chromatin immunoprecipitation experiments in vivo have shown that hSWI/SNF is present on the active c-myc promoter in proliferating T cells, but not the inactive promoter in resting T cells (10). hSWI/SNF is required for β-catenin-dependent activation of c-myc (11), suggesting that it may play a role in colorectal cancer and melanoma in which c-myc upregulation frequently results from uncontrolled activation of β-catenin (8). hSWI/SNF is also a required coactivator for Estrogen Receptor α (12), suggesting that it plays a role in the estrogen-dependent upregulation of c-myc, which has been proposed to be essential for growth of hormone-dependent human breast cancer cells (13). Under some conditions, hSWI/SNF promotes cell cycle arrest, together with a corresponding decrease in c-myc transcription (14–16). While the repressive effect on c-myc is likely to be a secondary effect of arrest, it may indicate a potential role for hSWI/SNF in c-myc repression as well as activation. Importantly, c-myc transcription is associated with chromatin alterations that may result from hSWI/SNF action. Under conditions where c-myc was transcriptionally inactive, low resolution mapping experiments indicated the presence of several positioned nucleosomes covering the c-myc promoter (17–19). Upon c-myc activation two of these nucleosomes situated over the P1 and P2 promoters (which are responsible for ~90% of all c-myc gene transcription (20)) become “disrupted,” meaning that MNase-accessible regions corresponding to linker DNA between nucleosomes could no longer be readily detected by indirect end labeling. One potential explanation for this apparent disruption is that nucleosomes have been moved away from their initially favored positions by hSWI/SNF.
Our prior studies using atomic force microscopy on polynucleosomal DNAs clearly demonstrated nucleosome repositioning by hSWI/SNF (21). In addition, we also found that hSWI/SNF action on polynucleosomal templates resulted in stable changes in restriction enzyme accessibility, causing the most highly nucleosome-occluded sites to become the most accessible and vice versa (21, 22). A similar effect was also seen for yeast SWI/SNF remodeling of a trinucleosomal template (23). While these studies hinted at a possible sequence-specificity in hSWI/SNF-dependent nucleosome repositioning, they provided only very low-resolution information about nucleosome positions. Furthermore, we have shown that hSWI/SNF forms abundant structurally-altered dinucleosomes (altosomes) on polynucleosomal templates, which are characterized by an unusual nuclease accessibility profile (22). Thus, it was possible that altosome formation, rather than repositioning of normal nucleosomes, could have been responsible for the observed changes in restriction enzyme accessibility due to hSWI/SNF action. Attempts to provide more detailed information about how DNA sequence affects nucleosome repositioning by human and yeast SWI/SNF, in our lab as well as others, have employed short-length linear DNA sequences with one or two nucleosomes (24–30). Unfortunately, these studies encountered the problem that nucleosomes are generally repositioned by SWI/SNF to the ends or over the edges of linear DNA fragments, regardless of the sequence used.
Our current study avoids both of the problems associated with earlier studies (altosome formation as well end-positioning effects) by using mononucleosome minicircles to examine the specificity of hSWI/SNF-directed nucleosome repositioning in the absence of DNA ends. Such minicircle templates were previously employed to examine the relationship between DNA topology and nucleosome structure (31), but had not been used for remodeling experiments until now. The results from two c-myc promoter sequence mononucleosome minicircles as well as a third 5S rDNA positioning sequence minicircle, indicate that hSWI/SNF moves mononucleosomes away from initially favored, nucleosome positioning sequences. These observations give new insights into the possible mechanisms by which human SWI/SNF regulates its target genes involved in cell growth, cell-cycle control and cellular differentiation, as well as into general mechanisms of transcriptional control by chromatin remodeling complexes.
Experimental Procedures
Creation of minicircles
C-myc promoter sequences were PCR amplified from human genomic DNA, ligated to CCGGAATTCCGG EcoRI linkers, and subcloned into pGEM 11 (Promega). Primers were, P1_upper TGCGATGATTTATACTCACAGGACAAGGATGCGGT, P1_lower TCGGGGGTCCTCAGCCGTCCAGAC, P1/P2_upper TCGGCTGCCCGGCTGAGTCTCCTC and P1/P2_lower ATTACTACAGCGAGTTAGATAAAGCCCCGAAAACCGG. The Xenopus Borealis 5S rDNA positioning sequence was PCR amplified from pXP10 (32), using the primers GGGGGAAGCTTGTGGAATTGTGAGCGGATAACAATTTCACACAGG (upper) and AACCTTATGTATCATACACATAC (lower). The product was digested with HindIII, at sites internal to the top primer and 54bp upstream of the bottom primer, and subcloned into pBluescript SK. To generate the template with ApaI ends, the 359bp HindIII fragment was circularized using T4 DNA ligase, cut with ApaI and subcloned into pGEM 11. Minicircle sequences were PCR amplified and radiolabeled using primers flanking the plasmid polylinkers and a 1:20 ratio of α32P dATP to cold dATP. The products were digested with EcoRI or HindIII, separated by a 1% agarose gel, excised, and eluted by a QIAquick gel extraction kit (Qiagen).
Minicircle mononucleosome assembly
Nucleosomes were assembled onto the 359bp linear DNA fragments by salt dilution using a 0.8:1 weight ratio of human HeLa cell core histones (33) to DNA, at room temperature, in 2M NaCl, 10mM Tris pH 7.5, 1mM EDTA, 0.1mg/ml BSA, 1mM DTT, and 0.2mM PMSF. Dilutions to 1.25, 0.9, 0.7, 0.6, 0.5, 0.4 and 0.3M NaCl were done by adding the same buffer without salt every 10 minutes. Mononucleosomes were purified from dinucleosomes and bare DNA by ultracentrifugation on a 5% to 30% glycerol gradient, essentially as per Imbalzano et al. (34). Briefly, samples were layered onto a 5ml linear gradient of glycerol gradient buffer (GGB) containing 50 mM Tris pH 7.5, 1 mM EDTA, 0.1mg/ml BSA, 0.2mM PMSF and 5 to 30% glycerol, and samples centrifuged at 35,000 RPM for 16 hrs at 4°C in a Beckman SW55 rotor. The gradient was fractionated into 200 μl aliquots, and peak mononucleosome fractions, identified by 4% polyacrylamide gel electrophoresis (PAGE) and autoradiography, were then exchanged into ligase buffer (NEB) by serial washes in a 10kD Millipore Centrifugal Filter. Ligation was performed at a DNA concentration below 0.3ng/μl (which favors minicircle formation over intermolecular ligation (31)) using T4 DNA ligase at room temperature for 1 hour. Ligated products were separated by 4% PAGE and the wet gel exposed to a Phosphorimager screen at 4° to identify all product locations. The M0 form (mononucleosomal “minicircle octamer”) was excised from the gel and eluted into 5% GGB overnight at 4° in the dark without shaking.
To form fully-closed-circular P1 minicircle mononucleosomes, the bare linear fragment was ligated in the presence of 0.3 – 0.45 μg/μl Ethidium Bromide, which favors formation of −1 supercoiled DNA minicircles (31). After electrophoresis and Phosphorimager analysis, the −1 supercoiled form was excised and eluted at 4°C in TE overnight, followed by concentration using a 10kD Millipore Centrifugal Filter. Minicircle mononucleosomes were then assembled and purified as above. To measure the percentage of assembled minicircle mononucleosomes that were nicked, samples were treated with 0.2% SDS in order to remove histones, before resolution of topological forms by 4% PAGE. To correct for bare DNA circles that were present as a result of dissociation during M0 purification, we subtracted the signal for the −1 supercoiled or nicked/-0 bare DNA forms that were present when the sample was separated by EMSA without SDS (e.g. Fig. 1B) from the signal for that form after SDS treatment. The percentage of nicked minicircle mononucleosomes in the sample was then calculated by dividing the corrected signal for the -0/nicked forms by the total corrected signal for the -0/nicked and −1 forms and multiplying by 100%.
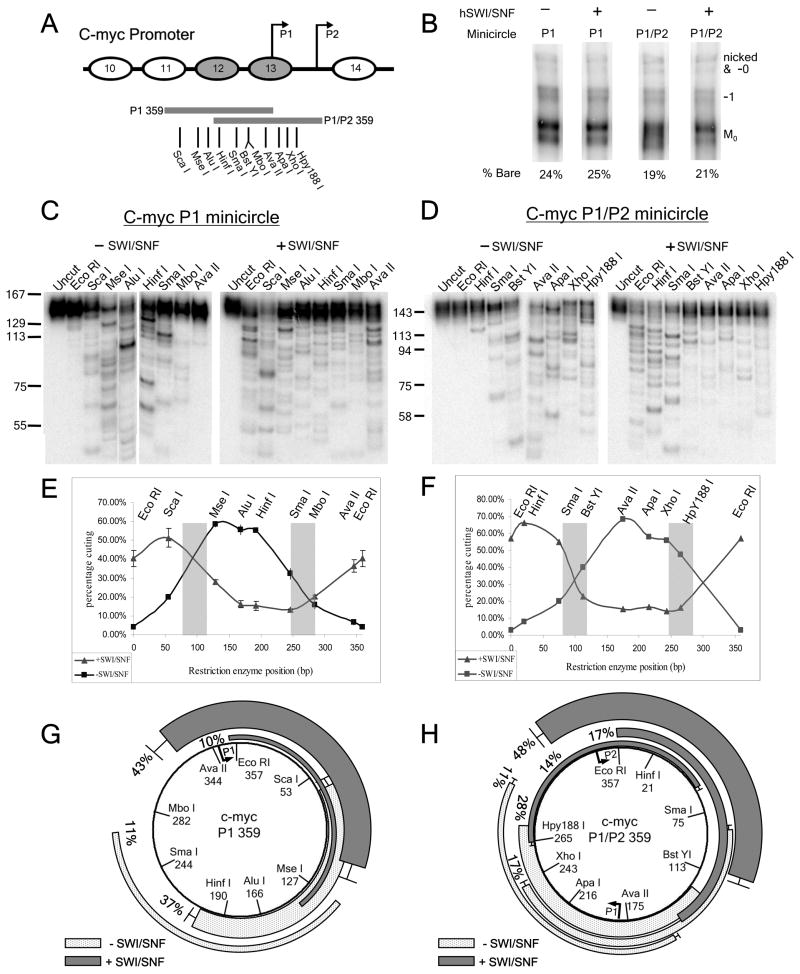
FIGURE 1. hSWI/SNF moves nucleosomes away from c-myc promoter positioning sequences.
(A) C-myc promoter map based on in vivo mapping results (17–19). Shaded ovals represent the approximate positions of nucleosomes present on the repressed promoter that are disrupted upon c-myc activation. The P1 and P1/P2 minicircle sequences are represented by gray bars, with the ocations of restriction sites indicated. (B) hSWI/SNF remodeling does not cause minicircle mononucleosome dissociation. Samples of hSWI/SNF-remodeled (+) and control (−) reactions on P1 and P1/P2 minicircles were treated with competitor DNA & polynucleosomes to eliminate hSWI/SNF binding (see Experimental Procedures), before 4% non-denaturing PAGE. The positions of the minicircle octamer (Mo) and bare DNA forms (−1 supercoiled and -0/nicked forms) are indicated on the right. The percentage of bare DNA was calculated as the signal due to bare DNA forms divided by the total amount of signal for the lane times 100%. Note that incubation in hSWI/SNF reaction buffer alone does not promote dissociation, since the percentage of bare DNA in the control reaction was the same as for a sample of the mononucleosome preparation immediately after gel isolation (data not shown). (C&D) Restriction enzyme digestion products for the 146 bp MNase fragments of the P1 and P1/P2 minicircle mononucleosomes with and without (+ or −) SWI/SNF remodeling. (E&F) Percent cutting graphs corresponding to the data in B & C. Shaded bars denote the approximate edges of c-myc nucleosomes 12 and 13 from the in vivo mapping studies. For B, the results show the average and standard error for two independent experiments. (G&H) Maps of each nucleosome position representing ≥10% of the P1 or P1/P2 minicircles. Light gray arcs; positions before hSWI/SNF treatment. Dark gray arcs; positions after hSWI/SNF remodeling. Bars at the ends of an arc indicate locations where overlapping nucleosome positions were within 10bp of each other.
SWI/SNF remodeling and MNase reactions
hSWI/SNF was purified from HeLa cells by immunoaffinity chromatography against the FLAG-tagged Ini1 subunit (35). 25ng of minicircle samples (~1.1nM) were reacted with 1.25μg hSWI/SNF (~6.3nM) in 100μl reactions containing 0.4μg/μl BSA, 65mM KCl, 0.5mM ATP, and 2.5mM MgCl2. For some reactions, wheat germ Topoisomerase I (Promega) was added at a concentration of 0.1 units/μl. The reactions were incubated at 30°C for 2.5 hours, and hSWI/SNF remodeling stopped by the addition of ADP to a concentration of 10mM. Time course studies, using EMSA to resolve different nucleosome positions after hSWI/SNF remodeling (similar to Figs. 3A & 3B), indicated that these remodeling conditions produce maximal changes in nucleosome positions after ~10 minutes, with no further changes apparent after longer incubation ((24), and data not shown). These observations, together with the observation that remodeled positions are the same regardless of nucleosome starting positions (Figs. 3 & 4), indicate that the remodeling conditions used here allow the repositioning reaction to reach an effective equilibrium, resulting in nucleosome positions that are most favored by hSWI/SNF. Note that, in preliminary studies on polynucleosomal templates, we see similar sequence-directed repositioning at a hSWI/SNF to nucleosome ratio of ~1:9, arguing that the effects seen in these minicircle experiments are not an artifact of the higher hSWI/SNF:mononucleosome ratio used to ensure complete remodeling (HS & GRS, unpublished observations). For MNase digestion reactions: the buffer was adjusted to 1mM MgCl2, 3mM CaCl2, 60mM KCl, 0.7μg/μl BSA, 0.1mM ATP, and 2mM ADP in a total volume of 660μl. The reactions were pre-warmed for several minutes at 30°C, digested with 0.2 units/μl micrococcal nuclease (MNase, Roche unit definition) for 5 minutes, and stopped by adjusting the buffer to 0.2% SDS and 15mM EDTA. MNase products were phenol extracted, ethanol precipitated in the presence of 50μg glycogen carrier, and separated by 4% PAGE. 146 +/− ~3bp mononucleosomal products were then excised, eluted by continuous shaking at 37°C in TE overnight, and concentrated using a 10kD Millipore Centrifugal Filter.
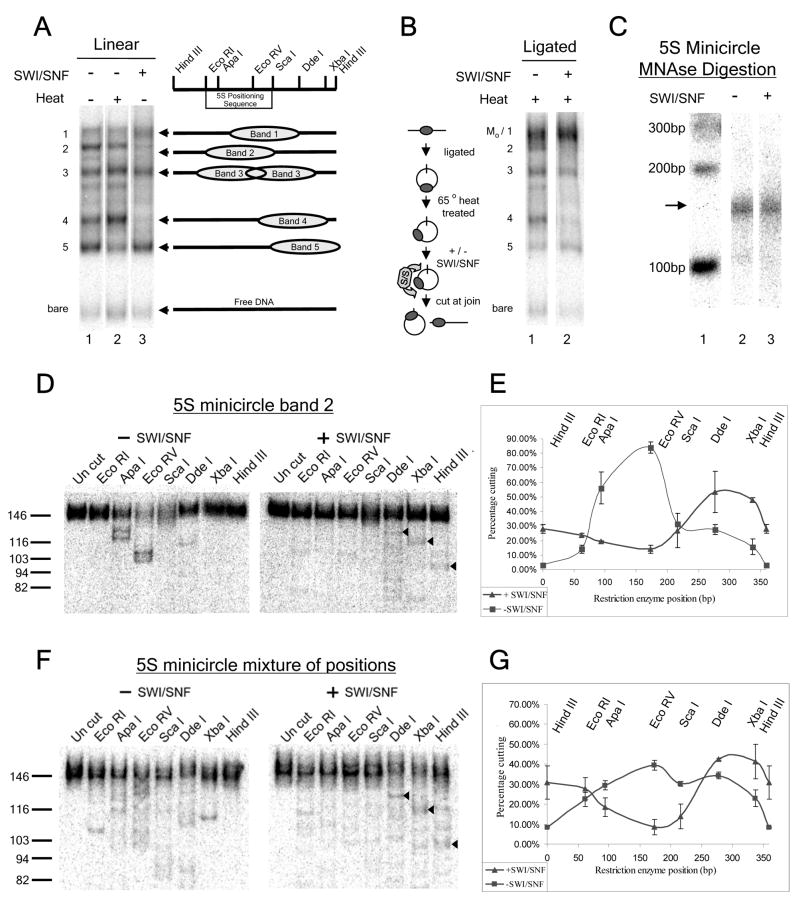
FIGURE 3. hSWI/SNF moves nucleosomes away from a 5S positioning sequence, and independent of starting position.
(A) Mononucleosomes assembled onto the linear 359 bp Xenopus borealis 5S rDNA template with HindIII ends were purified by gradient ultracentrifugation. These were incubated in the absence of hSWI/SNF (lane 1), the presence of SWI/SNF and ATP (lane 3), or heated to 65°C for 10 minutes, to promote movement to thermally-favored positions (lane 2). Numbers on left; the five major gel shift positions/bands present after assembly. Cartoon on right: the nucleosome positions corresponding to each assembly-favored gel shift band, as determined by elution from the gel and MNase/restriction enzyme mapping (data not shown) (B) Ligated templates were heat treated, followed by incubation without hSWI/SNF (lane 1) or with hSWI/SNF (lane 2). hSWI/SNF activity was inhibited by addition of competitor DNA and chromatin, followed by HindIII digestion to release linear products. Cartoon on left: Experimental outline. Mo indicates the position of the uncut minicircle mononucleosome (“minicircle octamer”), which comigrates with linear band 1. (C) 5S minicircle mononucleosomes ligated at the HindIII site were treated without hSWI/SNF (lane 2) or with hSWI/SNF (lane 3), followed by addition of ADP to stop remodeling before MNase digestion (see Experimental Procedures). MNase footprint products were resolved by 4% PAGE, and the wet gel exposed to a Phosphorimager screen for 30 minutes at 4°C to identify ~146 bp mononucleosomal footprint products (arrow on left), which were subsequently excised from the gel and eluted. Lane 1: 32P end-labeled 100 bp ladder. (D–G) 359bp Xenopus borealis 5S rDNA minicircles with (D) almost all histone octamers localized to the 5S positioning sequence (from ligation of band 2 mononucleosomes) or with (F) multiple octamer positions resulting from circularization of an even mixture of linear mononucleosomes isolated from bands 1 through 5, were treated with or without hSWI/SNF, and nucleosome positions mapped as described in Fig. 1. Arrowheads (◂) highlight strong bands observed after hSWI/SNF remodeling on both minicircles. (E&G) Percent cutting plots showing the averages and standard errors for the results shown in D & F as well as a duplicate experiment. For (D), the signal was increased by kinase labeling MNase fragments with γ32P-ATP before isolation of 146bp products. Some DNA contaminants in the glycogen carrier were also labeled, giving rise to copurifying ~146bp contaminant DNAs. We corrected for the percentage of signal due to these contaminants by subtracting the ~146bp signal remaining after digestion of the MNase products with all seven restriction enzymes at once. The presence of this contamination did not obscure the underlying result, as indicated by the similarity between the experiment shown in (D) and a duplicate experiment where fragments were not kinase labeled (error bars in (E)).
FIGURE 4.
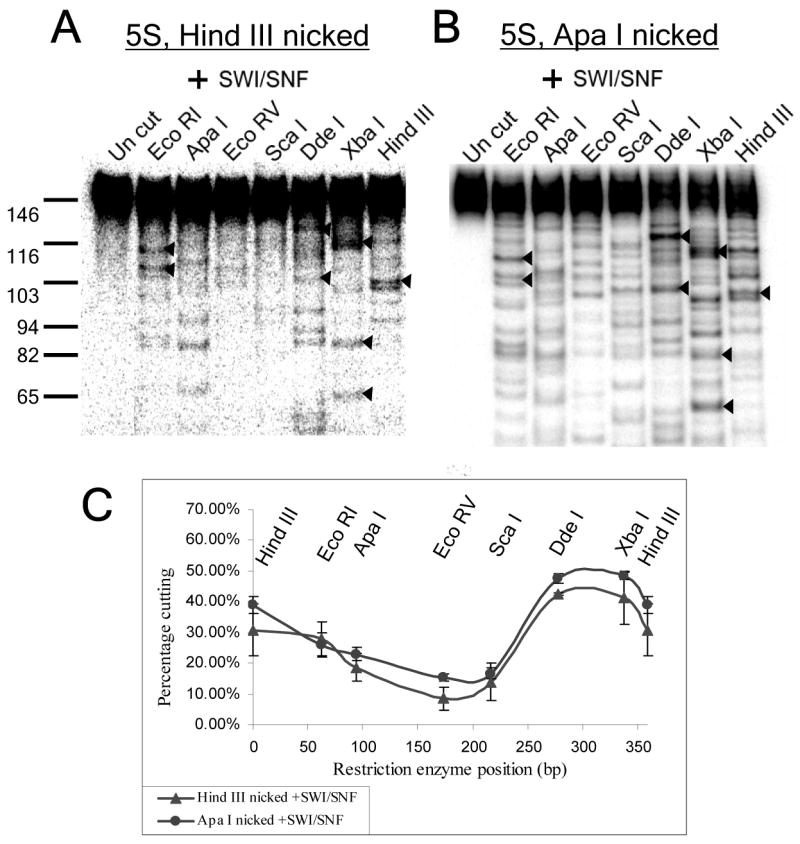
The distribution of remodeled nucleosomes is not altered by nucleosome starting position or the position of a DNA nick. (A) Restriction digestion pattern for hSWI/SNF-remodeled 5S minicircles, 33% nicked over the Hind III site of ligation. (B) Restriction digestion pattern hSWI/SNF-remodeled 5S minicircles formed by ligation of a 359bp mononucleosome with ApaI ends, and, hence, 28% nicked over the ApaI site of ligation, 94bp away from the HindIII site. Arrowheads (◂) indicate shared major bands. (C) Percentage cutting graphs showing the averages and standard errors for the results shown in A & B as well as one duplicate experiment.
Restriction enzyme digestion and analysis of nucleosome positions
Typically, for each restriction enzyme, >500 cpm of labeled ~146bp DNA was digested for 4hrs in 20μl reactions under supplier-specified ideal reaction conditions and using 10–20 units of each enzyme (enzymes were from New England Biolabs, with the exception of EcoRI, HinfI and EcoRV from InVitrogen and SmaI from Promega). The products were then separated by 8% PAGE along with a radiolabeled 100bp NEB DNA ladder, and the gels were dried. Reactions containing full-length 359bp linear DNA fragment digested with the same enzymes were also included as a control for complete restriction enzyme cleavage. The dried gels were exposed to a Phosphorimager screen, and quantitated using ImageQuant software. To determine percent cutting, the signal for all restriction enzyme-digested products (bands smaller than the 146bp MNase-digested input DNA) was divided by the total signal in the lane. To map individual nucleosome positions, band lengths were calculated by comparison to the ladder and/or digestion control lanes using FluorChem 8800 software. Restriction sites were chosen (spaced every ~50bp apart) such that any nucleosome position must cover at least two sites, which allows nucleosome positions to be mapped by comparing bands from adjacent restriction sites. To determine the fraction of nucleosomes occupying each position, the signal from the larger of each pair of bands in a lane (>73bp) was multiplied by 146 and divided by the length of the larger fragment. This calculation corrected for the intensity of the smaller band, simplifying the analysis and also accounting for fragments smaller than ~20bp which were not resolved by the gel. The percentage of nucleosomes in any position was then calculated by dividing this corrected intensity by the signal for the entire lane and multiplying by 100%.
Gel shift analysis of nucleosome positions resulting from assembly, hSWI/SNF or heat treatment
For linear 5S 359 bp mononucleosomes, standard remodeling reactions were incubated with or without hSWI/SNF for 1 hour at 30°C, or heated to 65°C for 10 minutes (to promote movement to thermally-favored positions). Remodeling was then stopped, and hSWI/SNF removed from the template, by addition of competitor DNA and chromatin (1μg each of HeLa cell polynucleosomes and unrelated, supercoiled plasmid DNA). The different nucleosome positions were then resolved by 5% PAGE. To analyze minicircle mononucleosomes, 5S 359bp linear mononucleosomes with HindIII ends were incubated with T4 DNA ligase in hSWI/SNF buffer for 1 hour, followed by heat treatment at 65°C for 10 minutes (to both inactivate the ligase and promote thermally-favored nucleosome positions). The ligated template was then incubated at 30°C for 1 hour with or without hSWI/SNF, followed by addition of 1μg each of competitor DNA and polynucleosomes to stop the remodeling reaction. Reactions were then digested with 10 units of HindIII at 30°C for 1 hr, followed by PAGE, to examine the nucleosome positions of the linear forms released by HindIII digestion. To determine the fraction of templates that remained circular, a sample of each reaction was treated with 2%SDS and proteinase K, followed by 5% PAGE to resolve circular and linear forms.
Results
Two 359bp DNA fragments from the P1 and P2 region of the hSWI/SNF-regulated c-myc gene promoter were PCR amplified and radiolabeled using a 1:20 ratio of α32P dATP to cold dATP. These fragments were centered over the approximate in vivo locations of two promoter nucleosomes that were previously shown to be “disrupted” upon c-myc activation (Nucleosome 12, P1 359, & nucleosome 13, P1/P2 359, in Fig. 1A, (17, 18)). These linear fragments with EcoRI ends were assembled into nucleosomes by salt dilution, and the purified mononucleosomes ligated to form minicircles (referred to as Mo, for “minicircle octamers”). The 359 bp length allows mononucleosome formation and also permits the DNA ends to bend back together and form a non-strained circle of normal B form DNA (31). To map nucleosome positions, the purified mononucleosome minicircles were digested with MNase in order to produce mononucleosome-size 146bp fragments. These fragments were purified by gel electrophoresis and then subjected to restriction enzyme digestion using enzymes with unique sites spaced ~50 bp apart throughout the DNA sequence.
The percentage of mononucleosome minicircles that had a nucleosome over any given restriction enzyme site can be determined by measuring the percentage of 146bp MNase fragments that are cut by each restriction enzyme. For example, the 146bp nucleosome fragments from the P1 minicircle, when digested with AluI, produced one strong band and several weaker bands below 146bp, which together represented 60% of the total DNA in the lane (Fig. 1C, “−SWI/SNF”). Percent cutting for each restriction enzyme can be graphed to give a curve representing the proportion of minicircles in each reaction that had a nucleosome covering each site (Fig. 1E & F, “−SWI/SNF”). Note that previous in vivo mapping studies provided information about the locations of linker regions to either side of c-myc promoter nucleosomes 12 and 13, at an estimated resolution of +/−20 bp (shaded bars in Figs. 1E and 1F, (17, 18)). The percent cutting values for the P1 and P1/P2 minicircles indicate that a high percentage of nucleosomes are assembled over approximately the same locations in vitro. This close correspondence between in vivo and in vitro nucleosome positions indicates that these sequences are functional nucleosome positioning sequences.
The specific locations of individual nucleosomes on the minicircles can be mapped to a resolution of +/−5 bp by measuring restriction fragment lengths and intensities for adjacent restriction sites. For example, 37% of the nucleosomes formed on the P1 minicircle give rise to the intense bands in the AluI lane at 104bp and in the HinfI lane at 135bp (Fig. 1C, “−SWI/SNF”). This and other major assembly positions of nucleosomes (≥10% of the total) are shown as light gray arcs in Figs. 1G and 1H. Minor positions representing 5–10% of nucleosomes also localized to the same region of the DNA, but for simplicity are not shown in Figs. 1G–H. In addition, while we could not reliably map bands representing less than 5% of the total, almost all of these weaker bands were found near the major bands. Well-characterized naturally-occuring nucleosome positioning sequences (NPSes), such as the 5S rDNA NPS, tend to result in several assembly-favored positions clustered around a few most highly-favored positions (36–40). Accordingly, our results are consistent with the presence of locally strong NPSes near the centers of the 359 bp c-myc P1 and P1/P2 promoter sequences.
hSWI/SNF moves nucleosomes away from positioning sequences on c-myc minicircles
Minicircle mononucleosomes were treated with hSWI/SNF and ATP under conditions that allow for the hSWI/SNF-driven repositioning reaction to reach effective equilibrium (see Experimental Procedures). Accordingly, these experiments do not measure the initial positions adopted after the first hSWI/SNF-driven movement event, but instead are designed to determine the DNA sequences at which hSWI/SNF intrinsically prefers to leave nucleosomes after complete remodeling. To eliminate any effect of remodeling during the MNase digestion reaction, hSWI/SNF activity was then inhibited by using a 20-fold molar excess of ADP, which competitively inhibits SWI/SNF ATPase function (22, 34, 41). For control reactions, minicircles were treated under equivalent conditions except that hSWI/SNF was omitted, or ADP was added to prevent remodeling before addition of SWI/SNF.
Strikingly, for both the P1 and P1/P2 minicircles, the percent cutting analysis showed that nucleosomes were almost entirely moved from their original positions by hSWI/SNF, and now covered sites on the opposite side of the minicircle (Figs. 1E & 1F “+SWI/SNF”). Fine-scale mapping indicated that hSWI/SNF remodeling results in movement of mononucleosomes to a few strong and several weak positions, which are all far removed from the assembly-preferred positions established by the Nuc 12 and Nuc 13 positioning sequences (Figs. 1G & 1H, dark gray bars). In no case was an assembly-preferred position present after remodeling on more than 5% of the minicircles. These results suggest that nucleosome repositioning by hSWI/SNF is strongly influenced by DNA sequence, and that hSWI/SNF may preferentially relocate nucleosomes away from positioning sequences. Note that, control experiments showed that this hSWI/SNF treatment did not increase the percentage of bare DNA minicircles, indicating that remodeling did not result in removal of histones from the template (Fig. 1B).
The final position of nucleosomes on minicircles is not a favored position for nucleosome assembly
Ligation of the linear 359bp fragment to form a minicircle could potentially generate a new nucleosome positioning sequence from the DNA on either side of the joint. If this was the case, then hSWI/SNF might simply be moving nucleosomes from the known positioning sequence to a stronger, newly-formed positioning sequence. While the fortuitous formation of new positioning sequences on both c-myc DNA minicircles seemed unlikely, we wanted to formally rule out this possibility. To do so, fully-closed-circular bare DNA minicircles were created by ligating the P1 linear DNA in the presence of ethidium bromide, which intercalates into the DNA backbone resulting in negative supercoiling of the ligated product DNA. The −1 supercoiled form was purified by PAGE, assembled by salt dialysis, and the mononucleosome minicircle isolated by elution from PAGE. When assembly-favored nucleosome positions on these minicircles were mapped, the major positions were very similar to those seen for the P1 minicircle mononucleosomes assembled before ligation, indicating that no strong positioning sequence was formed at the joint sequences (Compare “−SWI/SNF” results for the P1 template in Fig. 1 and Fig. 2). Assembly onto the circle did allow some low-abundance nucleosome positions to form over and around the EcoRI site (e.g. 20% cutting in Fig. 2B, “−SWI/SNF”). However, the great majority of nucleosomes were still observed to form at the same assembly-preferred positions observed for assembly onto the linear template in Fig. 1. Accordingly, these results confirmed that hSWI/SNF was not simply moving nucleosomes to a higher-affinity positioning sequence that was fortuitously formed at the minicircle ligation site, instead it confirms that the preference for nucleosome movement is away from the high-affinity positioning sequence on the minicircle.
FIGURE 2. hSWI/SNF moves nucleosomes away from c-myc positioning sequences on fully closed-circular minicircles.
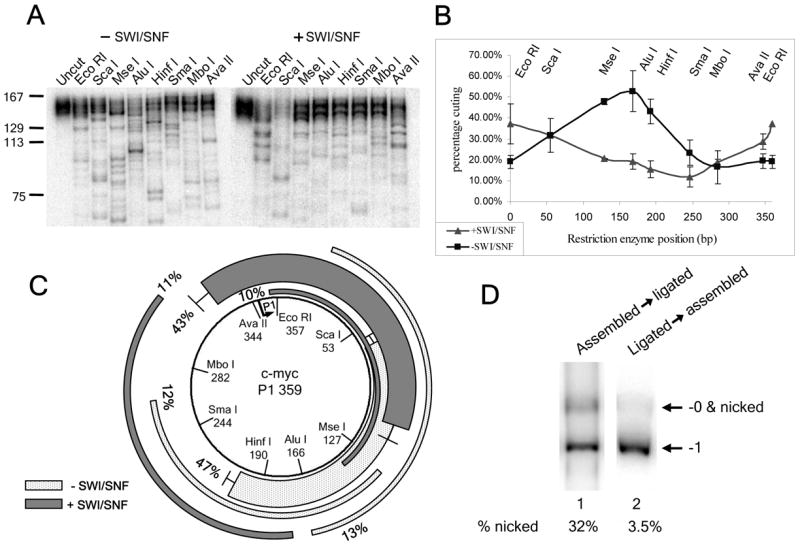
Mapping results for control and remodeled fully-closed circular –1 supercoiled P1 minicircles: (A) restriction digestion gels, (B) percentage cutting (showing the average values and standard error for two independent experiments) & (C) maps of major positions, as described for Fig. 1. (D) Minicircle mononucleosomes derived from assembly onto the linear P1 template (lane 1) or circular P1 template (lane 2) were treated with SDS before PAGE, to measure the percentage of nicked/and or -0 closed circular DNA forms (which co-migrate on the gel). Note that, this DNA was associated with gel-isolated minicircle mononucleosomes. Given that the histone octamer constrains one negative supercoil, there should be virtually no mononucleosomal -0 closed circles. Thus, under the conditions used here, the percentage of nicked templates is expected to be almost equal to the percentage of templates that migrate as nicked/-0, after subtracting non-SDS treated control signal (see Experimental Procedures).
The presence or location of a nick on minicircles does not affect nucleosome repositioning by hSWI/SNF
As a result of the inherent inefficiency of DNA ligase, when linear 359bp templates were circularized after mononucleosome assembly, a single DNA nick at the ligation site was left on about one third of the minicircles. This could be observed by SDS treatment, which removes histones from the DNA, followed by PAGE (Fig. 2D, lane 1, 32% nicked). By contrast, assembly onto closed-circular bare DNA yielded mononucleosome minicircles that were only 3.5% nicked (Fig. 2D, lane 2). Theoretically, the presence of a DNA nick might alter the positions favored after remodeling. For instance, a nick might block repositioning past it, resulting in nucleosomes stalled at specific positions to the left and right of the ligation site on nicked templates. To test whether this was the case, we compared the frequencies of individual nucleosome positions after remodeling on the 32% nicked (linear assembly) and 3.5% nicked (circular assembly) P1 templates (Figs. 1G and 2C). We found that the hSWI/SNF-favored positions were very similar in both cases, with the most noticeable difference being a position representing 11% of the total in Fig. 2C (leftmost dark grey bar) that was only 8% in the experiment shown in Fig. 1 (and thus did not make the 10% cut-off to be displayed in Fig. 1G). These results argue against any strong effects of a DNA nick on the final preferred locations of hSWI/SNF-remodeled nucleosomes. This is consistent with recent studies using nicked linear templates, which indicated that repositioning by hSWI/SNF and other remodeling complexes was not greatly effected by single DNA nicks (27, 30, 42, 43).
Repositioning of nucleosomes on minicircles by SWI/SNF occurs despite topological constraints
On closed-circular templates, twisting of the DNA comes at an energetic cost that is inversely proportional to the length of the DNA circle (44). As a result, chromatin remodeling reactions that produce unconstrained supercoils can be inhibited unless a topoisomerase is included in the reaction to relax the torsional strain. Specifically, on circular multinucleosomal templates, topoisomerase action was shown to be important for the ability of yeast and human SWI/SNF to promote the altosome-associated loss of nucleosome-constrained supercoils (22, 23, 41), and for yeast SWI/SNF to transiently increase restriction enzyme accessibility to nucleosome-occluded sites during ongoing ATP-hydrolysis (23). Intriguingly, we found that the effects of hSWI/SNF on minicircle mononucleosome positions was identical in reactions lacking or containing wheat germ topoisomerase I (TopoI), which relaxes both unconstrained positive and negative supercoils (Fig. 1 and data not shown). Even more strikingly, TopoI was not required for repositioning on a mononucleosome minicircle that was 97% closed circular (Fig. 2 “+SWI/SNF”). Control experiments indicated that no topoisomerase activities were associated with our purified hSWI/SNF (data not shown). Thus, these results indicate that stable hSWI/SNF-driven nucleosome repositioning is possible on topologically-constrained templates where generation of unconstrained supercoils has a high energetic cost. The apparent difference in toposomerase requirement between SWI/SNF-dependent repositioning (this work) and transient restriction enzyme accessibility (23) could potentially arise from differences in reaction conditions. For instance, it is possible that repositioning, like transient restriction enzyme accessibility, is also slowed down in the absence of TopoI, but that this was not detected under the equilibrium remodeling conditions used in our studies. Alternatively, our results may indicate that transient restriction enzyme accessibility does not arise solely from ongoing nucleosome repositioning (which would create moments where restriction sites are in accessible linker DNA), but also from the creation of a transient SWI/SNF-remodeled nucleosomal species whose formation is inhibited by torsional strain and which is not required for repositioning.
hSWI/SNF demonstrates directed nucleosome movement away from the 5S rDNA positioning sequence
Analysis of our c-myc minicircles indicated that hSWI/SNF may have an intrinsic preference to move nucleosomes away from positioning sequences. To further examine this positioning effect, we made use of the well-characterized positioning sequence from the somatic 5S rDNA of Xenopus Borealis (32). We assembled mononucleosomes onto a 359bp DNA fragment containing the 5S positioning sequence. Linear mononucleosomes with histone octamers at different positions were then separated by polyacrylamide gel electrophoresis (PAGE), in which fragments bearing a centrally-located nucleosome migrate more slowly than fragments bearing a nucleosome at the end of the DNA (Fig. 3A, lane 1, (24, 45)). Each of the five major mononucleosomal gel-shift positions were excised, eluted, and mapped by MNase and restriction enzyme digestion (Fig. 3A, cartoons on the right). The linear species with a nucleosome localized over the 5S positioning sequence (band 2 in Fig. 3A, (32)), was circularized, and the circular form purified. These 5S minicircle mononucleosomes were then subjected to remodeling by hSWI/SNF. In the control reaction, 88% of these minicircles had nucleosomes in two overlapping positions covering the 5S positioning sequence and separated by ~10bp (Figs. 3D & E “−SWI/SNF”). After remodeling by hSWI/SNF, it was observed that the majority of nucleosomes had been moved to a new location away from the positioning sequence, and that the area containing the positioning sequence had become the least occupied region of the minicircle (Fig. 3D & E, “+SWI/SNF”). Together with the previous c-myc minicircle data, these results suggest that hSWI/SNF has an intrinsic preference to move nucleosomes away from positioning sequences.
hSWI/SNF does not generate structurally-altered nucleosomes on minicircles
Previous studies from our lab showed that, on polynucleosomal templates, hSWI/SNF generates abundant “altosomes” consisting of structurally-altered dinucleosomes with altered histone-DNA contacts and reduced nucleosome-constrained negative supercoiling (22). One of the advantages of the mononucleosome minicircle system is that altosomes cannot be formed from a single nucleosome. Indeed, the characteristic ~220bp and ~80bp altosomal MNase products were not observed upon MNase digestion of hSWI/SNF-remodeled mononucleosome minicircles (Fig. 3C, and data not shown). Thus, use of mononucleosome minicircles has allowed us to examine the effects of DNA sequence on hSWI/SNF-directed nucleosome repositioning, uncomplicated by other hSWI/SNF remodeling effects.
hSWI/SNF-favored positions differ from thermally favored positions
Several studies have indicated that nucleosome positions established by salt dialysis in vitro can accurately represent in vivo nucleosomes positions. For instance, the sequence characteristics determined for high affinity nucleosomes generated by salt dialysis assembly onto yeast genomic, mouse genomic or random sequence DNA are very similar to those for naturally-occurring nucleosome positions from yeast and chicken genomic chromatin (see (46) for comparison of all these conditions). However, nucleosome positions established during salt-dialysis assembly can be altered by treatment at elevated temperatures (e.g. (24, 25, 45, 47–50)), suggesting that mammalian body temperature might promote some level of nucleosome redistribution in vivo. Thermal repositioning is most rapid at high temperatures (up to 65°C) and in the absence of divalent cations or linker histones (45, 48, 51, 52)). In general, thermal repositioning appears to result in redistribution between sites that are all at least moderately-favored during nucleosome assembly (causing increases in some moderately favored positions and decreases in some strongly favored positions). Evidence also suggests that thermal repositioning may sometimes favor movement of the histone octamer towards DNA ends (53).
Given the above characteristics of thermal repositioning, we did not expect that hSWI/SNF favored positions on minicircles (which are always at locations seen at a very low frequency after assembly) would be the same as thermally favored positions. To test this directly, we used EMSA analysis to compare thermally favored and hSWI/SNF-favored positions on both linear and circular mononucleosome templates. While this approach does not give detailed information about individual nucleosome positions, it is an efficient way to compare the effects of each treatment on the overall distribution of nucleosomes. As shown in Fig. 3A, lane 2, 65°C heat treatment of linear 359 bp 5S mononucleosomes induces thermal repositioning, resulting in decreases in bands 1, 2 & 5 and increases in bands 4 and 3, relative to the unheated control sample in lane 1. hSWI/SNF remodeling of the linear template gave positions that differed from heat treatment, resulting in loss of bands 2 and 4, and accumulation of band 5 nucleosomes at the end of the template (Fig. 3A, lane 3). Since DNA ends are known to alter the positions of hSWI/SNF products, and might also influence heat-treatment positions, we next compared the effects of these treatments on minicircle templates, in the absence of DNA ends. To do this, we performed a ligation and recutting experiment (as outlined in the cartoon in Fig. 3B). Briefly, linear mononucleosomes were ligated to form circles. These minicircle mononucleosomes were heat treated at 65°C to promote thermal repositioning to heat-favored sequences, as well as to inactivate the ligase. Next, these ligated templates were treated with ATP alone or ATP and hSWI/SNF. Remodeling was then stopped, and hSWI/SNF removed from the template, by addition of a vast excess of competitor chromatin and DNA (1 μg of each), followed by digestion with HindIII. On all templates where the histone octamer does not cover the HindIII site, this results in release of linear mononucleosomes, allowing relative nucleosome positions to be compared. For the reaction lacking hSWI/SNF (Fig. 3B, lane 1) heat treatment followed by HindIII digestion released a pattern of bands similar to the heat-treated linear sample, with bands 3 and 4 being strongest. This indicates that the same positions are preferred by heat treatment on both the linear and circular templates. To determine the percentage of templates where the mononucleosome blocked HindIII access, leaving the “Mo” circular form, a sample of the reaction was treated with SDS to remove histones, and circular and linear forms separated by PAGE (data not shown). This showed that 36% of the heat-treated, unremodeled templates remained circular, indicating that heat treatment also promoted movement of some octamers over the HindIII site. hSWI/SNF treatment of the ligated, heat-treated minicircles (Fig. 3B, lane 2) resulted in a very different pattern, in which all bands except band 1/Mo and band 5 were decreased (with the greatest decrease in the heat-favored band, band 4). The percentage of uncut templates was also increased to 52%. This increase in fast migrating band 5 (nucleosomes near the DNA edge) and nucleosomes over the HindIII site is consistent with the 5S minicircle mapping results shown in Fig. 3D & 3E, “+SWI/SNF”. Importantly, while some gel shift positions (e.g. band 3) were similar under both conditions, the overall pattern indicates that heat treatment and hSWI/SNF result in very different distributions of nucleosome positions on the 5S minicircle. These results indicate that hSWI/SNF does not move nucleosomes to low energy binding sites preferred either by thermal repositioning or by salt dialysis assembly.
The final positions of nucleosomes on minicircles are independent of the nucleosome starting positions
The mapping experiments in Figs. 1 and 3D all started with the majority of the nucleosomes localized to a specific region of each template (the apparent nucleosome positioning sequences). We wanted to determine whether nucleosomes would move to the same hSWI/SNF-remodeled positions if they started at different initial positions. To test this, we combined equal amounts of linear 5S mononucleosomes isolated from each of the gel-shift bands 1 through 5, followed by ligation and minicircle isolation. This created an even mixture of nucleosome starting positions (Fig. 3F & 3G “−SWI/SNF”, note the roughly equal cutting of all sites except for the HindIII ligation site). When this mixture was treated with hSWI/SNF, we found that the nucleosome positions were very similar to those seen after remodeling the minicircle with a nucleosome localized over the 5S positioning sequence (Compare Fig. 3F & G to Fig. 3D & E, “+SWI/SNF”). Similar results were also seen upon hSWI/SNF remodeling of minicircles formed from purified “band 4” 5S mononucleosomes, on which the histone octamer is shifted ~130bp away from the 5S positioning sequence (Fig. 3A, and data not shown).
To explore this issue further, we cut the 5S minicircle with ApaI, subcloned this linear fragment, and used the resulting vector to prepare a 359bp template with ApaI ends. Nucleosome assembly followed by ligation of this template resulted in mononucleosome minicircles that were identical in sequence, but 28% nicked at the ApaI site (95bp from the normal Hind III ligation site, see cartoon in Fig. 3A). Note that, since the 5S positioning sequence was not intact on the ApaI linear template, it was impossible for nucleosomes to be assembled over this preferred sequence. When this template was remodeled by hSWI/SNF, the restriction enzyme digestion pattern and percent cutting graphs were essentially identical to those seen for hSWI/SNF treatment of mononucleosome minicircles formed from the linear 5S template with HindIII ends, which was 33% nicked at the Hind III site (Fig. 4). This further indicates, together with the results in Fig. 2, that the presence and/or location of a single DNA nick does not greatly alter hSWI/SNF repositioning specificity. In addition, the combined results from Figs. 3D–G and Fig. 4 show that the same remodeled positions are observed regardless of nucleosome starting positions. This argues that the positions observed after remodeling do not represent initial repositioning events, for which remodeled positions might be expected to remain nearby starting positions. Instead, these results indicate that the observed repositioning of nucleosomes to DNA sequences away from assembly favored positions represents a hSWI/SNF-favored equilibrium state, which is independent of nucleosome starting positions.
Discussion
The question of how DNA sequence directs nucleosome repositioning by SWI/SNF complexes has long remained unanswered, largely due to the strong tendency of SWI/SNF complexes to move histone octamers to the ends of linear DNA fragments. Our results on mononucleosome minicircles provide the first evidence for sequence-directed nucleosome movement by any SWI/SNF-family remodeling complex, and indicate that hSWI/SNF moves nucleosomes away from assembly-favored positions established by nucleosome positioning sequences. They also indicate that hSWI/SNF-preferred positions differ from thermally-favored nucleosome positions. Together with previous studies showing that SWI/SNF action on polynucleosomes can stably increase restriction enzyme accessibility at sites normally covered by nucleosomes (21–23), our results suggest that nucleosome movement away from positioning sequences will be a general property of hSWI/SNF-remodeled chromatin.
We find that hSWI/SNF action on c-myc promoter minicircles directs nucleosome movement away from assembly-favored positions corresponding to the approximate locations of nucleosomes 12 and 13 from the repressed c-myc promoter in vivo. This suggests that sequence-driven nucleosome repositioning by hSWI/SNF contributes to the observed “disruption” of these two nucleosomes upon c-myc activation. The apparent “disruption” of these two nucleosomes might also arise from the conversion of the nucleosomes to an altered nucleosomal form or removal of nucleosomal histones from the DNA, activities that have also been demonstrated for human and yeast SWI/SNF complexes. However, of the different types of SWI/SNF remodeling activity, repositioning is likely to be one of the most important, since in vitro comparisons indicate that altered nucleosome formation and histone removal often affect a smaller fraction of nucleosomes or are slower than repositioning (1, 2, 22, 54).
One favored model for nucleosome repositioning argues that the translocation of a remodeling complex ATPase domain across the DNA phosphate backbone gives rise to a DNA loop or bulge on the nucleosomal surface, whose movement to the other side of the histone octamer results in repositioning (55–57). Our results demonstrate that nucleosome repositioning can occur on a topologically closed minicircle in the absence of topoisomerase action. This suggests that the formation of the initial DNA loop may be able to proceed without significant DNA twisting. Alternatively, the energy afforded by ATP hydrolysis may be high enough to allow the formation of a supercoiled DNA loop, despite the high energetic cost of the unconstrained supercoils that would also be generated on these minicircle templates (44).
Two recent studies have indicated that ~50% of yeast nucleosome positions are specified by a nucleosome positioning sequence code, indicating that the control of nucleosome positions by DNA sequence is an unexpectedly common event (46, 58). Our results suggest a model in which hSWI/SNF-activated gene promoters, such as c-myc, have been evolutionarily selected to encode nucleosome positioning sequences that place nucleosomes over important transcription factor binding sites. These nucleosome-occluded sites would then be opened up by hSWI/SNF-dependent repositioning away from these positioning sequences, when the complex is recruited to the promoter by transcriptional activators. Consistent with this model, we find that hSWI/SNF-directed nucleosome movement away from the nucleosome 13 positioning sequence uncovers the P1 promoter TATA box and initiation site (Fig. 1H), which might directly promote c-myc transcription. The same model might also explain how hSWI/SNF can function as a corepressor when recruited to other target gene promoters (e.g. by Rb (3, 7)): if default nucleosome positions facilitated the binding of transcriptional activators or blocked the binding of transcriptional repressors, then hSWI/SNF-directed repositioning away from these sequences would result in transcriptional repression. One recent study indicated that the yeast Isw2 remodeling complex repressed transcription by over-riding sequence-specified nucleosome positions that facilitated promoter accessibility (59). However, in contrast to our results, Isw2 remodeling did not appear to be affected by promoter DNA sequence, suggesting possible functional differences between ISWI and SWI/SNF class remodeling complexes. The idea that nucleosome positions can be functionally regulated by a combination of DNA sequence, SWI/SNF- and ISWI-class remodeling complexes, is also supported by a recent study showing that nucleosome positions on the yeast HIS3 gene differ considerably between wild-type, SWI/SNF- and ISW1- strains (60).
We find that certain mononucleosome positions are strongly favored after hSWI/SNF remodeling, and that these preferred locations are independent of nucleosome starting positions. This suggests that, in addition to directing movement away from positioning sequences, hSWI/SNF may have the additional property of placing mononucleosomes over specific hSWI/SNF-preferred sequences. It is unclear what makes a sequence a preferred location for hSWI/SNF-repositioned mononucleosomes. One hint comes from the observation that, in most cases, the positions to which hSWI/SNF moves nucleosomes are present at a low but measurable level after nucleosome assembly. For instance, in Fig. 2A “+SWI/SNF” EcoRI lane, two preferred positions are represented by strong bands of ~129 and ~113 bp. These same bands are present, but at greatly reduced levels, in the “−SWI/SNF” control. These observations suggest that hSWI/SNF moves nucleosomes away from positions that are strongly preferred during assembly to positions that are unfavorable during assembly, but still allowed. Accordingly, hSWI/SNF appears not to move histone octamers to sequences that are incompatible with the assembly of a normal nucleosome, but instead moves them to sequences that have some characteristics in common with nucleosome positioning sequences as well as some distinct, hSWI/SNF-preferred characteristics. It is hoped that ongoing studies comparing hSWI/SNF-favored positions on a variety of minicircle and polynucleosomal templates will provide further details about the nature of hSWI/SNF-favored sequences.
In summary, the use of mononucleosome DNA minicircles has provided an initial answer to the long-standing question of how DNA sequence directs nucleosome repositioning by hSWI/SNF. This work, however, is just a first step in understanding how repositioning by ATP-dependent remodeling complexes might affect DNA accessibility. Future application of the minicircle remodeling system developed here could provide insights into how transcription factor binding, variant core histones, linker histones or histone tail modifications might alter the intrinsic sequence-specificity of hSWI/SNF-directed nucleosome positioning. In addition this system could be used to compare hSWI/SNF repositioning specificity to that of other remodeling complexes, such as those in the ISWI and NURD families, some of which may function by actively reversing hSWI/SNF remodeling effects. Finally, detailed studies on polynucleosomes will be necessary to determine where hSWI/SNF-altered dinucleosomal products (altosomes) are generated, and how repositioning specificity is influenced by the presence of neighboring nucleosomes.
Acknowledgments
The authors would like to thank the National Cell Culture Center for FLAG-Ini1 HeLa cell culture, Brandi Davis and Kyle Havens for help with initial experiments, and Carol Kumamoto, Ananda Roy, Robert Kingston and Geeta Narlikar for helpful comments on the manuscript.
Footnotes
This work was funded by grants to GRS from the American Cancer Society (RSG-04-188) and the National Cancer Institute (K01CA88835).
hSWI/SNF, human SWI/SNF; MNase, micrococcal nuclease; NPS, nucleosome positioning sequence; PAGE, polyacrylamide gel electrophoresis; EMSA, electrophoretic mobility shift analysis; SDS, sodium dodecyl sulfate; GGB, glycerol gradient buffer; ISWI, imitation switch; NURD, nucleosome remodeling deacetylase complex; M0, minicircle octamers.
References
- 1.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandran A, Schnitzler G. Regulating transcription one nucleosome at a time: Nature and function of chromatin remodeling complex products. Recent Res Devel Mol Cell Biol. 2004;5:149–170. [Google Scholar]
- 3.Simone C. SWI/SNF: The crossroads where extracellular signaling pathways meet chromatin. J Cell Physiol. 2005;207:309–314. doi: 10.1002/jcp.20514. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. Epub 2005 Jul 7. [DOI] [PubMed] [Google Scholar]
- 5.Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–977. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- 6.Muller C, Leutz A. Chromatin remodeling in development and differentiation. Curr Opin Genet Dev. 2001;11:167–174. doi: 10.1016/s0959-437x(00)00175-1. [DOI] [PubMed] [Google Scholar]
- 7.Klochendler-Yeivin A, Muchardt C, Yaniv M. SWI/SNF chromatin remodeling and cancer. Curr Opin Genet Dev. 2002;12:73–79. doi: 10.1016/s0959-437x(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 8.Hermeking H. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets. 2003;3:163–175. doi: 10.2174/1568009033481949. [DOI] [PubMed] [Google Scholar]
- 9.Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- 10.Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 11.Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. Embo J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol. 2000;20:7541–7549. doi: 10.1128/mcb.20.20.7541-7549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 16.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 17.Albert T, Wells J, Funk JO, Pullner A, Raschke EE, Stelzer G, Meisterernst M, Farnham PJ, Eick D. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J Biol Chem. 2001;276:20482–20490. doi: 10.1074/jbc.M100265200. [DOI] [PubMed] [Google Scholar]
- 18.Pullner A, Mautner J, Albert T, Eick D. Nucleosomal structure of active and inactive c-myc genes. Journal of Biological Chemistry. 1996;271:31452–31457. doi: 10.1074/jbc.271.49.31452. [DOI] [PubMed] [Google Scholar]
- 19.Siebenlist U, Hennighausen L, Battey J, Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 20.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 21.Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, Lieber CM. Direct Imaging of Human SWI/SNF-Remodeled Mono- and Polynucleosomes by Atomic Force Microscopy Employing Carbon Nanotube Tips. Mol Cell Biol. 2001;21:8504–8511. doi: 10.1128/MCB.21.24.8504-8511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulyanova NP, Schnitzler GR. Human SWI/SNF generates abundant, structurally altered dinucleosomes on polynucleosomal templates. Mol Cell Biol. 2005;25:11156–11170. doi: 10.1128/MCB.25.24.11156-11170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavin I, Horn PJ, Peterson CL. SWI/SNF Chromatin Remodeling Requires Changes in DNA Topology. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran A, Omar M, Cheslock P, Schnitzler GR. Linker histone H1 modulates nucleosome remodeling by human SWI/SNF. J Biol Chem. 2003;278:48590–48601. doi: 10.1074/jbc.M309033200. [DOI] [PubMed] [Google Scholar]
- 25.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 27.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 28.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF Unwraps, Slides, and Rewraps the Nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 29.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 30.Aoyagi S, Hayes JJ. hSWI/SNF-catalyzed nucleosome sliding does not occur solely via a twist-diffusion mechanism. Mol Cell Biol. 2002;22:7484–7490. doi: 10.1128/MCB.22.21.7484-7490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prunell A, Alilat M, Lucia FD. Nucleosome structure and dynamics. The DNA minicircle approach. Methods Mol Biol. 1999;119:79–101. doi: 10.1385/1-59259-681-9:79. [DOI] [PubMed] [Google Scholar]
- 32.Hayes JJ, Wolffe AP. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proceedings of the National Academy of Sciences, USA. 1992;89:1229–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Utley RT, Owen-Hughes TA, Juan LJ, Cote J, Adams CC, Workman JL. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 34.Imbalzano AN, Schnitzler GR, Kingston RE. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 35.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 36.Becker PB. NEW EMBO MEMBER’S REVIEW: Nucleosome sliding: facts and fiction. Embo J. 2002;21:4749–4753. doi: 10.1093/emboj/cdf486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buttinelli M, Di Mauro E, Negri R. Multiple nucleosome positioning with unique rotational setting for the Saccharomyces cerevisiae 5S rRNA gene in vitro and in vivo. Proc Natl Acad Sci U S A. 1993;90:9315–9319. doi: 10.1073/pnas.90.20.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes and Development. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka S, Livingstone-Zatchej M, Thoma F. Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J Mol Biol. 1996;257:919–934. doi: 10.1006/jmbi.1996.0212. [DOI] [PubMed] [Google Scholar]
- 40.Dong F, Hansen JC, van Holde KE. DNA and protein determinants of nucleosme positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guyon JR, Narlikar GJ, Sullivan EK, Kingston RE. Stability of a Human SWI-SNF Remodeled Nucleosomal Array. Mol Cell Biol. 2001;21:1132–1144. doi: 10.1128/MCB.21.4.1132-1144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- 43.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark DJ, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. Embo J. 1991;10:387–395. doi: 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meersseman G, Pennings S, Bardbury EM. Mobile nucleosomes--a general behavior. EMBO Journal. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JPZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 48.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. Embo J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flaus A, Richmond TJ. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. Journal of Molecular Biology. 1998;275:427–441. doi: 10.1006/jmbi.1997.1464. [DOI] [PubMed] [Google Scholar]
- 50.Kang JG, Hamiche A, Wu C. GAL4 directs nucleosome sliding induced by NURF. Embo J. 2002;21:1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ura K, Hayes JJ, Wolffe AP. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO Journal. 1995;14:3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sera T, Wolffe AP. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Molecular and Cellular Biology. 1998;18:3668–3680. doi: 10.1128/mcb.18.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakaue T, Yoshikawa K, Yoshimura SH, Takeyasu K. Histone Core Slips along DNA and Prefers Positioning at the Chain End. Phys Rev Lett. 2001;87:078105-1–078105-4. doi: 10.1103/PhysRevLett.87.078105. [DOI] [PubMed] [Google Scholar]
- 54.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 56.Flaus A, Owen-Hughes T. Mechanisms for nucleosome mobilization. Biopolymers. 2003;68:563–578. doi: 10.1002/bip.10323. [DOI] [PubMed] [Google Scholar]
- 57.Becker PB. Nucleosome remodelers on track. Nat Struct Mol Biol. 2005;12:732–733. doi: 10.1038/nsmb0905-732. [DOI] [PubMed] [Google Scholar]
- 58.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 59.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 60.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol Cell Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]