Abstract
Mutation of the axial Met ligand of the type 1 copper site of amicyanin to Ala or Gln yielded M98A amicyanin, which exhibits typical axial type 1 ligation geometry but with a water molecule providing the axial ligand, and M98Q amicyanin which exhibits significant rhombic distortion of the type 1 site [Carrell, C. J., Ma, J. K., Antholine, W. E., Hosler, J. P., Mathews, F. S and Davidson, V. L., (2007) Biochemistry 46, 1900–1912]. Despite the change of the axial ligand, the M98Q and M98A mutations had little effect on the redox potential of the copper. The true electron transfer (ET) reactions from O-quinol methylamine dehydrogenase to oxidized native and mutant amicyanins revealed that the M98A mutation had little effect on kET, but the M98Q mutation reduced kET 45-fold. Thermodynamic analysis of the latter showed that the decrease in kET was due to an increase of 0.4 eV in the reorganization energy (λ) associated with the ET reaction to M98Q amicyanin. No change in the experimentally determined electronic coupling or ET distance was observed confirming that the mutation had not altered the rate determining step for ET and that this was still a true ET reaction. Nor is the basis for the increased λ the nature of the atom which provides the axial ligand since each uses an oxygen, from Gln in M98Q amicyanin and from water in M98A amicyanin. Comparison of the distance of the axial copper ligand from the equatorial plane that is formed by the other three copper ligands in isomorphous crystals of native and mutant amicyanins at atomic resolution indicate an increase in distance from 0.20 Å in the native to 0.42 Å in M98Q amicyanin, and a slight decrease in distance for M98A amicyanin. This correlates with the rhombic distortion caused by the M98Q mutation that is clearly evident in the EPR and visible absorption spectra of the protein, and suggests that the extent of rhombicity of the type 1 copper site influences the magnitude of λ.
Type 1 copper sites are found in a wide variety of electron transfer (ET)1 proteins, including cupredoxins in bacteria and plants, and multicopper proteins such as ascorbate oxidase and ceruloplasmin which are found in animals as well (1). The active site of type 1 copper proteins typically consists of three strong equatorial ligands, two nitrogens of two His and a sulfur of a Cys forming a trigonal plane, plus an additional axial ligand usually provided by the sulfur of a Met (1). In amicyanin from P. denitrificans the three strong equatorial copper ligands are provided by residues His53, His95 and Cys92, respectively, and the weak axial ligand is provided by Met98 (Figure 1) (2).
Figure 1.
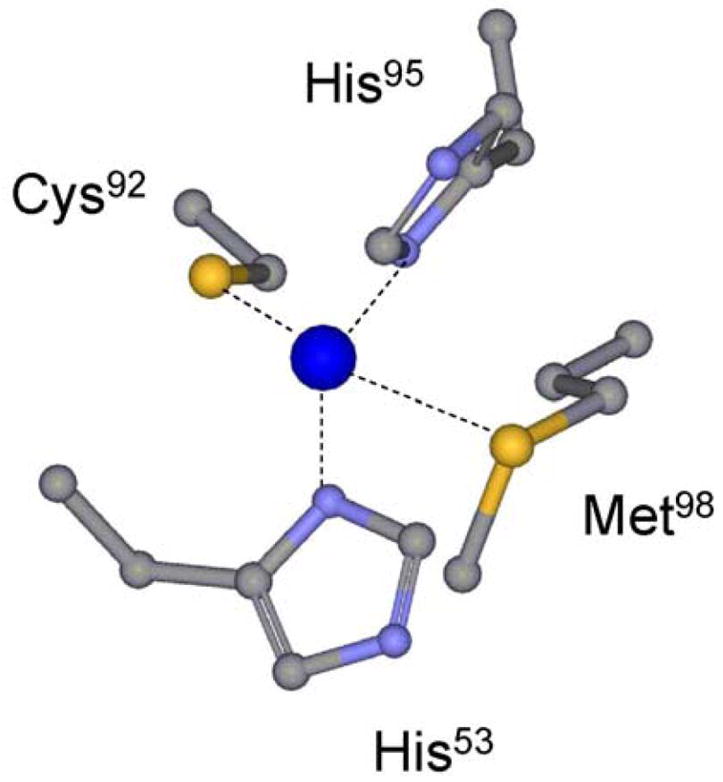
The type 1 copper center of native amicyanin.
Amicyanin is the electron acceptor for methylamine dehydrogenase (MADH) (3). MADH and amicyanin (4) from P. denitrificans form one of the best characterized physiologic ET complexes of proteins. X-ray crystal structures are available for the binary complex of MADH and amicyanin (5) and for a ternary protein complex (6), which includes cytochrome c-551i (7), the electron acceptor for amicyanin. It was demonstrated by single crystal polarized absorption microspectroscopy (8) and EPR spectroscopy (9) that in the crystalline state, these complexes can catalyze the oxidation of methylamine and subsequent ET from TTQ to copper, and from copper to heme. The steady-state kinetic parameters for methylamine-dependent cytochrome c-551i reduction by the MADH-amicyanin complex in solution have been characterized as well (10).
ET reactions can be classified as true, gated, or coupled (11, 12). In a true ET reaction, the rate-limiting reaction step is the ET event, and therefore, the observed rate and the true rate of ET (kET) are the same. In solution, the interprotein ET from the reduced O-quinol tryptophan tryptophylquinone (TTQ) cofactor of MADH to the oxidized type 1 copper site of amicyanin has been shown to be a true ET reaction (13–15). Analysis by ET theory (16) of the temperature and ΔGo-dependence of this reaction yielded values for the reorganization energy (λ) and electronic coupling (HAB) that are associated with the true ET reaction (14, 15).
It has been argued that the so-called rack-induced folding of type 1 copper proteins and protein-enforced constraints on the copper ligation of the type 1 copper geometry play an important role in reducing the λ for the ET reaction of the copper relative to that of copper in an unconstrained complex (17). Met98 was previously mutated to either Gln or Ala and crystal structures were obtained for the oxidized and reduced forms of M98A and M98Q amicyanins (18). M98A and M98Q amicyanins exhibit highly unusual type 1 copper sites. M98A amicyanin exhibits a typical axial type 1 ligation geometry but a mobile water rather than a rigid amino acid side-chain provides the axial ligand (18). M98Q amicyanin exhibits a rhombic distorted type 1 site (18). The ET reactions of these amicyanins with MADH were studied to determine the effects of these structural perturbations on the oxidation-reduction midpoint potential (Em) value, and the rate and ET parameters for their respective ET reactions. The only parameter that exhibited significant variation as a consequence of the mutation was the value of λ for the ET reaction to M98Q amicyanin. The results suggest that the extent of rhombicity of the type 1 copper site is an important determinant of λ.
EXPERIMENTAL PROCEDURES
Protein Purification
Previously described procedures were used to purify MADH (19) and wild-type amicyanin (4) from P. denitrificans (ATCC 13543). M98Q and M98A amicyanins were expressed in E. coli and purified from the periplasmic fraction and reconstituted with copper as described previously (18).
Redox Potential Determination
Em values of the amicyanin mutants were determined by spectrochemical titration as described previously (20). The reaction mixture contained 130 μM amicyanin in 50 mM BisTris propane (BTP) buffer at the indicated pH, at 25°C with potassium ferricyanide (520 μM) present as a mediator. The mixture was titrated by addition of incremental amounts of ascorbate, which was used as a reductant, and which had been previously adjusted to the set pH. The absorption spectrum of amicyanin was recorded at different potentials and the concentrations of oxidized amicyanin and reduced amicyanin were determined by comparison with the spectra of the completely oxidized and reduced forms. The data were analyzed according to eq 1 to determine Em values.
| (1) |
For determination of the Em values of native amicyanin and M98Q amicyanins in complex with MADH, the reaction mixture contained 20 μM amicyanin and 60 μM MADH in 0.01 M potassium phosphate, pH 7.5, at 25°C. Potassium ferricyanide (400 μM), quinhydrone (200 μM), and phenazine ethosulfate (40 μM) were present as mediators. The absorption spectrum of the complex was recorded at different potentials and the concentrations of the oxidized and reduced forms were determined by comparison with the spectra of the completely oxidized complex and a mixture of oxidized MADH with fully reduced amicyanin as described previously (20).
Kinetic Studies of Electron Transfer Reactions of Amicyanins
The rates of ET reactions from O-quinol MADH to oxidized amicyanins were determined using an On-Line Instruments (OLIS, Bogard, GA) RSM stopped-flow rapid scanning spectrophotometer as described previously (21). Experiments were performed in 10 mM potassium phosphate, pH 7.5. Prior to mixing, one stopped flow syringe contained reduced O-quinol MADH while the other contained oxidized amicyanin. Experiments were performed under pseudo-first-order conditions with a fixed concentration of 1 μM MADH and varied concentrations of excess amicyanin. The data are fit to the simple kinetic model in eq 2 using eq 3.
| (2) |
| (3) |
Analysis of Electron Transfer Reactions by Electron Transfer Theory
Values for k3 for each ET reaction were obtained at different temperatures and the temperature dependence of k3 was analyzed using eqs 4 and 5 where λ is the reorganization energy; HAB is the electronic
| (4) |
| (5) |
coupling, h is Plank’s constant, T is temperature, R is the gas constant, and ko is the characteristic frequency of nuclei (1013 s−1), which is the maximum ET rate when donor and acceptor are in van der Waals’ contact and λ=− λGo. The donor to acceptor distance is r, and ro is the close contact distance (3 Å). β is used to quantitate the nature of the intervening medium with respect to its efficiency to mediate ET. ΔGo is determined from ΔEm value using eq 6 where F is the Faraday constant and n is the number of electrons transferred. There is no significant vatiation in ΔGo over the temperature range used in these studies (11, 22).
| (6) |
Crystallographic calculations
Geometrical calculations were made using GEOMCALC of the CCP4 package (23). Structures used in these calculations were pdb entries 2OV0 for native amicyanin (0.75 Å resolution), 2IDQ for M98A amicyanin (0.90 Å resolution), 2IDT for M98Q amicyanin (1.00 Å resolution).
RESULTS
Redox Properties
We previously showed that the Em value of native amicyanin is pH dependent with a pKa value of 7.7, because reduced amicyanin exhibits a pH -dependent conformational change in which the His95 copper ligand rotates out of the coordination sphere of the copper when protonated (20). As such, if a mutation causes a change in Em value determined at a single pH in the pH-dependent region, one must distinguish whether it is due to a true electronic effect on the intrinsic Em value of the protein-bound copper or a change in the pKa of the Em value. It is desirable to obtain Em values from at least two values of pH, one in the pH-dependent region and one at pH 9.0 which is in the pH-independent region. At pH 9.0, amicyanin adopts the redox-active 4-coordinate geometry which mimics that which is maintained when amicyanin is in complex with MADH, even at neutral pH. That is because when amicyanin is in complex with MADH the redox and pH-dependent conformational change of His95 is restricted. Therefore the Em value of free amicyanin in the pH independent region (i.e., pH 9.0) approximates the Em value of amicyanin in complex with MADH which does not vary with pH in the physiological range (20).
Em values were determined for each mutant amicyanin (Table 1). The Em value of M98Q amicyanin at pH 7.0 is 43 mV less positive than that of native amicyanin. However, the Em value of M98Q amicyanin at pH 9.0 is only 11 mV less positive and within experimental error of that of native amicyanin. The Em value of M98Q amicyanin in complex with MADH at pH 7.5 (+225 ±1 mV) is essentially the same as that of native amicyanin in complex (+224 ±1 mV) (Figure 2). This indicates that the M98Q mutation has had minimal effect on the electronic properties of the copper site but has shifted the pKa for the Em value to a slightly more acidic value. The Em value of M98A amicyanin at pH 7.0 is nearly identical to that of native amicyanin. Unfortunately M98A amicyanin is unstable at higher pH (18). It was not possible to determine its Em value in the pH-independent region or in complex with MADH due to instability of the protein during these redox titrations. However, the fact that the Em value of M98A amicyanin at pH 7.0 is nearly identical to that of native strongly suggests that this mutation has had no effect on either the electronic properties or pH dependence of the Em value of the type 1 copper site. Thus, replacement of the axial Met sulfur ligand with an oxygen ligand provided by either water in M98A amicyanin or Gln in M98Q amicyanin has minimal effect on the intrinsic Em value of thetype 1 copper site.
Table 1.
Em values of native, M98A, and M98Q amicyanins
| Native amicyanin | M98A amicyanin | M98Q amicyanin | |
|---|---|---|---|
| Em at pH 7 (mV) | 294±7a | 297±1 | 251±4 |
| Em at pH 9 (mV) | 240±7a | NDb | 229±7 |
| Em in complex with MADH at pH 7.5 (mV) | 224±1 | NDb | 225±1 |
Taken from ref (20)
Not determined because of instability of this protein during the titration.
Figure 2.
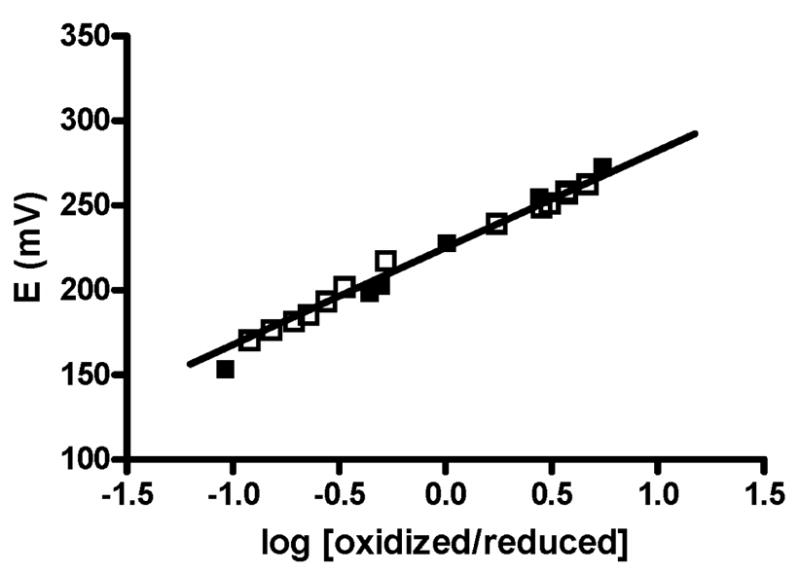
Spectrochemical redox titrations of native and M98Q amicyanins in complex with methylamine dehydrogenase. Titrations were performed as described under Experimental Procedures. Data for native amicyanin (solid squares) and M98Q amicyanin (open squares) are overlaid. The solid line is the fit of the data for the titration of M98Q amicyanin to equation 1.
ET from MADH to Amicyanin
The dependence on amicyanin concentration of the rate of the ET reaction from O-quinol MADH to wild-type and mutant amicyanins is shown in Figure 3. From these data it is possible to extract values of Kd and k3, the limiting first-order rate constant for the ET reaction, according to eq 2. Analysis of the data revealed that neither the M98A nor M98Q mutation had any significant affect on the Kd value for complex formation with MADH (Table 2). The rate of ET from O-quinol MADH to M98A is nearly identical to that of native amicyanin, but the rate of ET to M98Q amicyanin is 45-fold slower than that of native amicyanin. Given the similarities in the Em values of the native and mutant amicyanins, the much slower observed rate for M98Q amicyanin indicated that the mutation had affected an ET parameter other than the ΔGo value for the reaction. To determine the values of HAB and λ that are associated with these ET reactions, the temperature dependence of the k3 values for the ET reaction from O-quinol MADH to each amicyanin was analyzed.
Figure 3.
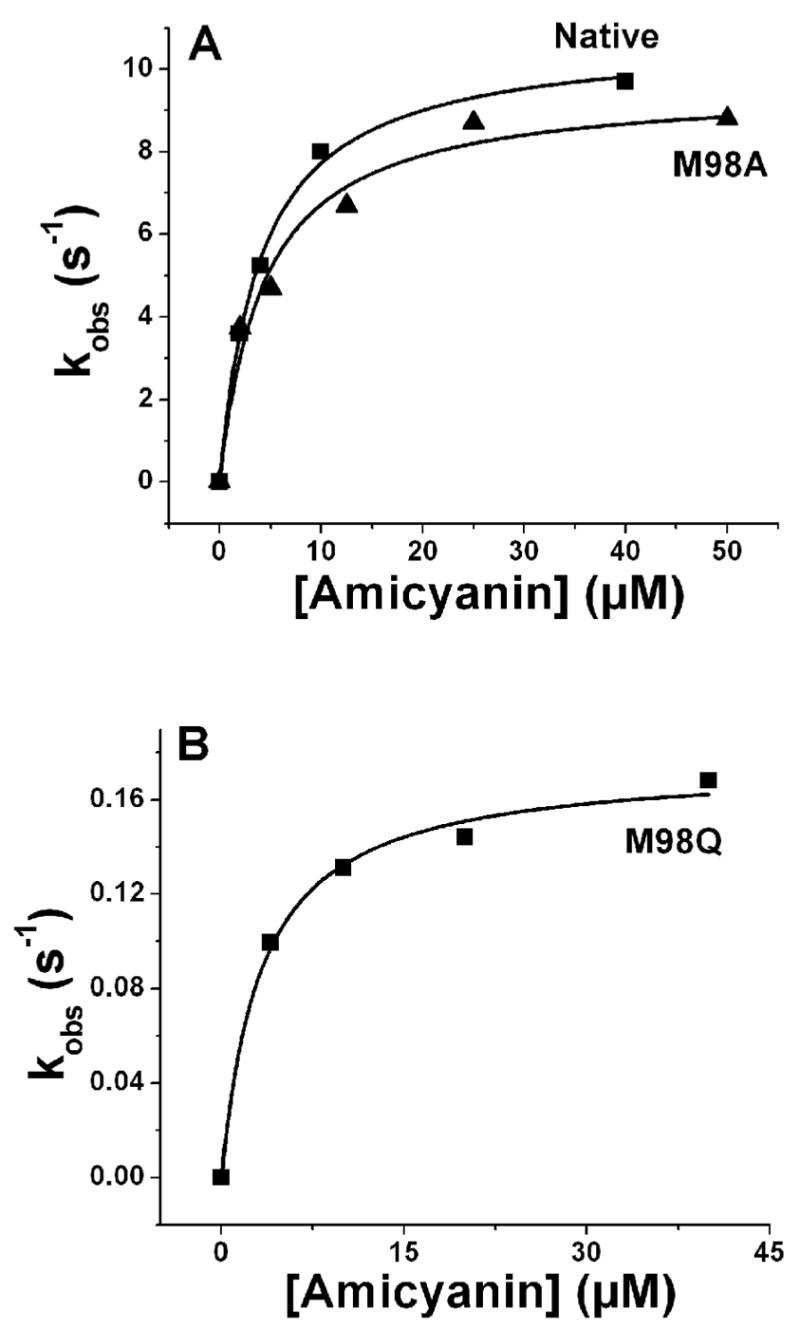
Dependence of the rate of the ET reaction from reduced O-quinol MADH on amicyanin concentration. Solid line represents fits of the data to eq 3.
Table 2.
Kinetic and ET parameters for the reactions of native and mutant amicyanins with O-quinol MADH
| Reaction Parameters | Native amicyanina | M98A amicyanin | M98Q amicyanin |
|---|---|---|---|
| k3 30°C (s−1) | 9.8 | 9.6 | 0.22 |
| Kd (uM) | 5 | 4 | 5 |
| ΔGo (kJ/mol) | −3.37±0.08 | −3.37±0.08 | −3.47±0.09 |
| λ (kJ/mol)b | 222±10 | 202±7 | 261±6 |
| λ (eV) | 2.30±0.10 | 2.09± 0.08 | 2.70±0.07 |
| HAB (cm−1) | 12±7 | 6.1±2 | 12±4 |
| r (Å) for β=1.0 Å−1 | 9.5 | 10.8±0.7 | 9.6±0.6 |
Taken from (14). In that reference a range of values of ΔGo was used to yield a range of values of λ because at that time it was uncertain as to whether it was most appropriate to use the Em value of free amicyanin or amicyanin in complex with MADH. The data in this table were recalculated using what is now known to be the appropriate ΔGo, as well as a λ value of 1.0 Å−1 which is generally accepted as reasonable for protein β sheets (41).
λ is sometimes expressed in units of kJ/mol and sometimes as eV so both values are given.
The reaction with M98A amicyanin was performed over a temperature range of 17 °C –40 °C (Figure 4A). Analysis of the temperature dependence of the rate of the reaction with M98A amicyanin according to eq 4 indicated that the values of λ and HAB for the ET reaction from O-quinol MADH to M98A amicyanin were within experimental error of those for the ET reaction from O-quinol MADH to native amicyanin (Table 2). Thus, the replacement of the Met98 ligand with a water did not influence the parameters for the ET reaction of the oxidized type 1 copper site.
Figure 4.
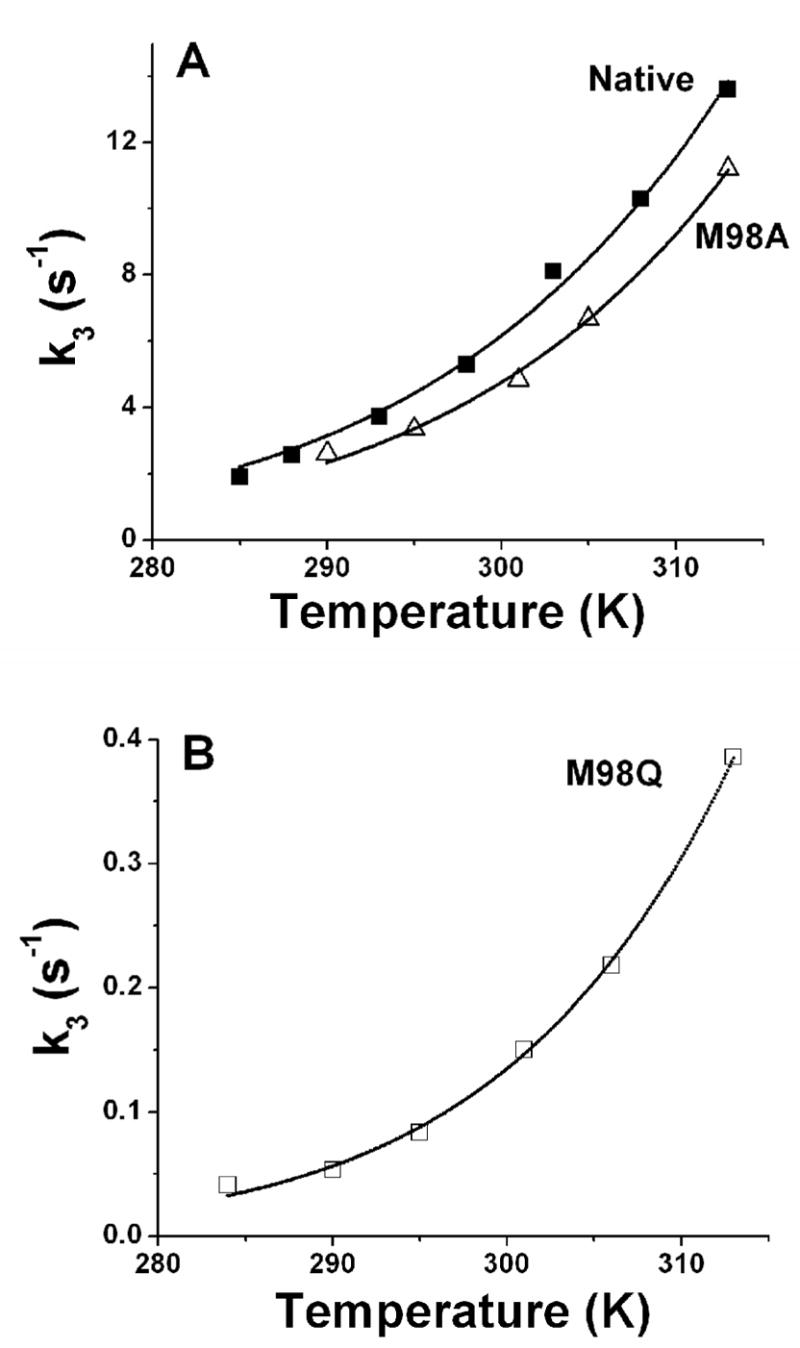
Dependence of the rate of the ET reaction from reduced O-quinol MADH to amicyanins on temperature. Solid lines represent fits of the data to eq 4.
The reaction with M98Q amicyanin was performed over a temperature range of 11 °C –40 °C (Figure 4B). Analysis of the temperature dependence of the rate of the reaction with M98Q amicyanin according to eqs 4 and 5 indicated that the values of HAB and ET distance (r) for the ET reaction from O-quinol MADH to M98Q amicyanin were the same as those for the reaction with native amicyanin, and consistent with this still being a true ET reaction. However, the value of λ was 0.4 eV (39 kJ/mol) greater than that for the reaction with native amicyanin. This accounts for the observed decrease in rate and indicates that the M98Q mutation has decreased kET specifically by increasing the reorganization energy for this true ET reaction.
DISCUSSION
Despite numerous studies it remains unclear exactly how the protein determines the Em value of the type 1 copper center. Em values range from +680 mV for rusticyanin to +184 mV for stellacyanin. It is tempting to think that the differences in Em values among type 1 copper proteins could be related solely to the fine-tuning by the axial ligands. Stellacyanin, which possesses an axial Gln ligand, exhibits a lower Em value than other type I copper proteins with an axial Met ligand (24). Mutation of the axial Gln to Met in stellacyanin increased the Em value by 160 mV (24). Conversely, mutation of the axial Met ligand to Gln in plastocyanin decreased the Em value by 35 mV (25). However, we observed no significant change in the Em value of either M98Q amicyanin or M98A amicyanin, in which the axial ligand is provided by a water molecule. The data presented here indicate that, in the absence of other factors, the identity and position of the axial ligand alone does not influence the Em value of amicyanin. Other factors that have been shown to influence the Em value of type I copper proteins include “rack-induced” constraints on metal binding (17), H-bonding pattern around the metal site, desolvation or hydrophobic effects, and electrostatic interactions. We have previously shown that mutation of Pro94 of amicyanin to Phe increases its Em value by 150 mV (26) despite there being no change in the copper ligand geometry (27). Inspection of the crystal structure revealed that this is due to formation of a H-bond between the amide nitrogen of Phe94 (not present in Pro94 in native amicyanin) and the sulfur of the Cys92 copper ligand (27). It was subsequently reported that mutation of Phe114 of azurin to Pro removed a H-bond to the Cys copper ligand and decreased the Em value by 60 mV (28). Rusticyanin possesses an axial Met ligand yet exhibits an Em value of +680 mV which is 390 mV greater than that of native amicyanin. This was attributed to highly hydrophobic residues in the immediate vicinity of the copper site which would preferentially stabilize the Cu(I) state (29, 30). Mutation of the axial Met ligand of azurin to hydrophobic residues (Ala, Ile, Val, or Leu) increased the Em value (31). This was attributed to the exclusion of water or other groups with electronegative ligand atoms from the metal site by these hydrophobic residues. In contrast, M98A amicyanin permits the presence of a water molecule in the axial position that apparently helps to stabilize Cu(II) to the same extent as Met98, and it maintains an Em value similar to that of native amicyanin. In summary, axial ligand distances and identity alone are poor predictors of the Em value of type 1 copper sites in proteins.
ET reactions can be classified as true, gated, or coupled (11, 12). In a true ET reaction, the rate-limiting reaction step is ET event, and therefore the observed rate and kET are the same. Excessively high values of λ and HAB are suggestive of a gated ET reaction (12, 32). The ET from O-quinol MADH to oxidized amicyanin exhibits a relatively large value of λ but has been shown to be a true ET reaction on the basis of analysis of the temperature dependence (14) and ΔGo-dependence (13, 15) of the rate of reaction. Essentially the same values of λ and HAB were obtained from each analysis. Furthermore, alteration of ΔGo by site directed mutagenesis of amicyanin caused changes in kET which were consistent with the predictions of ET theory (33). It is likely that the large magnitude of λ is more a consequence of the TTQ cofactor of MADH than of the copper center of amicyanin (34). The ET rate and values of λ and HAB for the reaction of M98A amicyanin with MADH are similar to those of native amicyanin. However, kET for the reaction with M98Q amicyanin is much less than for native amicyanin and analysis of the temperature dependence of the reaction indicates that this is correlated with an increase in λ of 0.4 eV. We have previously shown that a P52G mutation of amicyanin caused increases in both λ and HAB which were attributed to a change in kinetic mechanism that has caused the reaction to become gated (35). M98Q amicyanin is the first amicyanin mutant for which we have observed an increase in λ without concurrent changes in HAB. Thus, the effects of the M98Q mutation cannot be attributed to a change in kinetic mechanism, but rather indicate a true increase in λ for a true ET reaction.
In accord with the concept of “rack-induced” folding of type 1 copper proteins (17), the protein constraints allow very little change in structure on reduction of the copper center. This facilitates rapid ET by reducing λ. The most notable change in the structural and electronic properties of the type 1 site that is caused by the M98Q mutation, and not seen with other mutant amicyanins, is significantly increased rhombicity of the type 1 geometry. The rhombic nature of the type 1 site of M98Q amicyanin is clearly evident and defined by its atypical EPR and visible absorption spectra (Figure 5) (Table 3) (18). These features of the EPR and absorption spectra arise from changes in the electronic properties associated with the interaction between Cu(II) and the Cys ligand. The broad absorption band around 450 – 700 nm of amicyanin is assigned to the charge transfer from Sπ and Sσ of Cys92 to the dx2-y2 orbital of Cu(II). In native amicyanin, the interaction is predominantly π in character. When Gln replaces Met as the axial ligand, increasing competition by oxygen for the Cu d orbital results in the Cys92 charge transfer to dx2-y2 to becoming more σ in character, causing an increase in A462/A595 (18). However, this electronic property is not related solely to the use of O as an axial ligand since M98A amicyanin does not exhibit the increased rhombicity, or change in λ. The change in electronic properties is also not necessarily related to the ligand-Cu distances. Other mutants of the axial residue of type 1 copper proteins, such as M121Q azurin (36) and M148Q rusticyanin (37) display a similar increase in rhombicity without significant elongation of the ligand-Cu distance. It has been suggested that increase in the axial electronic interactions of copper involves contribution from dx2-y2, dxz, and dy orbitals (38). Therefore, the large rhombicity of the g-tensor which is reflected in the EPR spectrum may result from a small amount of Cu dz2 character in the ground-state wave function of the unpaired electron. Such deviations from the native electronic structure may result in an increase in inner sphere reorganization energy and account for the observed increase in λ for the ET reaction with MADH.
Figure 5.
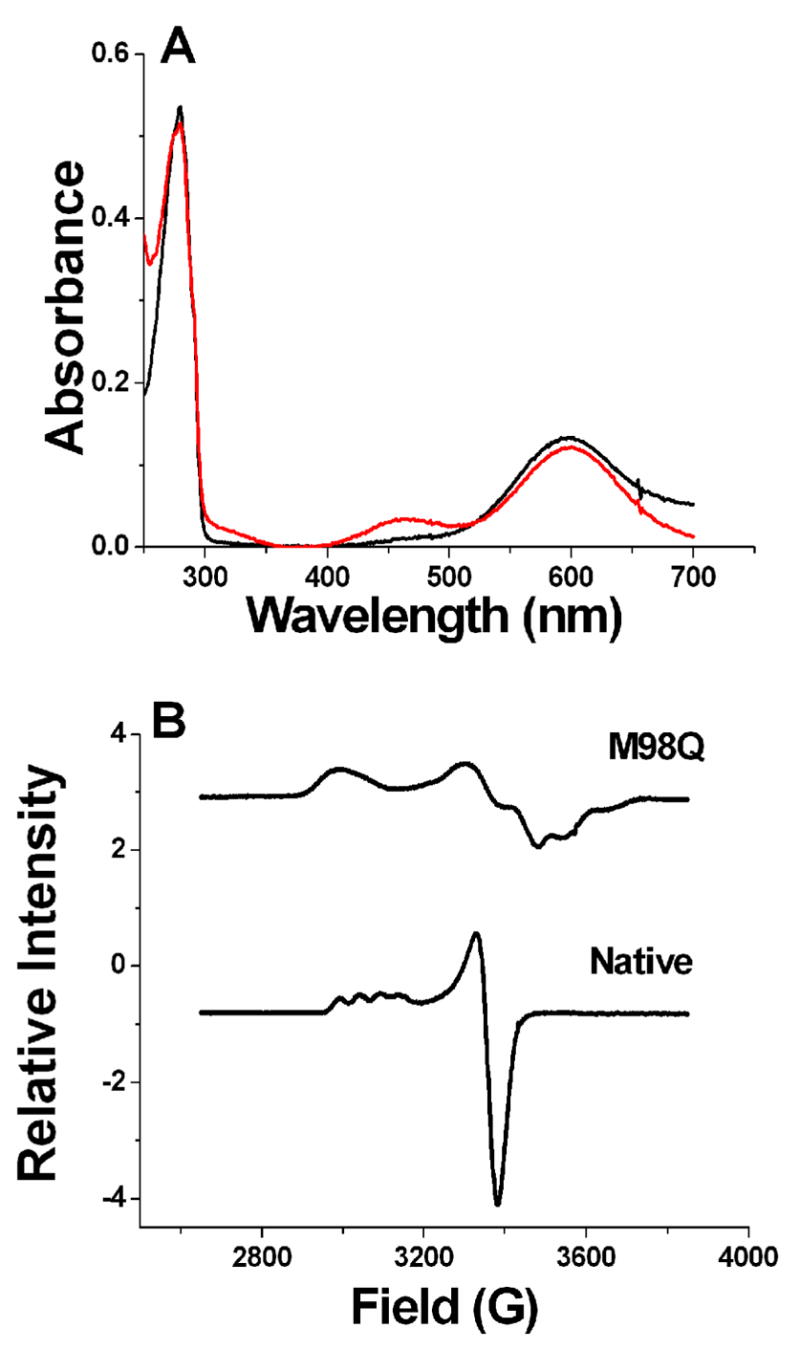
Spectroscopic properties of native and M98Q amicyanin. A. Visible absorption spectra of oxidized M98Q (red line) and native (black line) amicyanins. B. EPR spectra of native (lower) and M98Q (upper) amicyanins. The figure was adapted from data in reference (18).
Table 3.
Properties of native and mutant amicyanins
| Property | Amicyanina | ||
|---|---|---|---|
| Native | M98A | M98Q | |
| Cys92-Cu distance (Å) | 2.17 | 2.14 | 2.16 |
| Axial ligand-Cu distance (Å) | 3.07 | 2.41 | 2.12 |
| Trigonal plane-Cu distance (Å) | 0.20 | 0.14 | 0.42 |
| EPR gllb | 2.24 | - | 2.27 |
| EPR All (G)b | 53 | - | 23 |
| Absorbance ε464/ε595b | 0.11 | 0.12 | 0.17 |
| λ (eV) | 2.30±0.10 | 2.09± 0.08 | 2.70±0.07 |
Structures used to determine distances were pdb entries 2OV0 for native amicyanin (0.75 Å resolution), 2IDQ for M98A amicyanin (0.90 Å resolution), 2IDT for M98Q amicyanin (1.00 Å resolution). These structures are all isomorphous, in space group P21, and contain one molecule in the asymmetric unit. The crystal contacts are weak and very similar among them.
Taken from ref (18)
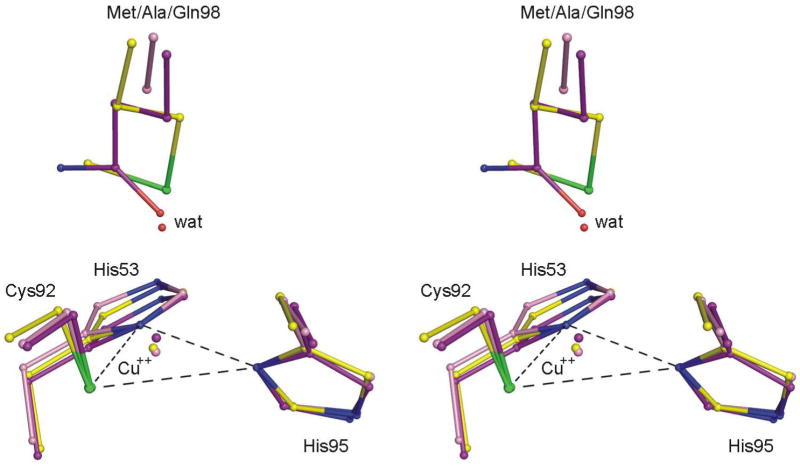
The rhombic distortion of the type 1 copper site in M98Q amicyanin is also evident from comparison of its structure with that of native amicyanin. To be sure that these relatively small differences in distance from copper to the equatorial plane were not an artifact of the crystal packing, isomorphous structures of native, M98Q and M98A amicyanins at atomic resolution were compared (Figure 6). This increased rhombicity is well correlated with a doubling (from 0.20 Å to 0.42 Å) of the distance of the copper atom to the equatorial plane defined by the His53 ND1, Cys92 SG and His95 ND atoms among the three isomorphous crystal structures of wild-type, M98A and M98Q amicyanins whose structures have been determined to resolution of 1.0 Å or below (Table 3). For the M98Q amicyanin oxidized structure determined at 1.0 Å resolution, the estimated standard deviations of the three coordination bonds from the Cys and the two His ligands range from 0.005 to 0.013 Å and for the native structure at 0.75 Å resolution, from 0.002 to 0.006 Å. Since the calculated distance from the copper to the equatorial plane in these structures is a linear combination of these distances it is estimated that their standard deviations are about 0.017 and 0.010 Å, respectively. This leads to an estimate of the precision of the difference between the copper-to-plane distances of about 0.020 Å, about 10-fold lower than the observed difference of 0.22 Å (Table 3). These structures are all isomorphous, in space group P21, and each contains one molecule in the asymmetric unit. The crystal contacts are weak and very similar among them. This ensures that the relatively small differences in relative distances are real and not a consequence of differences in crystal packing or resolution. For these reasons it is difficult to make valid comparisons of relative rhombicity with structures of other type 1 copper proteins in the literature having different resolutions and crystal packing constraints.
Figure 6.
Stereoview of the position of copper relative to the equatorial plane formed by ligands from Cys92, His 95 and His53 in native, M98A and M98Q amicyanins. The three structures are superimposed on the copper-coordinated ligand atoms ND1 of His53 and His95 and the SG of Cys92. These atoms are connected by dashed lines to illustrate the equatorial plane. The carbon and copper atoms of wild type, Ala98 and Gln98 are yellow, pink and purple, respectively. The sulfur, oxygens and nitrogens are green, red and blue, respectively.
There is very little information available on the correlation of the nature of the axial ligand of the type 1 copper site and the experimentally determined λ value associated with ET reactions of the host protein. It was recently reported that mutagenesis of the axial Met ligand of the type 1 copper in nitrite reductase to Gly or Thr increased the reorganization energy by 0.3 eV (39). Extensive quantum chemical calculations of the λ of type 1 copper proteins have been performed using model compounds. The effect of replacing the Met ligand with a Gln ligand, as naturally occurs in stellacyanin, was examined by comparing a Cu(Im)2(SCH3)(CH3CONH2) model with a Cu(Im)2(SCH3)(SCH3)2 model (40). The calculated inner sphere λ for the former (Gln) model was 90 kJ/mol (0.94 eV) compared to 62 kJ/mol (0.64 eV) for the latter model. This difference of approximately 0.3 eV from the computational study correlates very well with our experimentally-determined increase in λ of 0.4 eV and supports the idea that the introduction of Gln as an axial ligand of the type 1 copper in amicyanin is responsible for the increased λ. It is important to note that the structural basis for the increased λ for M98Q amicyanin is not simply the nature of the atom which provides the axial ligand. M98A amicyanin also uses an oxygen for the axial ligand. There is also no significant change in the distances and orientations of the three equatorial copper ligands.
The feature that most clearly distinguishes M98Q amicyanin from native and M98A amicyanin, as well as all other amicyanin mutants which we have studied, is the extent of rhombicity which is manifestly evident from its spectroscopic properties (Figure 5) (18). Ideally one would like to compare the extent of rhombic distortion in the oxidized and reduced amicyanins rather than relying solely on comparison of the oxidized forms. Unfortunately on reduction amicyanin becomes EPR silent and the visible absorption is lost so these techniques cannot be applied. Furthermore, because of differences in the crystals of oxidized and reduced amicyanin, a comparable precise measurement of the copper to equatorial plane for reduced amicyanin was not possible. We feel that the most reasonable explanation consistent with our data showing the increased λ for the true ET reaction of M98Q amicyanin is that it correlates with the rhombic distortion of the type 1 copper site. This is a rare example of a structure-function relationship that describes how the protein may influence the λ associated with a protein ET reaction. The results also begin to delineate the structural and physical properties that specifically modulate λ independent of Em value for a redox-active metalloprotein.
Footnotes
Abbreviations: MADH, methylamine dehydrogenase; TTQ, tryptophan tryptophylquinone; ET, electron transfer; HAB, electronic coupling; λ, reorganization energy; Em, oxidation-reduction midpoint potential.
This work was supported by NIH Grant GM-41574 (V.L.D.) and NSF Grant MCB0343374 (F.S.M.)
References
- 1.Adman ET. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 2.Cunane LM, Chen Z, Durley RCE, Mathews FS. X-ray crystal structure of the cupredoxin amicyanin from Paracoccus denitrificans, refined at 1.31 Å resolution. Acta Cryst. 1996;D52:676–686. doi: 10.1107/S0907444996001072. [DOI] [PubMed] [Google Scholar]
- 3.Davidson VL. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv Protein Chem. 2001;58:95–140. doi: 10.1016/s0065-3233(01)58003-1. [DOI] [PubMed] [Google Scholar]
- 4.Husain M, Davidson VL. An inducible periplasmic blue copper protein from Paracoccus denitrificans. Purification, properties, and physiological role. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 5.Chen L, Durley R, Poliks BJ, Hamada K, Chen Z, Mathews FS, Davidson VL, Satow Y, Huizinga E, Vellieux FM. Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin. Biochemistry. 1992;31:4959–4964. doi: 10.1021/bi00136a006. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Durley RC, Mathews FS, Davidson VL. Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i. Science. 1994;264:86–90. doi: 10.1126/science.8140419. [DOI] [PubMed] [Google Scholar]
- 7.Husain M, Davidson VL. Characterization of two inducible periplasmic c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986;261:8577–8580. [PubMed] [Google Scholar]
- 8.Merli A, Brodersen DE, Morini B, Chen Z, Durley RC, Mathews FS, Davidson VL, Rossi GL. Enzymatic and electron transfer activities in crystalline protein complexes. J Biol Chem. 1996;271:9177–9180. doi: 10.1074/jbc.271.16.9177. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari D, Di Valentin M, Carbonera D, Merli A, Chen Z-W, Mathews FS, Davidson VL, Rossi G-L. Electron transfer in crystals of the binary and ternary complexes of methylamine dehydrogenase with amicyanin and cytochrome c551i as detected by EPR spectroscopy. J Biol Inorg Chem. 2004 doi: 10.1007/s00775-003-0513-0. [DOI] [PubMed] [Google Scholar]
- 10.Davidson VL, Jones LH. Intermolecular electron transfer from quinoproteins and its relevance to biosensor technology. Anal Chim Acta. 1991;249:235–240. [Google Scholar]
- 11.Davidson VL. Unraveling the kinetic complexity of interprotein electron transfer reactions. Biochemistry. 1996;35:14035–14039. doi: 10.1021/bi961577p. [DOI] [PubMed] [Google Scholar]
- 12.Davidson VL. What controls the rates of interprotein electron-transfer reactions. Acc Chem Res. 2000;33:87–93. doi: 10.1021/ar9900616. [DOI] [PubMed] [Google Scholar]
- 13.Bishop GR, Davidson VL. Electron transfer from the aminosemiquinone reaction intermediate of methylamine dehydrogenase to amicyanin. Biochemistry. 1998;37:11026–11032. doi: 10.1021/bi980265e. [DOI] [PubMed] [Google Scholar]
- 14.Brooks HB, Davidson VL. Kinetic and thermodynamic analysis of a physiologic intermolecular electron-transfer reaction between methylamine dehydrogenase and amicyanin. Biochemistry. 1994;33:5696–5701. doi: 10.1021/bi00185a005. [DOI] [PubMed] [Google Scholar]
- 15.Brooks HB, Davidson VL. Free energy dependence of the electron transfer reaction between methylamine dehydrogenase and amicyanin. J Am Chem Soc. 1994;116:11201–11202. [Google Scholar]
- 16.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 17.Malmstrom BG. Rack-induced bonding in blue-copper proteins. Eur J Biochem. 1994;223:711–718. doi: 10.1111/j.1432-1033.1994.tb19044.x. [DOI] [PubMed] [Google Scholar]
- 18.Carrell CJ, Ma JK, Antholine WE, Hosler JP, Mathews FS, Davidson VL. Generation of novel copper sites by mutation of the axial ligand of amicyanin. Atomic resolution structures and spectroscopic properties. Biochemistry. 2007;46:1900–1912. doi: 10.1021/bi0619674. [DOI] [PubMed] [Google Scholar]
- 19.Davidson VL. Methylamine dehydrogenases from methylotrophic bacteria. Methods Enzymol. 1990;188:241–246. doi: 10.1016/0076-6879(90)88040-h. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Cunane LM, Chen Z, Durley RC, Mathews FS, Davidson VL. Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin. Biochemistry. 1998;37:17128–17136. doi: 10.1021/bi9817919. [DOI] [PubMed] [Google Scholar]
- 21.Bishop GR, Brooks HB, Davidson VL. Evidence for a tryptophan tryptophylquinone aminosemiquinone intermediate in the physiologic reaction between methylamine dehydrogenase and amicyanin. Biochemistry. 1996;35:8948–8954. doi: 10.1021/bi960404x. [DOI] [PubMed] [Google Scholar]
- 22.Bishop GR, Davidson VL. Intermolecular electron transfer from substrate-reduced methylamine dehydrogenase to amicyanin is linked to proton transfer. Biochemistry. 1995;34:12082–12086. doi: 10.1021/bi00037a052. [DOI] [PubMed] [Google Scholar]
- 23.CCP4. Collaborative Computational Project Number 4. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JS. Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Sci. 1998;7:1915–1929. doi: 10.1002/pro.5560070907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hibino T, Lee BH, Takabe T. Expression and characterization of Met92Gln mutant plastocyanin from Silene pratensis. J Biochem. 1995;117:101–106. doi: 10.1093/oxfordjournals.jbchem.a124693. [DOI] [PubMed] [Google Scholar]
- 26.Machczynski MC, Gray HB, Richards JH. An outer-sphere hydrogen-bond network constrains copper coordination in blue proteins. J Inorg Biochem. 2002;88:375–380. doi: 10.1016/s0162-0134(02)00364-1. [DOI] [PubMed] [Google Scholar]
- 27.Carrell CJ, Sun D, Jiang S, Davidson VL, Mathews FS. Structural studies of two mutants of amicyanin from Paracoccus denitrificans that stabilize the reduced state of the copper. Biochemistry. 2004;43:9372–9380. doi: 10.1021/bi049634z. [DOI] [PubMed] [Google Scholar]
- 28.Yanagisawa S, Banfield MJ, Dennison C. The role of hydrogen bonding at the active site of a cupredoxin: the Phe114Pro azurin variant. Biochemistry. 2006;45:8812–8822. doi: 10.1021/bi0606851. [DOI] [PubMed] [Google Scholar]
- 29.Hall JF, Kanbi LD, Strange RW, Hasnain SS. Role of the axial ligand in type 1 Cu centers studied by point mutations of met148 in rusticyanin. Biochemistry. 1999;38:12675–12680. doi: 10.1021/bi990983g. [DOI] [PubMed] [Google Scholar]
- 30.Walter RL, Ealick SE, Friedman AM, Blake RC, 2nd, Proctor P, Shoham M. Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability. J Mol Biol. 1996;263:730–751. doi: 10.1006/jmbi.1996.0612. [DOI] [PubMed] [Google Scholar]
- 31.Pascher T, Karlsson BG, Nordling M, Malmstrom BG, Vanngard T. Reduction potentials and their pH dependence in site-directed-mutant forms of azurin from Pseudomonas aeruginosa. Eur J Biochem. 1993;212:289–296. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
- 32.Davidson VL. Chemically gated electron transfer. A means of accelerating and regulating rates of biological electron transfer. Biochemistry. 2002;41:14633–14636. doi: 10.1021/bi026812k. [DOI] [PubMed] [Google Scholar]
- 33.Sun D, Davidson VL. Effects of engineering uphill electron transfer into the methylamine dehydrogenase-amicyanin-cytochrome c-551i complex. Biochemistry. 2003;42:1772–1776. doi: 10.1021/bi0271594. [DOI] [PubMed] [Google Scholar]
- 34.Sun D, Chen ZW, Mathews FS, Davidson VL. Mutation of αPhe55 of methylamine dehydrogenase alters the reorganization energy and electronic coupling for its electron transfer reaction with amicyanin. Biochemistry. 2002;41:13926–13933. doi: 10.1021/bi026654x. [DOI] [PubMed] [Google Scholar]
- 35.Sun D, Li X, Mathews FS, Davidson VL. Site-directed mutagenesis of proline 94 to alanine in amicyanin converts a true electron transfer reaction into one that is kinetically coupled. Biochemistry. 2005;44:7200–7206. doi: 10.1021/bi050288a. [DOI] [PubMed] [Google Scholar]
- 36.Hough MA, Hall JF, Kanbi LD, Hasnain SS. Structure of the M148Q mutant of rusticyanin at 1.5 A: a model for the copper site of stellacyanin. Acta Crystallogr Sect D Biol Crystallogr. 2001;57:355–360. doi: 10.1107/s0907444900019156. [DOI] [PubMed] [Google Scholar]
- 37.Romero A, Hoitink CW, Nar H, Huber R, Messerschmidt A, Canters GW. X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin. J Mol Biol. 1993;229:1007–1021. doi: 10.1006/jmbi.1993.1101. [DOI] [PubMed] [Google Scholar]
- 38.Coremans JWA, Poluektov OG, Groenen EJJ, Warmerdam GCM, Canters GW, Nar H, Messerschmidt A. The azurin mutant Met121Gln: A blue-copper protein with a strong axial ligand. J Phys Chem. 1996;100:19706–19713. [Google Scholar]
- 39.Wijma HJ, MacPherson I, Farver O, Tocheva EI, Pecht I, Verbeet MP, Murphy MEP, Canters GW. Effect of the methionine ligand on the reorganization energy of the type-1 copper site of nitrite reductase. J Am Chem Soc. 2007;129:519–525. doi: 10.1021/ja064763j. [DOI] [PubMed] [Google Scholar]
- 40.Olsson MH, Ryde U, Roos BO. Quantum chemical calculations of the reorganization energy of blue-copper proteins. Protein Sci. 1998;7:2659–2668. doi: 10.1002/pro.5560071220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray HB, Winkler JR. Long-range electron transfer. Proc Natl Acad Sci U S A. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]