Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a secreted protein that controls plasma LDL cholesterol levels by posttranslational regulation of the LDL receptor (LDLR). Previously, we showed that PCSK9 binds specifically to an EGF-like repeat (EGF-A) in LDLR and reroutes the receptor from endosomes to lysosomes rather than to the cell surface. Here, we defined the regions in LDLR and PCSK9 that are required for receptor degradation and examined the relationship between PCSK9 binding and LDLR conformation. Addition of PCSK9 to cultured hepatocytes promoted degradation of WT LDLR and of receptors lacking up to four ligand binding domains, EGF-B or the clustered O-linked sugar region. In contrast, LDLRs lacking the entire ligand binding domain or the β-propeller domain failed to be degraded, although they bound and internalized PCSK9. Using gel filtration chromatography, we assessed the effects of PCSK9 binding on an acid-dependent conformational change that happens in the extracellular domain of the LDLR. Although PCSK9 prevented the reduction in hydrodynamic radius of the receptor that occurs at a reduced pH, the effect was not sufficient for LDLR degradation. A truncated version of PCSK9 containing the prodomain and the catalytic domain, but not the C-terminal domain, bound the receptor but did not stimulate LDLR degradation. Thus, domains in both the LDLR and PCSK9 that are not required for binding (or internalization) are essential for PCSK9-mediated degradation of the LDLR.
Keywords: cholesterol, proprotein convertase, β-propeller, endocytosis
Plasma levels of LDL cholesterol are directly related to the risk of coronary atherosclerosis. The tight association between LDL levels and ischemic heart disease is illustrated by the fact that all monogenic forms of severe hypercholesterolemia are accompanied by premature coronary atherosclerosis. The major pathway for removal of LDL from the circulation is by LDL receptor (LDLR)-mediated endocytosis in the liver. Mutations in the LDLR (1), the ligand for the LDLR (apolipoprotein B-100) (2), or an LDLR adaptor protein (ARH/LDLRAP) (3) all cause hypercholesterolemia by reducing the clearance of circulating LDL. In 2003, a third form of autosomal-dominant hypercholesterolemia was identified that is caused by selected missense mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) (4).
PCSK9 is a 692-aa secreted glycoprotein composed of a 22-aa signal sequence, a prodomain, a catalytic domain that resembles the proteinase K family of subtilisin-like serine proteases, and a cysteine- and histidine-rich C-terminal domain that is unique to this member of the proprotein convertase family (5). In the endoplasmic reticulum, PCSK9 undergoes autocatalytic cleavage between residues 151 and 152 (5, 6), releasing the N-terminal prodomain, which remains noncovalently attached to the catalytic domain, physically shielding the catalytic triad as the protein transits through the secretory pathway (7, 8). PCSK9 circulates in the blood (9) and binds the extracellular domain of the LDLR on the cell surface, triggering receptor degradation (9, 10). Proteolytic activity of PCSK9 is not required for PCSK9 action on the LDLR (11, 12).
PCSK9 is expressed at the highest levels in the liver, small intestine, kidney, and brain (5) and is regulated by the same transcriptional machinery that modulates expression of the LDLR (13, 14). When cellular cholesterol levels decrease, sterol regulatory element binding protein-2 is activated (15), increasing the expression of both LDLR and PCSK9. The increased number of LDLRs on the cell surface of hepatoctyes promotes the clearance of circulating LDL. PCSK9 has the opposite effect on circulating cholesterol levels because its effect is to stimulate LDLR degradation (16–18). PCSK9 variants that cause hypercholesterolemia in humans are gain-of-function mutations (reviewed in ref. 19) that accelerate hepatic LDLR destruction (16–18).
The mechanism by which PCSK9 binding at the cell surface targets the LDLR for degradation is not understood. Cell-surface LDLRs are internalized via clathrin-coated pits and delivered to endosomes. In the low pH environment of the endosome, the LDLR undergoes a conformational change that promotes the release of the bound LDL (20), which is delivered to lysosomes. After releasing LDL in the endosome, the LDLR recycles to the cell surface (21). Usually, each LDLR undergoes multiple rounds of internalization and recycling. When PCSK9 binds the LDLR, the receptor traffics from the endosome to the lysosome, rather than being recycled (22). Interestingly, PCSK9 binds with a much higher affinity to the receptor at acidic pH (7, 12, 22, 23).
Previously, we mapped the region of the LDLR that binds PCSK9 (22). The LDLR has five major structural domains: the ligand-binding domain, which consists of seven tandem cysteine-rich, ligand binding repeats (R1-R7); an EGF precursor homology domain (24, 25), which includes two EGF-like repeats (EGF-A and -B), followed a six-bladed β-propeller (26) and another EGF-like repeat (EGF-C); an O-linked sugar domain; a transmembrane domain; and a 50-aa cytoplasmic tail that contains the sequences necessary for receptor internalization. PCSK9 binds specifically to EGF-A, the first EGF-like repeat in the EGF precursor domain (22). Here, we refine the structural determinants both in the LDLR and in PCSK9 that are required for PCSK9-mediated LDLR degradation.
Results and Discussion
PCSK9 Binding to the LDLR Interferes with an Acid-Associated Conformational Change in the Receptor Protein.
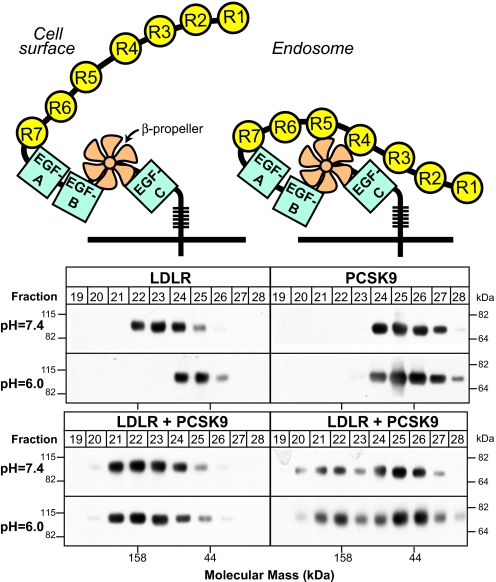
The deduced tertiary structure of the extracellular domain of the LDLR suggested a possible mechanism by which PCSK9 binding results in receptor degradation (Fig. 1). At neutral pH, the extracellular domain of the LDLR adopts an extended linear conformation, whereas at an acidic pH, the extracellular domain forms a hairpin structure such that ligand binding repeats 4 and 5 (R4 and R5) are bound to the β-propeller in the EGF precursor homology domain (20). The linear conformation would be expected to predominate when the receptor is on the cell surface, thus facilitating interactions between the ligand binding repeats and LDL. Upon transit to the endosome, the acidic pH of this organelle would favor release of the bound LDL (21) and formation of the hairpin structure, which may be required for recycling of the receptor to the cell surface. If PCSK9 binding to the receptor prevented formation of the hairpin structure, the LDLR-PCSK9 complex might be directed to the lysosome rather than the cell surface, leading to degradation of the receptor (20).
Fig. 1.
PCSK9 interferes with an acid-dependent conformational change in the LDLR. Purified extracellular domain of LDLR (60 μg) was incubated in the presence or absence of recombinant PCSK9 (160 μg) for 2 h at room temperature. The samples were centrifuged, and the supernatants were loaded onto Superose 12 10/300 GL columns equilibrated with Buffer A. The elution patterns of LDLR and PCSK9 were determined by SDS/PAGE and immunoblotting of the collected fractions. The LDLR and PCSK9 were detected using monoclonal antibodies (HL-1 and 15A6, respectively). The elution positions of the following protein standards are shown: γ-globulin (158 kDa) and ovalbumin (44 kDa).
To test this hypothesis, we examined the effects of PCSK9 on the conformation of the LDLR extracellular domain at a neutral and an acidic pH, as assessed by gel filtration (20). First, we confirmed that the apparent molecular mass of the LDLR decreases with a reduction in pH (Fig. 1, upper left). Changing the pH from 7.4 to 6.0 did not affect the elution of PCSK9 from the column (Fig. 1, upper right). This finding was consistent with crystallographic studies that derived essentially superimposable structures for PCSK9 over a wide pH range (4.6 to 10.5) (7, 8, 27).
Next, we examined the effect of pH on the elution of the LDLR-PCSK9 complex. In this experiment, the extracellular domain of the LDLR was incubated with a fourfold molar excess of purified, full-length PCSK9 at a pH of 7.4 for 30 min. The medium was then adjusted to a pH of 7.4 or 6.0 and the proteins were subjected to gel filtration. Fractions from the column were immunoblotted with anti-LDLR and anti-PCSK9 antibodies (Fig. 1, lower left and right, respectively). Addition of PCSK9 to the LDLR shifted the elution of the receptor to higher molecular weight fractions (Fig. 1, lower left), consistent with the formation of an LDLR-PCSK9 complex. In the presence of PCSK9, the LDLR eluted from the column in the same fractions at both pH 7.4 and pH 6.0. Thus, in the presence of PCSK9, no change in the hydrodynamic radius of the LDLR was apparent. As a control, incubation of the LDLR with BSA (160 μg) failed to alter the elution of the LDLR (data not shown). Taken together, these findings are consistent with PCSK9 preventing the acid-dependent conformational change in the LDLR.
Sequence Requirements for PCSK9-Mediated Degradation of the LDLR.
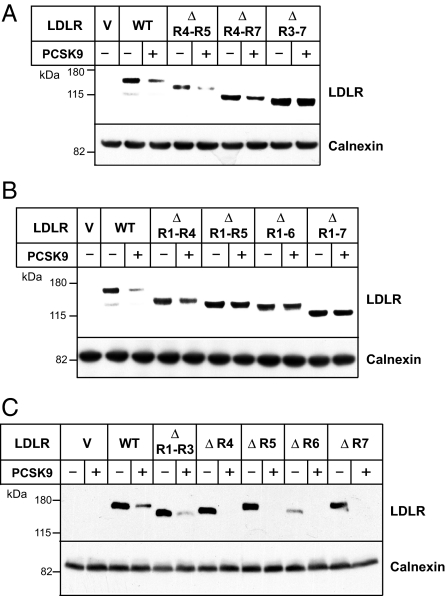
PCSK9 may act like a “doorstop” to block the association of R4 and R5 with the β-propeller domain, thereby preventing recycling of the receptor to the cell surface. To further test this model, we examined whether ligand-binding repeats R4 and R5 were required for PCSK9-stimulated degradation of the LDLR. Addition of PCSK9 to mouse hepatocytes (Hepa1c1c7) expressing LDLRs lacking R4 and R5 resulted in robust degradation of the LDLR (Fig. 2A). LDLRs lacking repeats 4 through 7 were also targeted for degradation, whereas PCSK9 failed to promote degradation of receptors lacking R3–R7.
Fig. 2.
Structural requirements in the ligand binding domain of the LDLR for PCSK9-mediated LDLR degradation. Hepa1c1c7 cells expressing HA-tagged WT or mutant LDLRs were incubated with 2.0 μg/ml purified PCSK9 (D374Y) for 4 h. Cells were lysed in 150 μl of lysis buffer, and analyzed by SDS/PAGE (8%) and immunoblotting as described in Methods. LDLR was detected with an anti-HA polyclonal antibody (A and B) or with a monoclonal antibody, HL-1 (C). Calnexin was detected by a polyclonal antibody. Similar results were obtained in two independent experiments.
Additional LDLR deletion constructs were generated to determine whether specific ligand-binding repeats were required for PCSK9-mediated degradation. Deletion of R1–R4 had little effect on degradation, whereas deletion of R1–R5, R1–R6, and R1–R7 largely abolished the effects of PCSK9 on LDLR levels (Fig. 2B). The observation that LDLRs lacking either R1–R4 or R4–R7 were efficiently targeted for degradation by PCSK9 indicated that no specific ligand-binding repeat was required. This conclusion was confirmed by deleting R1–R3 and by deleting R4, R5, R6, or R7 individually: receptors expressed from each of these constructs were efficiently degraded by PCSK9 (Fig. 2C). In contrast, the finding that PCSK9 had little effect on the cellular levels of receptors lacking R1-R5, or R3-R7 suggested that a minimum of three ligand-binding repeats were required for efficient PCSK9-mediated degradation of LDLRs.
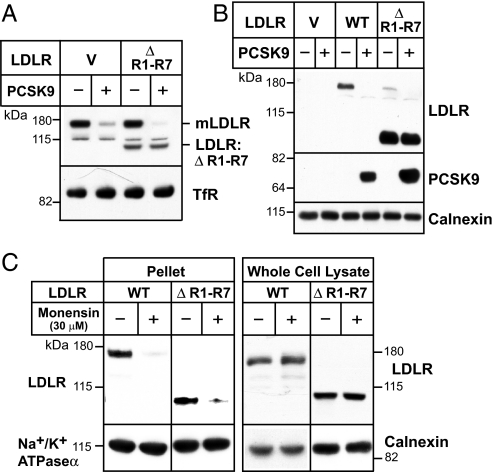
The failure of PCSK9 to promote degradation of LDLRs lacking five or more ligand-binding repeats might be a result of impaired binding of PCSK9 to the mutant receptors. When PCSK9 was added to cultured mouse hepatocytes expressing LDLR:ΔR1-R7, the levels of the endogenous mouse receptor (mLDLR) were reduced, whereas the levels of recombinant LDLR:ΔR1-R7 did not change (Fig. 3A). Two hours after the addition of PCSK9 to the medium, PCSK9 was detectable in cells expressing both the WT and mutant LDLR (Fig. 3B). Thus, PCSK9 bound to the mutant receptor, but failed to target the protein for degradation.
Fig. 3.
PCSK9 binding and internalization of LDLR:ΔR1-R7. (A and B). Experiments were performed as described in the legend to Fig. 2 except that antibody 3143 was used in A to detect LDLR. (C) Hepa1c1c7 cells expressing WT or mutant LDLR were treated with 30 μM monensin for 4 h. The cells were biotinylated exactly as described. Biotinylated proteins from the cell surface (pellet) and proteins from the whole cell lysate were analyzed by SDS/PAGE (8%) and immunoblotting. LDLR was detected using a polyclonal anti-HA antibody. Transferrin receptor (TfR) and Na+/K+-ATPase(α1) were detected with monoclonal antibodies and calnexin was detected with a polyclonal antibody.
PCSK9-mediated degradation requires internalization of the receptor (9). To determine whether LDLR:ΔR1-R7 was internalized, we tested whether the mutant receptor could be trapped inside the cell by monensin, which prevents acidification of endosomes and blocks recycling of receptors to the cell surface (28). The amount of receptor remaining on the cell surface after monensin treatment was monitored by labeling the cells with biotin. The cell surface proteins were precipitated using streptavidin and then immunoblotted for the LDLR. Monensin treatment was associated with a marked reduction in the amount of cell surface LDLR in cells expressing the WT or mutant protein. In contrast, monensin did not reduce the cell surface levels of the Na+/K+-ATPase (α1) receptor, a protein that does not undergo recycling through endosomes (Fig. 3C). As shown in previous studies, monensin did not reduce the amount of full-length LDLR in the whole cell lysates, indicating that the internalized LDLR remained sequestered in endosomes and was not degraded.
We concluded from these experiments that at least three ligand-binding repeats are required for PCSK9-mediated degradation of the LDLR. The role of these repeats could not be determined from these studies, but the repeats are not required for PCSK9 binding or receptor internalization.
Next we examined the effect of deleting four or five ligand binding repeats on the acid-dependent conformational change in the LDLR [supporting information (SI) Fig. S1]. If the conformational change is essential to the action of PCSK9, addition of PCSK9 should prevent this change in LDLRs lacking four ligand binding repeats (R1–R4), but not in receptors lacking five ligand binding repeats (R1–R5). No conformational change in the LDLR was detected with either form of the LDLR in the absence of PCSK9. Thus, preventing the pH-dependent conformational change in the LDLR is not sufficient to trigger degradation of the receptor.
Sequences in the EGF Precursor Homology Domain Required for PCSK9-Mediated Degradation of the LDLR.
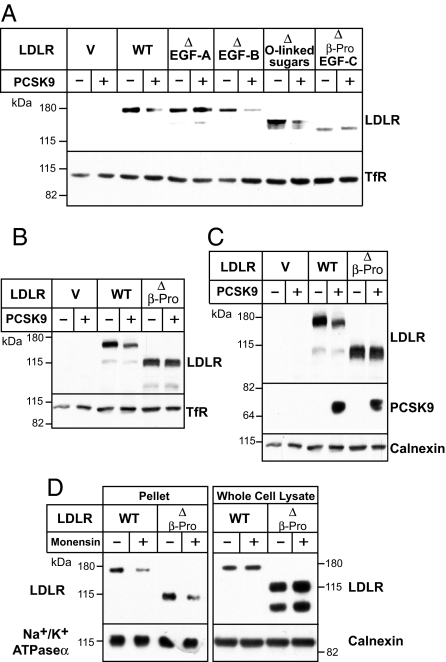
Next we determined which regions of the EGF precursor homology domain are required for PCSK9-associated LDLR degradation. As expected, no reduction in cellular LDLRs occurred in cells expressing receptors that did not contain the EGF-A repeat (Fig. 4A), because this repeat is required for PCSK9 binding to the LDLR (22). PCSK9 promoted degradation of the LDLR when either EGF-B repeat or the O-linked sugar region was removed. Deletion of EGF-C alone resulted in retention of the protein in the ER (22), so this construct was not studied further. Addition of PCSK9 failed to stimulate LDLR degradation if the β-propeller domain (YWTD repeats) plus EGF-C were deleted (Fig. 4A). Similarly, removal of the β-propeller domain prevented degradation (Fig. 4B). Thus, whereas LDLRs lacking R4 and R5 were efficiently degraded by PCSK9 (Fig. 2A), receptors that lacked the β-propeller domain were not (Fig. 4B).
Fig. 4.
Structural requirements within the EGF precursor homology domain of the LDLR for PCSK9-mediated degradation. (A–C) The experiments were performed as described in the legend to Fig. 2. (D) The experiments were performed exactly as described in the legend to Fig. 3C. Similar results were obtained in two additional independent experiments.
The inability of PCSK9 to promote degradation of the LDLR in the absence of the β-propeller domain might be caused by an inability of the mutant receptor to bind PCSK9 or to be internalized in hepatocytes. To distinguish between these possibilities, we performed experiments with monensin, as described earlier. PCSK9 bound LDLRs lacking the β-propeller domain (Fig. 4C), and the receptors were internalized (Fig. 4D). Moreover, examination of the whole cell lysates revealed no reduction in LDLR. These data demonstrated that the β-propeller domain (YWTD repeats), like the ligand binding repeats, is required for PCSK9-mediated degradation of the protein.
N-Terminal Domain of PCSK9 Binds LDLR but Does Not Promote Receptor Degradation.
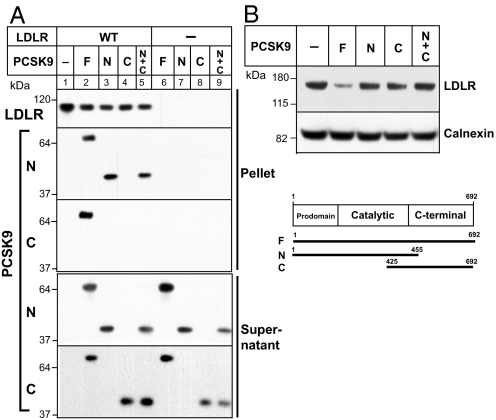
The PCSK9-binding site on LDLR was localized to the EGF-A domain (22). Cocrystallization of PCSK9 with the EGF-A and -B repeats revealed that the binding surface was between the N-terminal half of EGF-A and the catalytic domain of PCSK9 (29). To determine whether the C terminus of PCSK9 also binds the LDLR, we incubated the extracellular domain of the LDLR with recombinant PCSK9 fragments, including the full-length PCSK9 (F), the N-terminal two thirds of the protein (amino acid 1–454) comprising the prodomain and the catalytic domain (N), and the C-terminal domain of PCSK9 (amino acids 425–692) (C). The LDLR was immunoprecipitated and the pellet was size-fractionated and immunoblotted using antibodies that recognize the N terminus and the C terminus of PCSK9 (Fig. 5A). As expected, full-length PCSK9 was detected in the pellet using both anti-PCSK9 antibodies. The N terminus, but not the C terminus, of PCSK9 coprecipitated with the LDLR. When both the N- and C-terminal portions of PCSK9 were incubated with LDLR in the same reaction, only the N-terminal fragment bound to the receptor and was detected in the pellet.
Fig. 5.
Interaction of the extracellular domain of LDLR with WT and mutant PCSK9. (A) Coimmunoprecipitation of the extracellular domain of LDLR and PCSK9. A total of 1 μg of full-length (amino acids 1–692) (F), N-terminal (amino acids 1–454) (N), or C-terminal (amino acids 425–692) (C) PCSK9 was incubated overnight at 4°C with the extracellular domain of LDLR (1 μg) in the presence of an anti-LDLR polyclonal antibody (4548) and protein A agarose. After centrifugation, proteins were eluted from the pellets and subjected to electrophoresis on a 4–12% NuPage Bis-Tris gradient gel. (B) Degradation of LDLR by PCSK9 in Huh-7 cells. Cells were incubated in 1 ml of DMEM containing 5% (vol/vol) newborn calf lipoprotein-poor serum, 10 μg/ml cholesterol, 1 μg/ml 25-HC, and 5.0 μg/ml purified PCSK9 for 4 h. Cell lysates were subjected to immunoblotting using a monoclonal antibody to LDLR. PCSK9 was detected using monoclonal antibodies (15A6 or 13D3). Calnexin was detected by a polyclonal antibody.
These data confirm that the major association between PCSK9 and the LDLR involves sequences in the N-terminal region of the protein. To determine whether the N terminus of PCSK9 alone can promote LDLR degradation, we added PCSK9 fragments to cultured human hepatocytes (Huh-7 cells) and measured the levels of LDLR. Only addition of the full-length protein to the medium was associated with a reduction in LDLRs (Fig. 5B). Thus, although the N-terminal portion of PCSK9 can bind the receptor, this fragment cannot support degradation of the LDLR. Similarly, addition of the C-terminal fragment together with the N-terminal fragment did not support receptor degradation.
PCSK9 Prodomain and Catalytic Domain Are Degraded Coordinately in Hepatocytes.
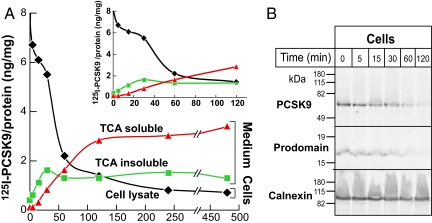
To determine the fate of PCSK9 after binding to the LDLR, we added 125I-radiolabeled PCSK9 to HepG2 cells and followed the metabolism of the enzyme. After 1-h incubation, the temperature was reduced to 4°C and the cells were washed to remove PCSK9 from the cell surface. Fresh medium was added and the total radioactivity in the cell lysates and trichloroacetic acid (TCA)-precipitable radioactivity in the medium were measured at intervals over the ensuing 8 h (Fig. 6A). We also monitored the amount of iodine released into the medium (i.e., TCA-soluble). Within 1 h, the amount of radioactive PCSK9 in the cell lysate was substantially reduced. The reduction in cellular PCSK9 was followed by an increase in TCA-soluble counts in the medium, indicating that PCSK9 was being degraded. The levels of TCA-insoluble protein in the medium increased over the first hour and then reached a plateau; this radioactivity may represent the release of residual PCSK9 from the cell surface into the medium.
Fig. 6.
Degradation of 125I-labeled PCSK9 in HepG2 cells. HepG2 cells were incubated in 2 ml of medium A plus 2.5 μg/ml 125I-labeled PCSK9 (600 cpm/ng) for 1 h. After washing, cells were incubated in medium A at 37°C. The amounts of 125I-labeled TCA-soluble and TCA-insoluble material were determined in the cells and medium at the indicated time intervals. (B) Cells were incubated with labeled PCSK9 (1 μg) for 3 h at 4°C and then 40 min at 37°C before adding stripping buffer. After the indicated times, 10% of the protein in the cell lysate and medium was subjected to immunoblotting as described. This experiment was repeated twice and the results were similar.
To determine whether the prodomain and the catalytic domain are degraded at the same rate, we incubated cells with labeled PCSK9 (DyLight 800) for 1 h and then washed the cells extensively at 4°C. Time courses for the disappearance of the prodomain and catalytic/C-terminal domain of PCSK9 are shown in Fig. 6B. No changes were apparent in the sizes of either the prodomain or the mature form of PCSK9 over the ensuing 2 h. The amount of the two protein segments decreased coordinately in the cells until only trace amounts of protein were visible at 120 min (Fig. 6B). No PCSK9 was detected in the medium (data not shown). These results are consistent with PCSK9 binding the LDLR and being internalized and degraded, possibly together with the receptor. PCSK9 did not undergo further processing or recycling.
Mechanistic Implications.
Several lines of evidence indicate that PCSK9 reroutes the LDLR from the recycling pathway to lysosomes, but the mechanism by which this rerouting occurs has not been molecularly defined. One possibility is that PCSK9 contains a lysosomal targeting signal that targets the LDLR for degradation. This model is not consistent with our observation that binding of PCSK9 to the LDLR is necessary, but not sufficient to promote degradation of the receptor; LDLRs lacking the ligand-binding domain or the β-propeller domain can bind PCSK9 and enter the endosome, but are not subject to PCSK9-mediated degradation (Figs. 2 and 4).
A second possibility is that PCSK9 binding prevents the LDLR from adopting a conformation required for recycling. Naturally occurring mutations in the β-propeller domain disrupt recycling of the LDLR and are associated with rapid turnover of the receptor (1, 25). Crystallization of the extracellular portion of the LDLR at acidic pH revealed a physical association between R4 and R5 with the β-propeller domain (20), and we previously suggested that PCSK9 binding may prevent this pH-dependent conformational change (19, 22). The gel filtration studies presented here indicate that PCSK9 does prevent the LDLR from assuming a different conformation at acidic pH (Fig. 1), but the finding that R4 and R5 are not required for PCSK9-mediated degradation of the receptor suggests that the doorstop model may be an oversimplification. The observation that deletion of four ligand binding repeats disrupts the acid-dependent change in LDLR conformation (Fig. S1) but does not prevent PCSK9-mediated degradation of the receptor (Fig. 2) provides further evidence that disrupting the conformational change in LDLR is not sufficient to explain PCSK9 action.
PCSK9 binding to EGF-A may disrupt the interface that forms between the last ligand binding repeat (R7) and EGF-A. Previously, Blacklow and colleagues (30) demonstrated that the structural orientation of these two modules did not change significantly with reductions in pH. This interface serves as a rigid elbow that allows the ligand binding repeats to associate with the β-propeller domain when the pH decreases. Mutations that disrupt the architecture of this region have reduced acid-dependent ligand release (30), and it remains possible that PCSK9 binding may interfere with the structure of this region.
It also is possible that PCSK9 associates with more than one region of the LDLR, although we failed to detect association of PCSK9 with the LDLR in cells expressing receptors lacking EGF-A, either at pH 7.4 or 6.0 (data not shown).
Our data are most consistent with a model in which PCSK9 either prevents the receptor from interacting with another protein that is required for receptor recycling or promotes the association of the receptor with protein(s) that signal lysosomal delivery. Such an interacting protein may bind the prodomain or the C-terminal region of PCSK9, two otherwise conserved domains of unknown function. The prodomain of PCSK9 appears not to undergo a secondary cleavage event and remains tightly attached to the catalytic domain after PCSK9 is secreted, which distinguishes it from other proprotein convertases. Furthermore, there is no evidence that the prodomain is further processed, either in the circulation or after it is internalized with the LDLR (Fig. 6). The similarity in the time course of degradation of the prodomain and the catalytic domain of PCSK9 is consistent with these two regions remaining tightly associated until they are degraded, presumably in the lysosome. The functional role of the C-terminal domain of PCSK9 also remains to be defined. This large, ≈240-aa, cysteine- and histidine-rich domain appears to be quite unrestrained by the catalytic domain (27), and may provide a site for interaction with other proteins involved in PCSK9 trafficking. Our finding that the C-terminal domain does not bind the LDLR, but is essential for its degradation, is consistent with this notion.
Materials and Methods
Materials.
Lipofectamine 2000 was obtained from Invitrogen. Culture medium and PBS were purchased from Mediatech and FBS was obtained from Atlanta Biologicals. Complete EDTA-free protease inhibitors were purchased from Roche, and all other reagents were obtained from Sigma-Aldrich unless otherwise indicated.
The extracellular domain of the LDLR (amino acid 1–699) used in these experiments contained a six-histidine residue tag at the C terminus, and was purified exactly as described (20). The recombinant full-length, N-terminal, and C-terminal human PCSK9 contained a FLAG tag at the C terminus and was purified from HEK-293S cells (9). PCSK9 was labeled using a DyLight 800 Antibody Labeling Kit (Pierce Biotechnology) according to the manufacturer's protocol, and the proteins were visualized using the Odyssey infrared imaging system (LI-COR Biosciences). PCSK9 was radioidinated with 125I using the monochloride method (31).
The following antibodies were used: HL-1, a monoclonal antibody to the linker sequence between R4 and R5 of the LDLR (32); 3143, an anti-LDLR polyclonal anti-serum directed against the C-terminal 14 aa of the LDLR (33); 4548, a polyclonal antiserum against full-length LDLR (34); 13D3 and 15A6, monoclonal antibodies developed against full-length PCSK9 that recognize epitopes in the catalytic domain and the C-terminal regions of PCSK9, respectively (9); 228B, a polyclonal antibody against full-length PCSK9 (9); an antihemagglutinin polyclonal antibody (Invitrogen); TfR, an antitransferrin receptor monoclonal antibody (Invitrogen); a polyclonal antibody against calnexin (Invitrogen); and an anti-Na+K+-ATPaseα1 monoclonal antibody (Santa Cruz Biotechnology).
Gel Filtration of LDLR.
Gel filtration chromatography was performed on an AKTA purifier system (GE Healthcare). Purified extracellular domain of LDLR (60 μg) was incubated in 20 μl of Hepes buffer (50 mM Hepes, pH 7.4; 150 mM NaCl; 2 mM CaCl2) for 30 min at room temperature in the presence or absence of purified PCSK9 (160 μg) and then incubated in 500 μl of buffer A (50 mM Tris-Maleate, 150 mM NaCl, 2 mM CaCl2, 0.5% Tween-20, pH 7.4 or 6.0) for 2 h at room temperature on a rotator. The samples were centrifuged at 16,000 × g for 1 min and the supernatants were injected onto a Superdex 200 10/300 GL or a Superose 12 10/300 GL column (GE Healthcare) that had been equilibrated with the same buffer. The eluate was collected in 0.5-ml fractions. The retention time of LDLR and PCSK9 was determined by SDS/PAGE and immunoblotting of the collected fractions. Gel filtration protein markers, including thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) were separated under the same conditions and detected using a UV detector.
Site-Directed Mutagenesis.
Recombinant expression vectors containing the full-length LDLR cDNA linked to the SV40 early promoter (pLDLR2) (35) or the coding region driven by the human cytomegalovirus immediate early region promoter (pLDLR17) were used to generate mutant forms of the LDLR, including deletions in the ligand binding domain (36), the EGF precursor homology domain (25), and the clustered O-linked sugar domain (37). A pShuttle-RSV vector containing WT or mutant forms of the LDLR cDNA under the control of the rous sarcoma virus (RSV) promoter (pRSV-LDLR) was made from PCR fragments. A 421-bp amplification fragment containing the RSV promoter from pLenti6/V5-D-Topo (Invitrogen) was ligated into the KpnI site in the pShuttle promoterless vector (Stratagene). The WT or mutant LDLR cDNA was then amplified by PCR using pLDLR2 or pLDLR17 as templates and inserted into the vector pCRII-Topo (Invitrogen). The resulting construct was ligated into pShuttle-RSV after digestion with KpnI and XbaI, generating pRSV-LDLR. The presence of the desired mutation and the integrity of each construct were verified by DNA sequencing.
Degradation of LDLR by PCSK9.
The mouse hepatoma cell line Hepa1c1c7 was cultured in MEMα medium (Invitrogen) containing 10% (vol/vol) FBS at 37°C in a 7% CO2 incubator and seeded (4 × 105 cells per well) in six-well dishes in 2 ml of culture medium. After 24 h, cells were transfected with expression plasmids containing cDNAs for WT or mutant LDLR and pShuttle vector (1.5–2.0 μg per well) using Lipofectamine 2000 according to the manufacturer's protocol. Forty-eight hours after transfection, cells were washed twice with PBS, incubated in 1 ml of MEMα medium containing 5% (vol/vol) newborn calf lipoprotein-poor serum, 10 μg/ml cholesterol, 1 μg/ml 25-hydroxycholesterol, and 2.0 μg/ml purified PCSK9-D374Y for 4 h. Cells were then washed twice with ice-cold PBS, lysed in 150 μl of lysis buffer (50 mM Tris, 150 mM NaCl, 1.5 mM MgCl2, 1% [vol/vol] Triton). Whole-cell lysate protein extracts were then analyzed by SDS/PAGE (8%) and immunoblotted.
Monensin Treatment and Biotinylation of LDLR.
Hepa1c1c7 cells were transfected with plasmids expressing WT or mutant LDLR (1.5 to 2.0 μg per well) using Lipofectamine 2000. After 48 h, cells were treated with 30 μM monensin for 4 h. Cell surface proteins were biotinylated exactly as described (22). The cells were lysed in 150 μl of lysis buffer and then subjected to centrifugation at 15,000 rpm for 5 min. A total of 50 μl of the cell lysate was saved and 90 μl of the lysate was added to 60 μl of a 50% slurry of Neutravidin-agarose (Pierce) and 410 μl of lysis buffer. The mixture was rotated overnight at 4°C. After centrifugation at 3000 × g for 5 min, the pellets were washed in lysis buffer three times for 10 min at 4°C. The cell surface proteins were eluted from the beads by adding 1× SDS loading buffer (31 mM Tris·HCl, pH 6.8, 1% SDS, 12.5% glycerol, 0.0025% bromophenol) and incubated for 5 min at 90°C. Proteins were then analyzed by SDS/PAGE (8%) and immunoblotted.
Coimmunoprecipitation of Purified LDLR Extracellular Domain (Amino Acids 1–699) and PCSK9.
Purified WT or mutant PCSK9 (1 μg) and extracellular domain of LDLR (1 μg) were incubated in 500 μl of immunoprecipitation buffer (PBS, 1 mM CaCl2, 1% Tween-20 and protease inhibitors) for 2 h at 4°C on a rotator. An anti-LDLR polyclonal antibody (4548) and protein A agarose (Replicon) were added and the mixture was incubated overnight at 4°C. After centrifugation at 3,000 × g for 5 min, the supernatants were removed and retained. The pellets were washed in immunoprecipitation buffer three times for 10 min at 4°C, and the proteins were eluted using 1× SDS loading buffer containing 5% 2-mercaptoethanol (pellets). Proteins were then size fractionated on 4–12% Bis-Tris gradient gels (Invitrogen) and visualized by immunoblotting.
PCSK9 Degradation Assays.
HepG2 cells were seeded in 60-mm dishes (4 × 105 cells per dish) in DMEM (glucose, 1 g/liter) containing 5% (vol/vol) FBS. After 48 h, the medium was switched to DMEM plus 5% (vol/vol) human lipoprotein-poor serum (medium A). After 48 h, the cells were washed with PBS and incubated in 1 ml of medium A containing 2.5 μg/ml 125I-radiolabeled PCSK9 (600 cpm/ng) for 1 h. The medium was removed and the cells were washed for 5 min at 4°C with ice-cold Buffer B [PBS, 0.1 mM CaCl2, 1 mM MgCl2, 0.5% BSA (wt/vol)]. The cells were rewashed for 5 min with PBS and then incubated in 2 ml of medium A for 8 h at 37°C. The amounts of 125I-labeled TCA-soluble and TCA-insoluble material were determined in the cells and medium at the indicated time intervals (31).
Cells were incubated at 4°C for 1 h, washed once with Medium A, and 0.5 μg/ml of labeled PCSK9 (DyLight 800) was added to the medium. After 3 h at 4°C, the cells were washed twice with ice-cold Buffer B and once with PBS. The cells were then incubated at 37°C for 40 min in Medium A and washed twice with Buffer B, and the medium was replaced with stripping buffer (100 mM Na 2-mercapto-ethanesulfonate, 50 mM Tris, pH 8.6, 100 mM NaCl, 1 mM EDTA, 0.2% BSA) before being washed twice with Buffer B. The cells were incubated in serum-free medium and both medium and cells were collected at 0, 5, 15, 30, 60, and 120 min. The cells were washed with PBS and harvested in lysis buffer. The medium was concentrated using Microcon centrifugal filter (Ultra YM-3). A total of 10% of the cell lysate (30 μg of protein) and medium (200 μl) were subjected to SDS/PAGE and immunoblotting. The proteins were visualized using the LI-COR Odyssey infrared imaging system.
Supplementary Material
Acknowledgments.
We thank David Russell and Jay Horton for helpful discussions, and Christina Zhao and Natalie Toto for excellent technical assistance. This work was supported by National Institutes of Health Grant P01 HL 20948.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806312105/DCSupplemental.
References
- 1.Goldstein J, Hobbs H, Brown M, Scriver C, Beaudet A, Sly W, Valle D. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw Hill; 2001. pp. 2863–2913. [Google Scholar]
- 2.Innerarity TL, et al. Familial defective apolipoprotein B-100: A mutation of apolipoprotein B that causes hypercholesterolemia. J Lipid Res. 1990;31:1337–1349. [PubMed] [Google Scholar]
- 3.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 4.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 5.Seidah NG, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): Liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naureckiene S, et al. Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch Biochem Biophys. 2003;420:55–67. doi: 10.1016/j.abb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham D, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 8.Piper DE, et al. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure (London) 2007;15:545–552. doi: 10.1016/j.str.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Lagace TA, et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron J, et al. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum Mol Genet. 2006;15:1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- 11.McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J Biol Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem J. 2007;406:203–207. doi: 10.1042/BJ20070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of LDL receptor protein by proprotein convertase subtilisin/kexin type 9a (PCSK9) in mouse liver. J Biol Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 18.Lalanne F, et al. Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. J Lipid Res. 2005;46:1312–1319. doi: 10.1194/jlr.M400396-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudenko G, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 21.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DW, et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 23.Fisher TS, et al. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J Biol Chem. 2007;282:20502–20512. doi: 10.1074/jbc.M701634200. [DOI] [PubMed] [Google Scholar]
- 24.Südhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: A mosaic of exons shared with different proteins. Science. 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CG, et al. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 26.Springer T. An extracellular B-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 27.Hampton EN, et al. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc Natl Acad Sci USA. 2007;104:14604–14609. doi: 10.1073/pnas.0703402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu SK, Goldstein JL, Anderson RG, Brown MS. Monensin interrupts the recycling of low density lipoprotein receptors in human fibroblasts. Cell. 1981;24:493–502. doi: 10.1016/0092-8674(81)90340-8. [DOI] [PubMed] [Google Scholar]
- 29.Kwon HJ, Lagace TA, McNutt MC, Horton JD, Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc Natl Acad Sci USA. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beglova N, Jeon H, Fisher C, Blacklow SC. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell. 2004;16:281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 32.van Driel IR, Davis CG, Goldstein JL, Brown MS. Self-association of the low density lipoprotein receptor mediated by the cytoplasmic domain. J Biol Chem. 1987;262:16127–16134. [PubMed] [Google Scholar]
- 33.Russell DW, et al. Domain map of the LDL receptor: Sequence homology with the epidermal growth factor precursor. Cell. 1984;37:577–585. doi: 10.1016/0092-8674(84)90388-x. [DOI] [PubMed] [Google Scholar]
- 34.Beisiegel U, et al. Immunologic cross-reactivity of the low density lipoprotein receptor from bovine adrenal cortex, human fibroblasts, canine liver and adrenal gland, and rat liver. J Biol Chem. 1981;256:4071–4078. [PubMed] [Google Scholar]
- 35.Yamamoto T, et al. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 36.Esser V, Limbird LE, Brown MS, Goldstein JL, Russell DW. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J Biol Chem. 1988;263:13282–13290. [PubMed] [Google Scholar]
- 37.Davis CG, et al. Deletion of clustered O-linked carbohydrates does not impair function of low density lipoprotein receptor in transfected fibroblasts. J Biol Chem. 1986;261:2828–2838. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.