Abstract
The Walker A and B motifs of nucleotide binding domains (NBDs) of Cdr1p though almost identical to all ABC transporters, has unique substitutions. We have in the past shown that Trp326 of Walker B and Cys193 of Walker A motifs of N-terminal NBD of Cdr1p have distinct roles in ATP binding and hydrolysis, respectively. In the present study, we have examined the role of a well conserved Asp327 in the Walker B motif of the N-terminal NBD which is preceded (Trp326) and followed (Asn328) by atypical amino acid substitutions and compared it with its equivalent well conserved Asp1026 of the C-terminal NBD of Cdr1p. We observed that the removal of the negative charge by D327N, D327A, D1026N, D1026A and D327N/D1026N substitutions, resulted in Cdr1p mutant variants that were severely impaired in ATPase activity and drug efflux. Importantly, all the mutant variants showed characteristics similar to those of wild type with respect to cell surface expression and photoaffinity drug analogue [125I] IAAP and [3H] azidopine labeling. While Cdr1p D327N mutant variant showed comparable binding with [α-32P] 8-azido ATP, Cdr1p D1026N and Cdr1p D327N/D1026N mutant variants were crippled in nucleotide binding. That the two conserved carboxylate residues Asp327 and Asp1026 are functionally different was further evident from the pH profile of ATPase activity. Cdr1p D327N mutant variant showed ∼40% enhancement of its residual ATPase activity at acidic pH while no such pH effect was seen with Cdr1p D1026N mutant variant. Our experimental data suggest that Asp327 of N-terminal NBD has acquired a new role to act as a catalytic base in ATP hydrolysis, a role normally conserved for Glu present adjacent to the conserved Asp in the Walker B motif of all the non-fungal transporters.
One of the most clinically significant mechanisms of azoles resistance in the pathogenic fungi, C. albicans is an over expression of the drug efflux pumps encoding genes CDR1 and CDR2 belonging to the ABC (ATP-Binding Cassette) (1-7) and CaMDR1 belonging to MFS (Major Facilitator Superfamily) transporters (8-10). Among the ABC transporters, high level of expression of CDR1 invariably contributes to an increased efflux of fluconazole and thus corroborates its direct involvement in drug efflux (6, 11, 12). Hence, Cdr1p has not only acquired significant clinical importance but is also considered an important target in any design of strategies to combat antifungal resistance (7, 13, 14).
Typically like other ABC proteins, the functional form of Cdr1p consists of two hydrophilic nucleotide binding domains (NBDs) located at the cytoplasmic surface of the membrane that harness energy from ATP hydrolysis and two hydrophobic transmembrane domains (TMDs) that are thought to form the translocation pathway for drug efflux. These NBDs of the ABC proteins contain three conserved motifs required for nucleotide binding and hydrolysis: the Walker A and Walker B motifs (15) and the ABC signature sequence (16). ATP hydrolysis and substrate transport is dependent on the cooperativity between NBDs. The Walker A motif (GX4GK/CT/S) forms the phosphate binding loop, or P-loop, which wraps around the phosphate chain of ATP within this motif and makes extensive hydrogen bonding with β-phosphate (17). The Walker B motif (I(Hy)4D) provides a carboxylate residue that coordinates and stabilizes the Mg2+, which is a mandatory cofactor in the hydrolysis of ATP (18). Recent crystal structure of the stable MJ0796-E171Q dimer an ABC transporter of Methanococcus jannaschii, describes the 3D structure of the nucleotide binding pocket wherein it is proposed that NBDs form a symmetrical dimer in which two ATP molecules are sandwiched between the Walker A and Walker B motif of one NBD and the signature motif of other NBD (19). The deduced 3D structure has established that the signature motifs of NBDs physically contribute to the active site by forming hydrogen bonds with ribose and γ-phosphate of ATP (19). Although significant structural and functional differences are likely to exist among different ABC proteins, it has been proposed that because of the conserved nature of domains, the overall structure of NBD sites of ABC proteins is probably very similar (2). Unlike most other ABC transporters, the NBDs of C. albicans and all other fungal transporters of the super family have unique positioning of a typical amino acid Cys193 in the Walker A, Trp326 and Asn328 in and adjacent to Walker B motifs of N-terminal NBD respectively (20). Interestingly, the C-terminal NBD of Cdr1p and other ABC fungal transporters possesses conserved motifs which are essentially identical to non fungal transporters (20). To begin defining the functional significance of the conserved substitutions in N-terminal NBD of Cdr1p, we have recently demonstrated that the replacement of Cys193 with Ala gravely impaired the ATP hydrolysis without affecting its ability to bind the nucleotide (21, 22). On the other hand, substitution of Trp326 with Ala resulted in the loss of ATP binding to purified N-terminal NBD (NBD-512) (23). A mutagenesis screen of another conserved residue underscored the importance of the highly conserved, putative catalytic carboxylate residue Asp327 of the Walker B motif in the purified N-terminal NBD of Cdr1p. The substitution of Asp327 with Asn yielded mutant variant protein with strongly impaired ATPase activity, while it showed comparable nucleotide binding to that of wild type protein. In the crystal structure of HisP, the ATPase subunit of an ABC transporter Histidine Permease from Salmonella typhimurium, the negatively charged side chain of this equivalent Walker B Asp residue (Asp178) hydrogen bonds a water molecule in proximity to the γ-phosphate of ATP. This water molecule has been proposed to replace Mg2+ in the crystal structure (24), implying that Asp178 and its equivalent in P-gp (Asp555/Asp1200) coordinate Mg2+ (25). However, our initial results suggest that equivalent Asp residue (Asp327) in the Walker B motif of N-terminal NBD of Cdr1p unlike in other non-fungal ABC transporters, may not be involved in Mg2+ coordination (23).
In this study, the precise role of Asp327 of N-terminal NBD in ATP binding and hydrolysis is further explored and is compared with its counter part Asp1026 of C-terminal NBD. Substitution of both Asp residues resulted in severely impaired ATPase activity and drug transport. Importantly, both mutant variants showed characteristics similar to those of wild type with respect to photoaffinity drug analogues of prazosin, [125I] IAAP (iodoarylazidoprazosin) and of dihydropyridine, [3H]-azidopine labeling, but showed differential labeling with [α-32P] 8-azido-ATP. While Cdr1p D1026N and Cdr1p D327N/D1026N was severely impaired in nucleotide binding, indicating a direct conventional role of Asp1026 in Mg2+ coordination, mutant variants Cdr1p D327N or Cdr1p D327A elicited nucleotide binding comparable to that of wild type Cdr1p. Our results show that because of replacement of conserved Glu with Asn at 328 position in N-terminal NBD of Cdr1p and as well as other fungal ABC transporters, the conserved Asp327 of the Walker B motif of N-terminal NBD has acquired a new role to act as a catalytic base for ATP hydrolysis.
Experimental Procedures
Materials
Ribonucleotides (ATP, ADP), protease inhibitors: PMSF, leupeptin, pepstatin A, aprotinin, TLCK, TPCK and drugs: miconazole, cycloheximide, anisomycin, ketoconazole, itraconazole, R6G, DTT, oligomycin and other molecular grade chemicals were obtained from Sigma Chemical Co (St. Louis, Mo). Oligonucleotides used in this study, as listed in Table 1 were commercially procured from Sigma Genosys, Inc. Fluconazole was kindly provided by Ranbaxy Laboratories (New Delhi, India). Anti-GFP monoclonal antibody and [α-32P] 8-azido ATP (15-20 Ci/mmol) were purchased from BD Biosciences Clontech (Palo Alto, CA) and Affinity Labeling Technologies, Inc. (Lexington, Ky.), respectively. [3H] azidopine (60 Ci/mmol) was obtained from Amersham Biosciences (Arlington Heights, III). The radio labeled [125I] IAAP (2,200 Ci/mmol) was procured from Perkin Elmer Life Sciences (Boston, Mass.).
Table 1. List of oligonucleotides used in this study.
| Name | Sequence |
|---|---|
| D327A/F | 5′-CTAATATCCAATGTTGGGCAAATGCCACTAGAGGG-3′ |
| D327A/R | 5′-CCCTCTAGTGGCATTTGCCCAACATTGGATATTAG-3′ |
| D327N/F | 5′-CTAATATCCAATGTTGGAATAATGCCACTAGAGGG-3′ |
| D327N/R | 5′-CCCTCTAGTGGCATTATTCCAACATTGGATATTAG-3′ |
| D327E/F | 5′ -CTAATATCCAATGTTGGGAAAATGCCACTAGAGGG -3′ |
| D327E/R | 5′ –CCCTCTAGTGGCATTTTCCCAACATTGGATATTAG -3′ |
| N328E/F | 5′-TATCCAATGTTGGGAGAATGCCACTAGAGGGTTAGA-3′ |
| N328E/R | 5′-TCTAACCCTCTAGTGGCATTCTCCCAACATTGGATA-3′ |
| N328A/F | 5′-TATCCAATGTTGGGAGCATGCCACTAGAGGGTT-3′ |
| N328A/R | 5′-AACCCTCTAGTGGCATGCTCCCAACATTGGATA- 3′ |
| D1026N/F | 5′-CCTAAATTGTTGTTATTCTTAAACGAACCAACTTCAGGGTTA 3′ |
| D1026N/R | 5′-TAACCCTGAAGTTGGTTCGTTTAAGAATAACAACAATTTAGG 3′ |
| D1026A/F | 5′-CCTAAATTGTTGTTATTCTTAGCCGAACCAACTTCAGGGTTA 3′ |
| D1026A/R | 5′-TAACCCTGAAGTTGGTTCGGCTAAGAATAACAACAATTTAGG 3′ |
| E1027Q/F | 5′-ATTGTTGTTATTCTTAGACAAACCAACTTCAGGGTTAGA 3′ |
| E1027Q/R | 5′-TCTAACCCTGAAGTTGGTTTGTCTAAGAATAACAACAAT- 3′ |
| E1027A/F | 5′-ATTGTTGTTATTCTTAGAGCAACCAACTTCAGGGTTAGA 3′ |
| E1027A/R | 5′-TCTAACCCTGAAGTTGGTTGCTCTAAGAATAACAACAAT- 3′ |
Media chemical and strains
Plasmids were maintained in Escherichia coli DH5α. E. coli was cultured in Luria-Bertani medium (Difco, BD Biosciences, NJ, USA) to which ampicillin was added (0.1 mg/ml). The yeast strains were cultured in YEPD broth (Bio101, Vista, CA, USA) or SD-ura- (Bio101). Table 2 lists all the strains used in this study.
Table 2. List of strains used in this study.
| Name | Description | Reference |
|---|---|---|
| Yeast strains | ||
| AD1-8u- | MATa pdr1-3 hisG ura3 Δyor1::hisG Δsnq2::hisG Δpdr10::hisG Δpdr11::hisG Δycf1::hisG Δpdr15::hisG | (27) |
| PSCDR1-GFP | AD1-8u- cells harboring CDR1-GFP ORF integrated at PDR5 locus | (26) |
| VRCG D327N | CDR1-GFP cells carrying D327N mutation in CDR1 ORF and integrated at PDR5 locus | (23) |
| VRCG D327A | CDR1-GFP cells carrying D327A mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG D327E | CDR1-GFP cells carrying D327E mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG D1026N | CDR1-GFP cells carrying D1026N mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG D1026A | CDR1-GFP cells carrying D1026A mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG D327N/D1026N | CDR1-GFP cells carrying D327N/D1026N mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG N328E | CDR1-GFP cells carrying N328E mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG N328A | CDR1-GFP cells carrying N328A mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG E1027Q | CDR1-GFP cells carrying E1027Q mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG E1027A | CDR1-GFP cells carrying E1027A mutation in CDR1 ORF and integrated at PDR5 locus | This study |
| VRCG N328E/E1027Q | CDR1-GFP cells carrying N328E/E1027Q mutation in CDR1 ORF and integrated at PDR5 locus | This study |
Methods
Site-specific mutagenesis
Site directed mutagenesis was performed using quick-change mutagenesis system as described previously (26). The mutations were introduced into plasmid pPSCDR1-GFP according to manufacturer's instructions, and the mutated plasmid pPSCDR1-GFP linearized with XbaI was used to transform AD1-8u- cells by the lithium acetate transformation protocol exploiting uracil prototrophy (26, 27).
Immunodetection of Cdr1-GFP
Plasma membranes (PM) were prepared from Saccharomyces cerevisiae cells grown in YEPD to late exponential phase, as described previously (26). The Western blot analysis was carried out using anti-GFP monoclonal antibody (1:5000 dilution) and anti-Pma1p polyclonal antibody (1:1000 dilution) as described previously (26).
Assay of drug susceptibility, Rhodamine 6G efflux and ATPase activity
The drug susceptibilities, rhodamine 6G efflux and ATPase activity of S. cerevisiae cells, harboring wild type Cdr1p and its mutant variants were determined as described earlier (23, 26-28).
Confocal Microscopy and Flow cytometry
Confocal imaging and flow cytometric (FACS) analysis of the Cdr1p and its mutant variants carrying S. cerevisiae cells was performed with Bio-Rad confocal microscope (MRC 1024) with 100X oil immersion objective and FACSort flow cytometer (Becton-Dickson Immuno cytometry systems, San Jose, Calif.) as described previously (26).
Photoaffinity labeling
Photoaffinity labeling of Cdr1p and its mutant variants with 0.5 μM [3H]-azidopine (specific activity 60 Ci/mmol) and 7.5 nM [125I] IAAP (2200 Ci/mmol) and with 10 μM [α-32P] 8-azido ATP (7.5 μCi/nmol) was performed with PM proteins (30-50 μg) as previously described previously (26).
Homology modeling of NBDs of Cdr1p
Since no structural data is currently available for NBDs of Cdr1p, the model of CDR1-NBD catalytic dimer along with the ligands (ATP and Mg2+) was generated by homology modeling, to verify the experimental findings. The crystal structure of dimeric E171Q mutant of MJ0796, an ABC transporter from M. jannaschii (Protein Data Bank (PDB) code 1L2T) (19) was used as a template for modeling dimer. The target sequence was then aligned using ClustalX version 1.83 (29,30). The alignment was manually edited so that the loop regions involved with ATP were not influenced by the template structure but by the ATP and metal ligand. The model was built using the program Modeller8v2 (31). An ensemble of 50 structures was generated and the one with the lowest objective function (energy) was chosen for further analysis. Model evaluation was done using PROCHECK v.3.5.4 (32).
Routine Procedures
Protein concentrations were determined by bicinchonic acid method (33). SDS-PAGE was carried out according to Laemmli (34) using the Mini-PROTEAB II gel and Electro transfer system (Bio-Rad).
Results
Cdr1p mutant variants with conserved amino acid substitutions in the Walker B motif of NBDs are properly expressed and surface localized
Asp327 of N-terminal NBD and Asp1026 of C-terminal NBD in the Walker B motif of Cdr1p or its equivalent in other ABC transporters are well conserved (Figure 1). To probe whether Asp327 has acquired a new role because of its neighborhood conserved substitutions and to compare it's functioning with its counter part Asp1026 in the Walker B motif of C-terminal NBD, we made point mutations of these residues. Thus, we constructed the mutant variants D327N, D327A, D1026N, D1026A and D327N/D1026N of Cdr1p. Making the equivalent mutations in both the nucleotide sites of Cdr1p allowed us to examine their roles in ATP catalysis.
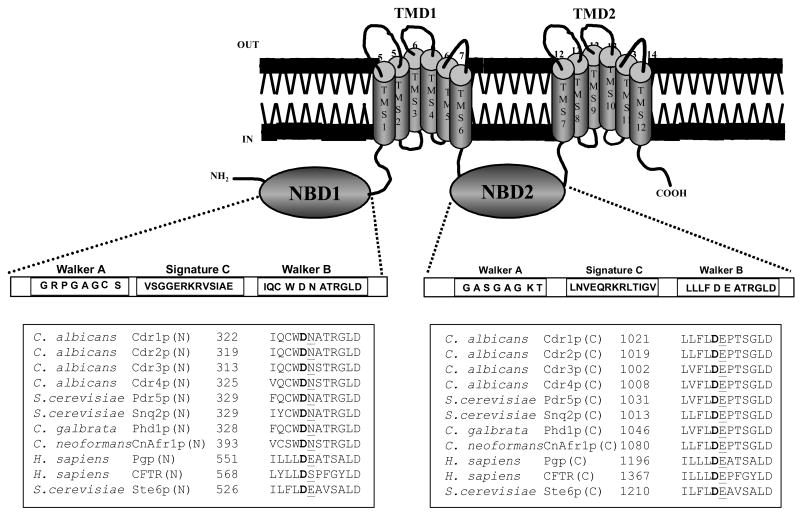
Figure 1. Topology of Cdr1p and sequence alignment of Walker B and extended Walker B motifs from various ABC transporters.
The sequence alignment of Walker B and extended Walker B motifs residues in NBDs with those from other nucleotide binding domains of some known ABC transporters. The conserved Asp and Asn residue within and adjacent the Walker B motifs, respectively, of fungal NBDs and the equivalent Asp/Glu in non-fungal NBDs is shown in bold and underlined.
The mutant variants, designated as Cdr1p D327N, Cdr1p D327A, Cdr1p D1026N, Cdr1p D1026A, and Cdr1p D327N/D1026N were constructed by site directed mutagenesis and stably over-expressed from genomic PDR5 locus in a S. cerevisiae mutant AD1-8u-, as a heterologous hyper-expression system (35). We also tagged green fluorescent protein (GFP) gene at the C-terminal end of CDR1, which was over-expressed as a fusion protein (26). The host AD1-8u- was derived from a Pdr1-3 mutant strain with a gain of function mutation in the transcription factor Pdr1p, resulting in constitutive hyper-induction of the PDR5 promoter (27). Single copy integration of each transformant at the PDR5 promoter was confirmed by Southern hybridization (data not shown). The wild type Cdr1p and its mutant variants were expressed at equivalent levels as evident from the Western blot analysis of the PM fraction of cells expressing these mutant variant proteins (Figure 2A). All the mutant variants and wild type Cdr1p were properly cell surface localized as was confirmed by confocal microscopy and FACS analysis (Figure 2B).
Figure 2. Membrane localization and expression profile of wild type and carboxylate residue mutant variant Cdr1p.
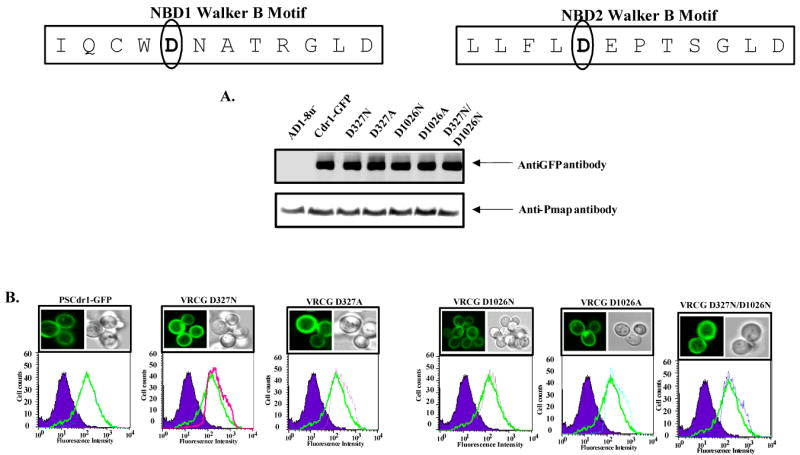
Boxed panel at the top shows the sequence of Walker B and extended Walker B motifs of N-terminal and C-terminal NBDs of Cdr1p. (A) PM of wild type and mutant variant proteins expressing cells were prepared and their immunodetection was done as described earlier (26).
(B) Fluorescence imaging (upper panel) by a confocal microscope showing membrane localization of Cdr1-GFP (Cdr1p) and its mutant variant proteins expressing cells. Flow cytometry (lower panel) of S. cerevisiae expressing Cdr1p and its mutant variants. The histogram derived from the cell quest program depicts fluorescence intensities for AD1-8u- (control) (purple filled area), PSCdr1-GFP (solid green line) for each panel and other extra line represent that respective Cdr1p mutant variant expressing cells.
Mutation of conserved carboxylate residues in the Walker B motif of NBDs resulted in abrogation of drug resistance, drug transport and ATPase activity
We examined the effect of these mutations on drug sensitivities of the cells expressing wild type Cdr1p and its mutant variants by two independent drug susceptibility assays viz., MIC80 (minimum inhibitory concentration for 80% inhibition in growth) and spot assays (26). MIC80 assays revealed that the host strain (AD1-8u-) was expectedly hypersensitive to all the tested drugs when compared to the growth control (without drug). On the contrary, for the cells expressing wild type Cdr1p, substantial growth in the presence of drugs was observed. As compared to the host strain (AD1-8u-), MIC80 for cells expressing wild type Cdr1p was considerably higher (MIC80 64 μg/ml for fluconazole, 16 μg/ml for anisomycin, 2.0 μg/ml for miconazole, 1.0 μg/ml for ketoconazole, 4.0 μg/ml for Itraconazole and 0.5 μg/ml for cycloheximide) (Figure 3A). Interestingly, cells expressing Cdr1p mutant variants Cdr1p D327N, Cdr1p D327A, Cdr1p D1026N, Cdr1p D1026A, and Cdr1p D327N/D1026N behaved like host strain (AD1-8u-) and remained hypersensitive to the tested drugs. Accordingly, they displayed extremely low MIC80 values (Figure 3A). The results of spot assays also corroborated MIC80 results (Figure 3B).
Figure 3.
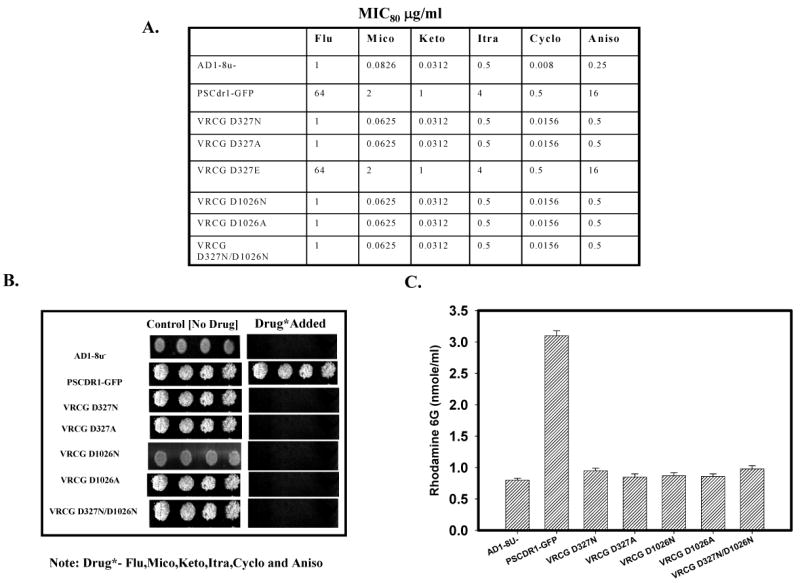
(A) MIC80 values of cells expressing Cdr1p and its mutant variants in presence of different drugs tested: Drug susceptibility (MIC80) of S. cerevisiae cells expressing wild type and mutant variants of Cdr1p, MICs were determined by microdilution method as described previously ‘(26). (B) Drug resistance profiles of the wild type and mutants determined by spot assay. It was done as per the protocol described earlier (26). The concentration of the drugs used : fluconazole (5 μg/ml), anisomycin (1.0 μg/ml), miconazole (0.5 μg/ml), ketoconazole, itraconazole and cycloheximide (0.15 μg/ml). (C) Rhodamine 6G efflux by the wild type Cdr1p and its mutant variant proteins expressing cells: The R6G efflux was measured as described previously (28) The values are mean ± SD (error bars) for three independent experiments.
To check if the observed drug sensitivity by Cdr1p carboxylate mutant variants expressing cells was associated with impaired extrusion ability of the cell, we performed rhodamine 6G (R6G) efflux assay (described under ‘Experimental Procedures’) which is a good indicator of Cdr1p activity (28). Strikingly, cells expressing these Walker B mutant variants Cdr1p D327N, Cdr1p D327A, Cdr1p D1026N, Cdr1p D1026A, and Cdr1p D327N/D1026N were severely impaired in efflux function (Figure 3C). It is apparent that the disruption of either NBD results in a nonfunctional protein and neither the N-terminal nor the C-terminal ATP site can function independently of each other.
The ability of conserved Walker B mutant variants of Cdr1p to hydrolyze ATP was also measured as the oligomycin-sensitive release of Pi from Mg-ATP as described under ‘Experimental Procedures’. PM preparations from cells expressing wild type Cdr1p elicited an ATPase activity of ∼50-55 nmoles of Pi released per min per mg of protein. None of the mutant variant proteins displayed ATPase activity above the host AD1-8u-control (discussed later; Figure 5A). These data suggest that the lack of transport function of mutant variant proteins (Figure 3C) is attributed to the impaired ATPase activity and that both NBDs need to be functional to generate an active transporter.
Figure 5.
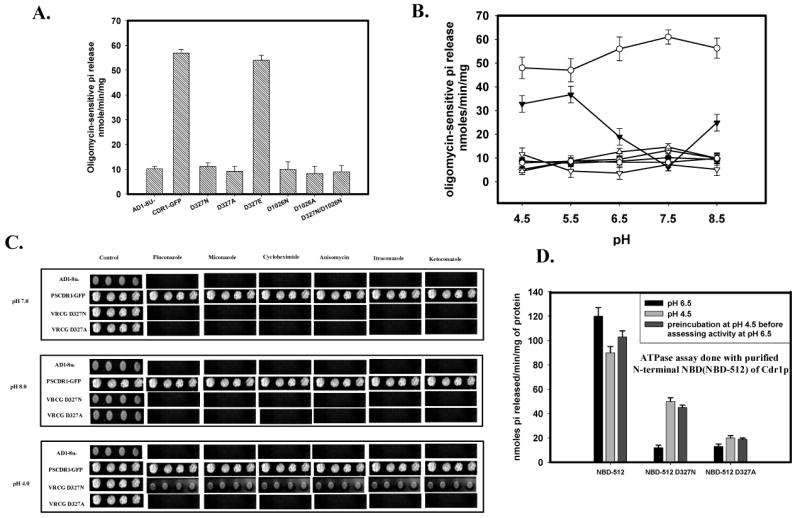
(A) ATPase activity of Cdr1p with its mutant variants: ATPase activity of PM fraction of cells expressing the wild type Cdr1p and its mutant variants were assayed as described previously (26). The values were the mean ± standard deviation of three independent experiments. (B) pH dependence of ATPase activity of Cdr1p and its mutant variants: ATPase activity of PM fractions expressing wild type Cdr1p and its mutant variants were assayed at different pH. Scattered plot represents AD1-8u- (●) PSCdr1-GFP (○), VRCG D327N (▼), VRCG D327A (▽), VRCG D1026N (◊), VRCG D1026A (
 ) and VRCG D327N/D1026N (Δ). (C) Reversal of drug resistance at low pH by VRCG D327N strain: Reversal of drug resistance by VRCG D327N strain at different pH was corroborated by spot assay by growing the cells at different pH in presence of drug; Flu (0.5 μg/ml), Mico (0.125 μg/ml), Keto (0.0156 μg/ml), Itra (0.125 μg/ml), Cyclo (0.0625μg/ml) and Aniso (0.5 μg/ml). (D) ATPase activity of isolated N-terminal NBD (NBD-512) of Cdr1p and its mutant variant proteins: ATPase activity of NBD-512 and its mutant variant proteins at pH 6.5 was assayed after pre-incubation of the protein at low pH (4.5).
) and VRCG D327N/D1026N (Δ). (C) Reversal of drug resistance at low pH by VRCG D327N strain: Reversal of drug resistance by VRCG D327N strain at different pH was corroborated by spot assay by growing the cells at different pH in presence of drug; Flu (0.5 μg/ml), Mico (0.125 μg/ml), Keto (0.0156 μg/ml), Itra (0.125 μg/ml), Cyclo (0.0625μg/ml) and Aniso (0.5 μg/ml). (D) ATPase activity of isolated N-terminal NBD (NBD-512) of Cdr1p and its mutant variant proteins: ATPase activity of NBD-512 and its mutant variant proteins at pH 6.5 was assayed after pre-incubation of the protein at low pH (4.5).
Drug binding remains unaffected in the Walker B mutant variants of NBDs
The drug binding properties of the wild type and mutant variant proteins were assessed by photoaffinity labeling with radio labeled drug substrate analogues of dihydropyridine ([3H]-azidopine) and of prazosin ([125I] iodoarylazidoprazosin; [125I] IAAP). These photoaffinity analogues have been successfully used earlier to study drug-binding sites on Cdr1p (26). PM preparations derived from yeast cells expressing wild type Cdr1p or its mutant variants Cdr1p D327N, Cdr1p D327A, Cdr1p D1026N, Cdr1p D1026A and Cdr1p D327N/D1026N were labeled with [3H]-azidopine or [125I] IAAP (as described under ‘Experimental Procedures’), subjected to SDS-PAGE and autoradiography. As expected, no binding of these analogues was observed in the host AD1-8u- negative control while an efficient labeling of [3H]-azidopine (Figure 4A) and [125I] IAAP (Figure 4B) was seen with all the mutant variant proteins, comparable to that of wild type protein. Photoaffinity labeling of these drug analogue with wild type and its mutant variant protein was specific as [125I] IAAP labeling can be competed out by Cdr1p drug substrates such as nystatin (Figure 4B). These data suggest that mutations made in the Walker B motifs of the N- and C-terminal NBDs do not affect the ability of the transporter to recognize the drug substrates analogues.
Figure 4.
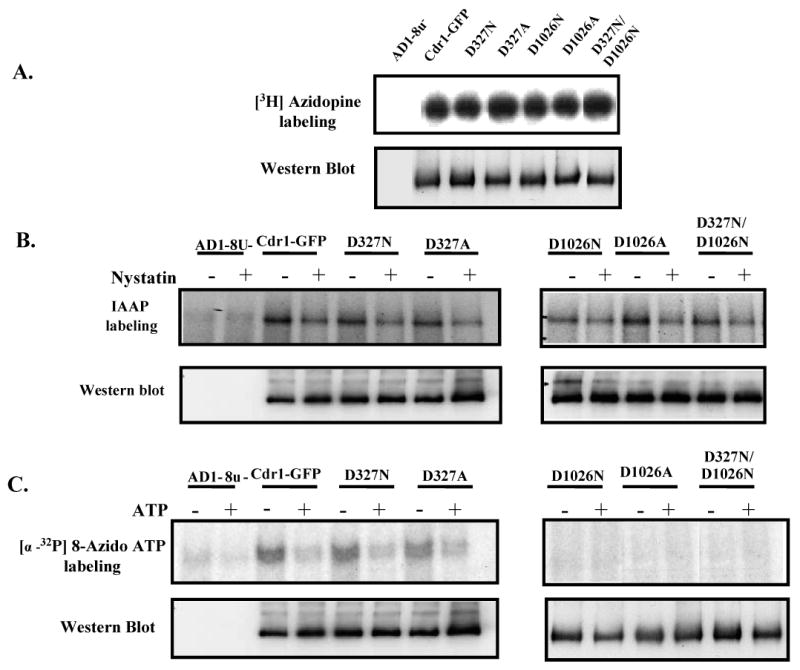
(A) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [3H]-azidopine: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were incubated with 0.5 μM [3H]-azidopine (60 Ci/mmol) for 5 min under subdued light. The samples were processed and analyzed as described in ‘Experimental Procedures’ and loaded as AD1-8u- (control; lane 1), Cdr1-GFP (lane 2), Cdr1p D327N (lane 3), Cdr1p D327A (lane 4), Cdr1p D1026N (lane 5), Cdr1p D1026A (lane 6) and Cdr1p D327N/D1026N (lane 7). (B) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [125I]-IAAP: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were incubated with 7.5 nM [125I]-IAAP (2200 Ci/mmol) in presence and absence of 2 μM Nystatin (+ lane) as described in ‘Experimental Procedures’. (C) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [α-32P] 8-azido ATP: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were incubated with 10 μM [α-32P] 8-azido ATP 7.5 μCi/nmole at 4°C and competed with 10 mM cold ATP (+ ATP lane) as described in ‘Experimental Procedures’. In the Fig. 4A, B and C, lower panel shows the immunoblotting using anti-GFP antibody to ensure an equal loading of wild type Cdr1p and its mutant variants in all the lanes.
Walker B mutant variants of NBDs differ in their ability to bind [α-32P] 8-azido ATP
To further examine the cause of impaired drug transport and ATPase activity, we explored the effect of single or double mutations in the Walker B motif on ATP binding in general, and in particular exploring if a mutation in either ATP site would allow nucleotide binding and hydrolysis at the other native site. For this, we examined [α-32P] 8-azido ATP labeling binding of Cdr1p and its mutant variants, which we had earlier shown to bind efficiently to Cdr1p (26). PM preparations were photoaffinity –labeled with 10 μM of [α-32P] 8-azido ATP in the presence of 8 mM MgCl2 as described under ‘Experimental Procedures’. An equivalent amount of PM derived from AD1-8u- host was used as a control, and as expected, no binding of [α-32P] 8-azido ATP was observed in this case. Membranes expressing wild type Cdr1p and its mutant variants were, however, labeled with [α-32P] 8-azido ATP and the labeling of the analogue could be competed out with molar excess of cold ATP (Figure 4C). This labeling was strictly dependent upon Mg2+, as the presence of 2 mM EDTA completely eliminated the labeling (data not shown). While the [α-32P] 8-azido ATP labeling in membranes expressing Cdr1p D1026N and its double mutant variant Cdr1p D327N/D1026N were severely affected, there was no significant difference in labeling between wild type Cdr1p and Cdr1p D327N or Cdr1p D327A. It should imply that the ability to bind ATP to the C-terminal ATP sites is reduced significantly in the protein containing a mutation in Mg2+ binding site (D1026N), whereas binding to N-terminal ATP site with D327N mutation is not compromised.
D327 is involved in ATP catalysis
The photoaffinity labeling of Cdr1p mutant variants with [α-32P] 8-azido ATP suggested that Asp1026 of Walker B motif of C-terminal NBD is implicated in ATP binding or Mg2+ coordination like its proposed conventional role in other NBDs (18). However, despite being crucial for ATP hydrolysis, Asp327 in the Walker B motif of N-terminal NBD does not appear to be involved in Mg2+ coordination (Figure 5A). To further explore the role of Asp327, we examined the pH dependence of ATPase activity of the wild type and its mutant variants to highlight different roles of the two-conserved Asp. As shown in Figure 5B, ATPase activity of Walker B mutant variant Cdr1p D1026N does not show any pH dependence, while the pH profile of enzyme activity of Cdr1p D327N mutant variant (retaining around 10% residual activity) was drastically different. Of note, Cdr1p D327N mutant variant regains ∼40% ATPase activity at acidic pH (4.5-5.5) (Figure 5B), whereas its Ala substitution i.e., Cdr1p D327A does not show similar enhancement of ATPase activity at low pH. No similar restoration of ATPase activity at acidic pH (4.5-5.5) could also be seen with Cdr1p D1024N or Cdr1p D1026A mutant variants (Figure 5B). Interestingly, the restoration of ATPase activity by cells expressing only Cdr1p D326N mutant variant could also partially restore resistance to tested drugs when grown at low pH while that was not the case with cells expressing Cdr1p D326A mutant variant (Figure 5C). Furthermore, as evidence by the MIC80 values (Figure 3A) and ATPase activity (Figure 5A), the mutant variant Cdr1p D327E also behaved very much like the wild type protein, confirming the importance of the carboxylate residue at the 327 position.
Isolated N-terminal Nucleotide Binding Domain D327N (NBD-512 D327N) confirms pH dependence
To further confirm that the observed pH dependence was indeed the result of unique placement of critical residues in the N-terminal NBD of Cdr1p, we used purified N-terminal NBD (NBD-512) protein (21). Indeed, purified NBD-512 D327N mutant variant protein also showed restoration of ATPase activity at low pH (4.5), which was not the case with NBD-512 D327A mutant variant protein (data not shown). Interestingly, when purified native NBD-512 and mutant variant NBD-512 D327N were exposed to low pH prior to assessing their ATPase activities at the normal pH of 6.5, the mutant variant protein retained enhanced activity (Figure 5D).
The substitution of conserved Asn328 resulted in selective loss of function, but unlike Cdr1p D327N do not show pH dependence
In all non-fungal ABC transporters, a very well conserved residue Glu adjacent to conserved Asp (equivalent to Asp327) of the Walker B motif has been implicated to function as a catalytic base in ATP catalysis cycle (35-39). However, all fungal ABC transporters including Cdr1p lack this conserved Glu residue adjacent to the Walker B core at N-terminal NBD, which is replaced by Asn (Figure 1). Given the importance of conserved Glu residue in the catalytic cycle of non-fungal ABC transporters, we explored the role of Asn328 in Cdr1p. To investigate this, Asn328 of N-terminal NBD and Glu 1027 of C-terminal NBD at equivalent position, adjacent to the core Walker B motif of Cdr1p (Figure 6) were substituted. The mutant variants designated Cdr1p N328E, Cdr1p N328A, Cdr1p E1027Q, Cdr1p E1027A and Cdr1p N328E/E1027Q were constructed and expressed in S. cerevisiae, mutant host strain (AD1-8u-). Substitution of these highly conserved residues did not affect cell surface expression of Cdr1p as determined by FACS and confocal analysis (Figure 6A). However, all these mutant variants were abrogated in their ability to efflux R6G (Figure 6B). This loss of efflux was the result of impaired ATPase activity in these mutant variants (Figure 6F). Interestingly, binding of photoaffinity drug analogues, [3H]-azidopine (Figure 6C), [125I] IAAP (Figures 6D) and of ATP analogue [α-32P] 8-azido ATP to these mutant variants remains unaffected (Figures 6E). Importantly, neither of these mutant variants showed pH dependence of ATPase activity as shown by Cdr1p D327N mutant variant (Figure 6F, Inset).
Figure 6.
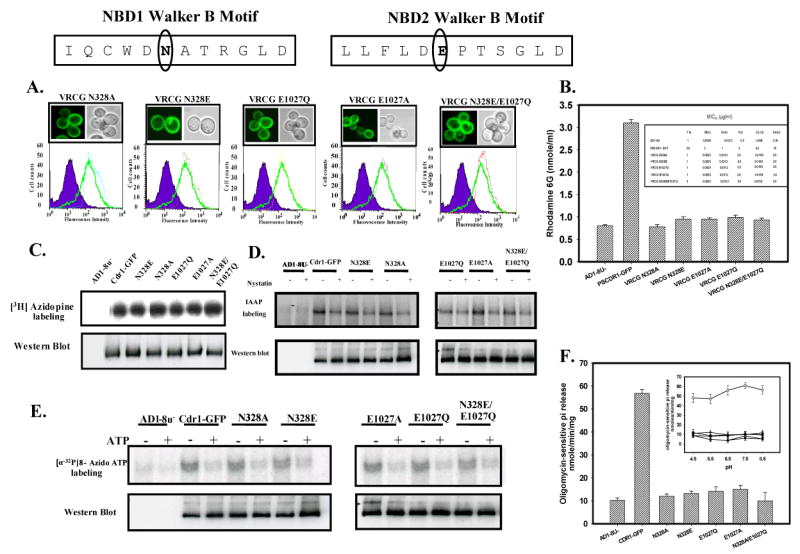
(A) Membrane localization and expression profile of wild type Cdr1p and mutant variants: Boxed panel at the top shows the sequence of Walker B and extended Walker B motifs of N-terminal and C-terminal NBDs of Cdr1p. Fluorescence imaging (upper panel) and Flow cytometry (lower panel) of S. cerevisiae expressing Cdr1p and its mutant variants has been done as mentioned in legends of Fig. 2B. (B) Rhodamine 6G efflux by the wild type Cdr1p and its mutant variants expressing cells: Inset shows the table of MIC80 values of wild type Cdr1p and its mutant variants expressing cells for the drug tested. (C) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [3H]-azidopine: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were photoaffinity labeled with 0.5 μM [3H]-azidopine as mentioned in legends of Fig. 4A. Loading pattern is AD1-8u- (control; lane 1), Cdr1-GFP (lane 2), Cdr1p N328E (lane 3), Cdr1p N328A (lane 4), Cdr1p E1027Q (lane 5), Cdr1p E1027A (lane 6) and Cdr1p N328E/E1027Q (lane 7). (D) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [125I]-IAAP: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were incubated with 7.5 nM [125I]-IAAP (2200 Ci/mmol) in absence or presence of 2 μM Nystatin (+ lane) as described in ‘Experimental Procedures’. (E) Photoaffinity labeling of wild type Cdr1p and its mutant variants with [α-32P] 8-azido ATP: The PM fraction (30 μg) of cells expressing the wild type Cdr1p and its mutant variants were incubated with 10 μM [α-32P] 8-azido ATP 7.5 μCi/nmole at 4°C and competed with 10 mM cold ATP (+ ATP lane) as described in ‘Experimental Procedures’. In the panel C, D and E, lower panel shows the immunoblotting using anti-GFP antibody to ensure an equal loading of wild type Cdr1p and its mutant variants in all the lanes.
(F) Comparison of ATPase activity of Cdr1p with its mutant variants: ATPase activity of PM fraction of cells expressing the wild type Cdr1p and its mutant variants were assayed as described under ‘Experimental Procedures’; the values represent the average ± standard deviation of three independent experiments. Inset shows ATPase activity of Cdr1p and its mutant variants at different pH. Scattered plot represents AD1-8u- (●) PSCdr1-GFP (○), VRCG N328E (□), VRCG N328A (◆), VRCG E1027Q (▲), VRCG E1027A (Δ) and VRCG N328E/E1027Q (♦).
Homology modeling of nucleotide binding domains (NBDs) of Cdr1p
In the absence of high-resolution structural data for NBDs of Cdr1p and to validate experimental data, we generated a homology model based on the dimeric E171Q mutant structure of a known homologue, MJ0796 an ABC transporter of M. jannaschii. The crystallographic and functional evidence for ABC transporters indicates that residues from both NBDs contribute to the nucleotide-binding site. Therefore, it is sensible to refer to ATP binding to sites rather than to individual NBDs (19). By homology modeling, we mapped the sequence of the Cdr1p N- and C-terminal NBDs onto the two subunits of the dimeric E171Q mutant of the MJ0796. Analysis of the ATP-binding pocket shows that ATP is present at the interface of the sandwich dimer formed between the Walker A and B motifs of N-terminal NBD and the signature motif of C-terminal NBD (Fig. 7A). In all the 50 models generated, Asp327 is predicted to form an integral part of the ATP binding pocket and could be a prime candidate, directly involved in catalysis at N-terminal ATP binding site (Figure 7B).
Figure 7. Atomic model of the N-and- C-terminal NBD dimer of Cdr1p.
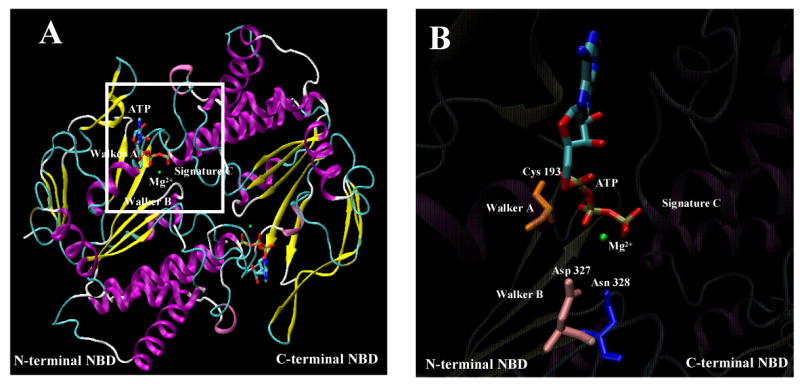
(A) Homology based model of N-and-C-terminal NBD of Cdr1p. The structure of N- and C-terminal NBD was generated by Modeller8v2 program (19,31). Structural diagrams were produced using Visual Molecular Dynamic (VMD) software (29), where α-helices are represented as ribbons and strands as arrows. The ATP molecule is shown in licorice format and the atoms, i.e., carbon, nitrogen, oxygen, phosphorus, sulfur, and magnesium ion, are represented in cyan, blue, red, tan, yellow, and green, respectively. Functionally important sequence motifs of NBDs are labeled. (B) A blown-up view of the N-terminal nucleotide binding pocket of the dimeric model of Cdr1p highlighting the close positioning of ATP and Mg2+ with Asp327 (pink), Asn328 (blue), Trp326 (green) and Cys193 (orange) residue side chain.
Discussion
Several aspects of the mechanisms of ABC drug transporters remain unresolved. Some questions such as what are the various steps in the catalytic cycle, role of NBDs with respect to ATP binding and hydrolysis, and the nature and type of signal generated at NBDs by ATP hydrolysis which is transduced to the drug binding sites in TMDs to mediate drug efflux, are poorly understood (1). Fungal ABC transporters pose additional challenges since they have typical amino acid substitutions in conserved motifs of NBDs which suggest possibility of mechanistic differences in ATP catalysis cycle (20). In an attempt to understand the molecular basis of ATP hydrolysis mediated by major ABC multidrug transporter of C. albicans, Cdr1p, we have demonstrated that the uncommon and atypical conserved Trp326 in the Walker B motif of N-terminal NBD is crucial for ATP binding (23). We have also shown that another uniquely replaced residue Cys193 in the Walker A motif of N-terminal NBD is critical for ATP hydrolysis (22). Interestingly, reports from non-fungal ABC transporters suggest that residues present at equivalent positions of Trp326 have no demonstrable role in ATP catalysis while the residue present at equivalent position of Cys193 is indispensable for ATP binding and hydrolysis (16, 40, 41). In the present study because of the unique placement of residues like Trp326 of Walker B motif and Asn328 adjacent to Walker B motif of N-terminal NBD of Cdr1p, we explored if the role of the highly conserved Asp327 of N-terminal NBD in any way has also changed as compared to its equivalent residue Asp1026 of C-terminal NBD which lies within well conserved Walker B motif. Of note in P-gp and other ABC transporters, residues equivalent to Asp327 and Asp1026 are involved in Mg2+ coordination (25, 42).
We observed that mutations in the Walker B region (D327N, D327A, D1026N, D1026A and D327N/D1026N) showed equivalent cell surface expression levels of Cdr1p but resulted in abrogated efflux of R6G in all the Cdr1p mutant variants. Single point mutations introduced in either of the two NBD sites of Cdr1p (D327N and D1026N) impaired the capacity of mutant variant protein to confer resistance to all the tested drugs. Mutations D327N, D1026N as well as the double mutations D327N/D1026N in the Walker B motif of Cdr1p showed severely decreased ATPase activity though there was no effect on their ability to recognize the substrate per se as confirmed by photoaffinity drug analogue [3H] azidopine and [125I] IAAP labeling. These results establish that both Asp327 and Asp1026 are critical for ATP hydrolysis at their respective positions and their substitution results in mutant variants which are unable to power drug efflux without affecting drug binding.
Our photoaffinity labeling experiments with [α-32P] 8-azido ATP illustrated the functional asymmetry of Asp327, Asp1026 and of the two NBD sites therein. For example while Cdr1p D327N, showed comparable binding of [α-32P] 8-azido ATP to the wild type protein, such was not the case with Cdr1p D1026N and it's double mutant Cdr1p D327N/D1026N variant protein which showed severely impaired labeling. The inability of Cdr1p D1026N mutant variant to efficiently bind [α-32P] 8-azido ATP even though N-terminal NBD site was intact highlights the differences in the role of the two residues. The photoaffinity labeling data further imply that unlike at C-terminal NBD with D1026N, ATP binding per se is not the major cause of dysfunction at N-terminal NBD site with D327N mutation, ATP binding per se is not the major cause of dysfunction, but the steps after initial binding are probably affected. In a recent in vivo study, the relative contribution of both the N – and C- terminal NBDs in ATP hydrolysis, drug resistance and drug transport activity of wild type Cdr1p was examined wherein unique Cys193 of Walker A of N-terminal NBD (C193K) and a conserved Lys 901 of Walker A of C-terminal NBD (K901C) were replaced (43). As compared to the Cdr1p C193K, the cells expressing Cdr1p K901C mutant variant became relatively more susceptible to drugs. This clearly suggested that the two NBDs of Cdr1p are functionally asymmetric (43). Based on sequence analyses, it was earlier suggested that N-terminal NBD, of Pdr5p (homologue of Cdr1p) of S. cerevisiae is probably unable to perform ATP hydrolysis and that transporter might function with only one of its two NBDs (20). Interestingly, in case of P-gp, which is a close homologue of Cdr1p, the two NBDs are partially interchangeable (25). On the other hand in prokaryotic ABC transporters such as histidine permease of S. typhimurium, both NBDs are functionally identical and contribute equally to the proteins activity (44). The functional asymmetry of NBDs in Cdr1p was also illustrated in our recent study where swapping of NBDs resulted in nonfunctional Cdr1p chimeras and thus suggested that the two NBDs are non exchangeable (45). Taken together, results confirm that the two potential ATP binding sites of Cdr1p are not identical. These observations are also consistent with a model in which the binding of nucleotide at one site is indispensable for hydrolysis at the other site, originally proposed by Senior and colleagues for P-gp (46).
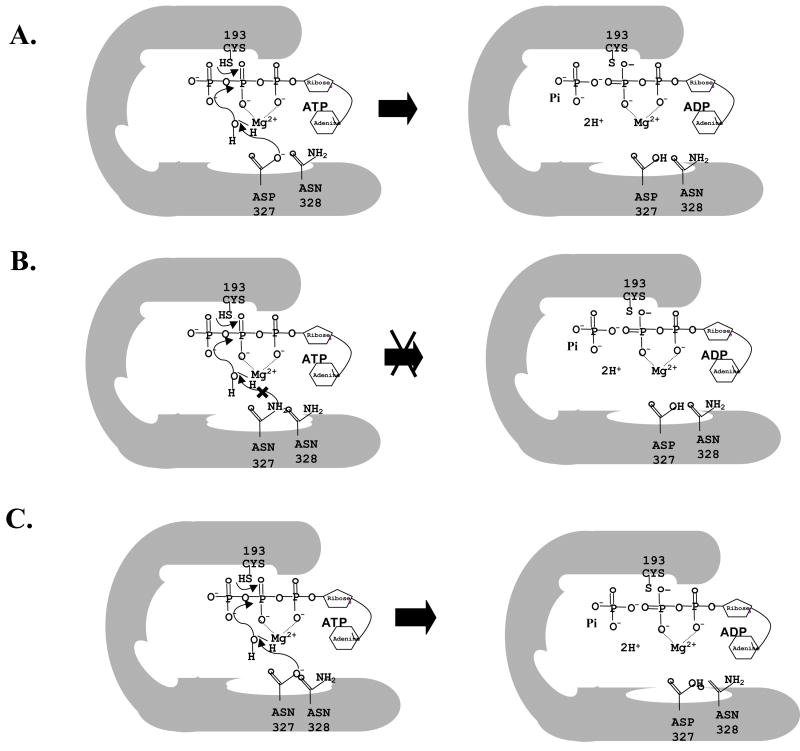
The photoaffinity labeling data further imply that at N-terminal NBD site with D327N mutation in the Walker B motif of Cdr1p, ATP binding per se is not the major cause of dysfunction, but the steps after initial binding are probably affected. That Asp327 and Asp1026 are important for ATP hydrolysis was apparent but that they have acquired different roles became clear, when we monitored pH dependence of ATP hydrolysis. While ATPase activity of Walker B mutant variant Cdr1p D1026N does not show any pH dependence, Cdr1p D327N mutant variant showed an enhancement in activity at acidic pH. This partial restoration of ATPase activity at acidic pH by Cdr1p D327N mutant variant expressing cells was sufficient to partially power drug transport since otherwise hypersensitive Cdr1p D327N mutant variant expressing cells could grow in the presence of tested drugs. The restoration of ATPase activity at low pH in Cdr1p D327N mutant variant and not in Cdr1p D327A mutant variant suggests that Asp327 probably acts as a catalysis base. At physiological pH of 7.5, Asp327 of Cdr1p would exist in its deprotonated form, fully capable of abstracting proton from a water molecule during ATP hydrolysis (Figure 8A). On the contrary, at the same pH, replaced Asn would be unable to abstract proton. Our observation that Cdr1p D327N mutant variant does not show any enhancement in ATPase activity at pH 7.5 supports such an assumption (Figure 8B). However, at acidic pH (4.5), solvent accessible Asn is expected to be hydrolyzed, become Asp, which could easily abstract proton from a water molecule and thus promote ATP catalysis (Figure 8C). Interestingly, mutation of Asp327 to Glu was functional and was able to power the drug efflux like wild type protein, demonstrating an exquisitely sensitive requirement for the carboxyl side chain at 327 position. Our results with purified domain mutant variant protein NBD-512 D327N further confirmed this pH dependence of Asp327. Interestingly, prior exposure of NBD-512 D327N mutant variant to low pH (4.5) was sufficient to enable it to hydrolyze ATP even at pH 6.5. The exposure of NBD-512 D327N mutant variant protein to acidic pH would allow Asn to become Asp, which once hydrolyzed, could now abstract proton at pH 6.5. Our results strongly suggest that unlike in other non-fungal ABC transport the conserved Asp327 in the Walker B motif has acquired a new role where it is not involved in nucleotide binding per se but rather serves as a catalytic base at N-terminal ATP site of Cdr1p and is involved in the abstraction of proton from water molecule. The conserved Asp1026 of C-terminal NBD, however, continues to perform its traditional established role of Mg2+ coordination in ATP catalysis. Consistent with experimental data, N-and C-terminal NBD heterodimer model shows that indeed Asp327 forms an integral part of the ATP binding pocket. Thus, in addition to extensive biochemical data, homology modeling of NBDs also demonstrates the critical role of Asp327 in ATP hydrolysis.
Figure 8. A hypothetical model depicting the N-terminal active site of Cdr1p.
(A) N-terminal active site of wild type Cdr1p at pH 7.5 (B) N-terminal active site of Cdr1p D327N mutant variant at pH 7.5 (C) N-terminal active site of Cdr1p D327N mutant variant at low pH 4.5.
Conventionally, the residue equivalent to Asp327 is part of the conserved Walker B motif found in many non-fungal different ATPase such as the ABC proteins, including transporters and DNA repair proteins such as Rad50 and MutS as well as helicase (47). It is known to be essential for chelating the Mg2+ bound to the nucleotide and is shown to be essential for activity (18). The very well conserved residue next to this Asp in non-fungal ABC transporters is Glu that points directly towards the phosphate of the nucleotide bound in the active site (48). It has been proposed for other proteins that in this position, Glu serves as catalytic base to activate a catalytic water molecule for nucleophilic attack on the γ-phosphate (19, 38, 24). For example, mutation of the Glu residue to Ala or Gln, in M. jannaschii ABC transporters, MJ0796 and MJ1267 results in the loss of ATPase activity, supporting the hypothesis that this residue is required for ATP hydrolysis (38). In the case of Bacillus subtilis ABC transporter, BmrA, substitution of this Glu with other amino acids resulted in entrapment of the nucleotide in triphosphate form in the active site, also indicating disruption of the hydrolysis reaction (36). In P-gp, the mutation of this Glu, however, indicated occlusion of the nucleotide in the diphosphate form, thus suggesting that the residue plays a role in ADP release and turnover, rather than in catalysis (49, 18). It would appear that the presence of Asn328 next to well-conserved Asp327 (instead of Glu) in the Walker B motif of N-terminal NBD of Cdr1p and in other fungal ABC transporters has necessitated Asp327 to acquire the role of a catalytic base in ATP hydrolysis.
Based on the role of Glu adjacent to the Walker B motif of NBDs of other non-fungal ABC transporters, we argued that the replacement of Asn with Glu (N328E) mutation in N-terminal NBD of Cdr1p could improve ATP hydrolysis. Contrary to expectation, this mutation (N328E) exhibited severely impaired ATPase and drug transport activity (Figure 6B). Interestingly, this mutant variant unlike Cdr1p D327N did not exhibit any pH dependence of ATPase activity (Figure 6F). These observations suggest that though Asn328 is important for the functionality of Cdr1p yet, unlike its substituted residues in other ABC transporters, it does not directly participate in ATP hydrolysis.
In conclusion, our study provides an instance of a functional residue migration or hopping during evolution of NBDs. Active site variation might reflect the evolutionary optimization of the catalytic efficiency of fungal ABC transporters. We show that a highly conserved Asp residue at position 327 in the Walker B motif of N-terminal NBD of a major ABC drug transporter Cdr1p has acquired a new role to act as a catalysis base. Thus the unique placement of Cys193, Trp326 and Asn328 in the conserved motifs of Cdr1p definitely has functional significance. Since such unique placements are also extended to other fungal ABC transporters, it strongly points towards their evolutionary relevance. However, what additional advantage such changes have brought to this class of proteins is not apparent. One needs to wait for 3D structure of fungal ABC transporters to highlight the mechanistic differences with other well-conserved proteins of this super family. Considering that many fungal ABC transporters belong to human pathogenic fungi, it would be interesting to examine if these unique changes have provided any extra advantage to their survival in the host environment.
Acknowledgments
We thank R.D. cannon for the gifts of plasmid and strains. We thank Ranbaxy Laboratories Ltd, New Delhi, India for providing fluconazole. We are also grateful R. Serrano for the kind gift of PM-ATPase antibodies. We thank Dr. Andrew M. Lynn for his help in generating atomic dimeric model of Cdr1p.
The work presented in this paper has been supported in parts by grants from Department of Biotechnology (DBT/PR3825/Med/14/488(a)/2003), Council of Scientific and Industrial Research (38(1122)/06/EMR-II), Department of Science and Technology (SR/SO/BB-12/2004), and European Commission, Brussels (QLK2-CT-2001-02377). V. R., M.G. and S.S.1 acknowledge the University Grants Commission, India for support in the form of Junior and Senior Research Fellowships. S.S.3 and S.V.A. acknowledge “Intramural program of the National Institute of Health, National Cancer Institute, and Centre for Cancer Research” for funding.
Abbreviations
- ABC
ATP binding cassette
- ATP
adenosine 5′-triphosphate
- ADP
adenosine 5′-diphosphate
- ATPase
adenosine-5′-triphosphatase
- PDR
pleiotropic drug resistance
- EDTA
ethylenediaminetetraacetic acid
- kDa
kilodalton(s)
- NBD
nucleotide binding domain
- PCR
polymerase chain reaction
- PAGE
polyacrylamide gel electrophoresis
- SDS
sodium dodecyl sulfate
- TMD
trans membrane domain
- TMS
trans membrane segment
- GFP
green fluorescent protein
- ORF
open reading frame
- PM
plasma membrane
- FACS
fluorescence associated cell sorting
- FLU
fluconazole
- MIC
miconazole
- CYCLO
cycloheximide
- ANISO
anisomycin
- KETO
ketoconazole
- ITRA
itraconazole
- R6G
rhodamine 6G
- [125I] IAAP
[125I] iodoarylazidoprazosin
References
- 1.Holland IB, Cole PC, Kuchler K, Higgins CF. ABC proteins from Bacteria to Man. Academic Press; San Diego, CA: 2003. [Google Scholar]
- 2.Walmsley MB, Mckeegan KS, Walmsley AR. Structure and function in efflux pumps that confer resistance to drugs. Biochem J. 2003;376:313–338. doi: 10.1042/BJ20020957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderone RA. Candida and Candidiasis. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- 4.Prasad R, Snehlata P, Smriti . Drug resistance in yeasts- an emerging scenario. In: Poole RK, editor. Advances in Microbial Physiology. First. Academic Press; London: 2002. pp. 155–201. [DOI] [PubMed] [Google Scholar]
- 5.White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increased azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernaez ML, Gil C, Pla J, Nombela C. Induced expression of the Candida albicans multidrug resistance gene CDR1 in response to fluconazole and other antifungals. Yeast. 1998;14:517–526. doi: 10.1002/(SICI)1097-0061(19980430)14:6<517::AID-YEA250>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: Characterisation of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 8.White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta V, Kohli AK, Krishnamurthy S, Puri N, Aalamgeer SA, Panwar SL, Prasad R. Identification of mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance and its in vitro transcriptional activation. Curr Genet. 1998;34:192–199. doi: 10.1007/s002940050385. [DOI] [PubMed] [Google Scholar]
- 10.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Zheng LX, Lentz BR. A slight asymmetry in the transbillayer distribution of lysophosphatidylcholine alters the surface properties and poly(ethylene glycol)-mediated fusion of dipalmitoylphosphatidylcholine large unilamellar vesicles. Biochemistry. 1996;35:12602–12611. doi: 10.1021/bi960168q. [DOI] [PubMed] [Google Scholar]
- 12.Niimi M, Niimi K, Takano Y, Holmes AR, Fischer FJ, Uehara Y, Cannon RD. Regulated overexpression of CDR1 in Candida albicans confers multidrug resistance. J Antimicrob Chemother. 2004;54:999–1006. doi: 10.1093/jac/dkh456. [DOI] [PubMed] [Google Scholar]
- 13.Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, McLnerney T, Gaskell S, Oliver SG, Cash P, Brown AJ. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
- 14.Prasad R, Gupta N, Gaur M. Pathogenic Fungi. Caister Academic Press; Nortfolk, U.K.: 2004. Molecular basis of antifungal resistance; pp. 357–414. [Google Scholar]
- 15.Walker JE, Sarsate M, Runswick M, Gay NJ. Distantly related sequences in the a - and b -subunits of ATP synthase, myosin, kinases and other ATP- requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunit/domain. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 17.Moody JE, Thomas PJ. NBD interactions during the mechanochemical reaction cycle of ATP-Binding Cassette transporter. J Bioenerg Biomembr. 2005;37:475–479. doi: 10.1007/s10863-005-9494-8. [DOI] [PubMed] [Google Scholar]
- 18.Urbatsch IL, Julien M, Carrier I, Rousseau ME, Cayrol R, Gros P. Mutational analysis of conserved carboxylate residues in the nucleotide binding sites of P-glycoprotein. Biochemistry. 2001;39:14138–14149. doi: 10.1021/bi001128w. [DOI] [PubMed] [Google Scholar]
- 19.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter dimer formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nature Genetics. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 21.Jha S, Karnani N, Dhar SK, Mukhopadhayay K, Shukla S, Saini P, Mukhopadhayay G, Prasad R. Purification and characterization of the N-terminal nucleotide binding domain of an ABC drug transporter of Candida albicans: uncommon cysteine 193 of Walker A is critical for ATP hydrolysis. Biochemistry. 2003;42:10822–10832. doi: 10.1021/bi0345900. [DOI] [PubMed] [Google Scholar]
- 22.Jha S, Karnani N, Lynn AM, Prasad R. Covalent modification of cysteine 193 impairs ATPase function of nucleotide-binding domain of a Candida drug efflux pump. Biochem Biophys Res Commun. 2003;310:869–875. doi: 10.1016/j.bbrc.2003.09.094. [DOI] [PubMed] [Google Scholar]
- 23.Rai V, Shukla S, Jha S, Komath SS, Prasad R. Functional characterization of N-terminal nucleotide binding domain (NBD-1) of a major ABC drug transporter Cdr1p of Candida albicans: uncommon but conserved Trp326 of Walker B is important for ATP binding. Biochemistry. 2005;44:6650–6661. doi: 10.1021/bi0474160. [DOI] [PubMed] [Google Scholar]
- 24.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 25.Hrycyna CA, Ramachandra M, Germann UA, Wu Cheng P, Pastan I, Gottesman MM. Both ATP sites of human P-glycoprotein are essential but not symmetric. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- 26.Shukla S, Saini P, Smriti, Jha S, Ambudkar SV, Prasad R. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot Cell. 2003;2:1361–1375. doi: 10.1128/EC.2.6.1361-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura K, Niimi M, Niimi K, Holmes AR, Yates JE, Decottignies A, Monk BC, Goffeau A, Cannon RD. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob Agents Chemother. 2002;45:3366–3374. doi: 10.1128/AAC.45.12.3366-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maesaki S, Marichal P, Bossche HV, Sanglard D, Kohno S. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J Antimicrob Chemother. 1999;44:27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- 29.Humphrey W, Dalke A, Schulten K. VMD-Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Protein. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 33.Sapan CV, Lundblad RL, Price NC. Colorimetric protein assay techniques. Biotechnol Appl Biochem. 1999;29:99–108. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 35.Carrier I, Julein M, Gros P. Analysis of catalytic carboxylate mutants E552Q and E1197Q suggests asymmetric ATP hydrolysis by the two Nucleotide binding domains of P-glycoprotein. J Biol Chem. 2003;42:12875–12885. doi: 10.1021/bi034257w. [DOI] [PubMed] [Google Scholar]
- 36.Orelle C, Dalmas O, Gros P, Di Pietro A, Jault JM. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J Biol Chem. 2003;278(47):47002–8. doi: 10.1074/jbc.M308268200. [DOI] [PubMed] [Google Scholar]
- 37.Yuan YR, Blecker S, Martsinkevich O, Millen L, Thomas PJ, Hunt JF. The crystal structure of the MJ0796 ATP-binding cassette; Implications for the structural consequences of ATP hydrolysis in the active site of an ABC transporter. J Biol Chem. 2001;276:32313–32321. doi: 10.1074/jbc.M100758200. [DOI] [PubMed] [Google Scholar]
- 38.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang R, McBride A, Hou YX, Goldberg A, Chang XB. Nucleotide dissociation from NBD1 promotes solute transport by MRP1. Biochim Biophys Acta. 2005;1668:248–261. doi: 10.1016/j.bbamem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE. Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris and characterization of nucleotide binding properties of wild type, Walker A, and Walker B mutant proteins. J Biol Chem. 1999;274:34711–34718. doi: 10.1074/jbc.274.49.34711. [DOI] [PubMed] [Google Scholar]
- 41.Saveanu L, Daniel S, Ender PM. Distinct functions of the ATP Binding Cassettes of transporters associated with antigen processing. J Biol Chem. 2001;276:22107–22113. doi: 10.1074/jbc.M011221200. [DOI] [PubMed] [Google Scholar]
- 42.Muller M, Bakos E, Welker E, Varadi A, Germann UA, Gottesman MM, Morse BS, Roninson IB, Sarkadi B. Altered drug-stimulated ATPase activity in mutants of the human multidrug resistance protein. J Biol Chem. 1996;271:1877–1883. doi: 10.1074/jbc.271.4.1877. [DOI] [PubMed] [Google Scholar]
- 43.Jha S, Dabas N, Karnani N, Saini P, Prasad R. ABC multidrug transporter Cdr1p of Candida albicans has divergent nucleotide-binding domains which display functional asymmetry. FEMS Yeast Res. 2004;5:63–72. doi: 10.1016/j.femsyr.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Nikaido K, Ames GF. One intact ATP-binding subunit is sufficient to support ATP hydrolysis and translocation in an ABC transporter, the histidine permeases. J Biol Chem. 1999;274:26727–26735. doi: 10.1074/jbc.274.38.26727. [DOI] [PubMed] [Google Scholar]
- 45.Saini P, Gaur NA, Prasad R. Chimera of the ABC drug transporter Cdr1p reveal functional indispensability of transmembrane domains and nucleotide binding domains, but transmembrane segment 12 is replaceable with the corresponding homologous region of the non-drug transporter Cdr3p. Microbiology. 2006;152:1559–1573. doi: 10.1099/mic.0.28471-0. [DOI] [PubMed] [Google Scholar]
- 46.Urbatsch IL, Sankaran B, Bhagat S, Senior AE. Both P-glycoprotein nucleotide-binding sites are catalytically active. J Biol Chem. 1995;270:26956–26963. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- 47.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters, unifying concepts from high resolution structure. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 48.Geourjon C, Orelle C, Steinfels E, Blanchet C, Deleage G, Di Pietro A, Jault JM. A common mechanism for ATP hydrolysis in ABC transporter and helicase family. Trends Biochem Sci. 2001;26:539–544. doi: 10.1016/s0968-0004(01)01907-7. [DOI] [PubMed] [Google Scholar]
- 49.Sauna ZE, Muller M, Peng XH, Ambudkar SV. Importance of the conserved Walker B glutamate residues, 556 and 1201, for the completion of the catalytic cycle of ATP hydrolysis by human P-glycoprotein (ABCB1) Biochemistry. 2002;41:13989–14000. doi: 10.1021/bi026626e. [DOI] [PubMed] [Google Scholar]