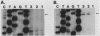Abstract
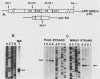
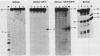
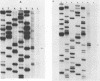
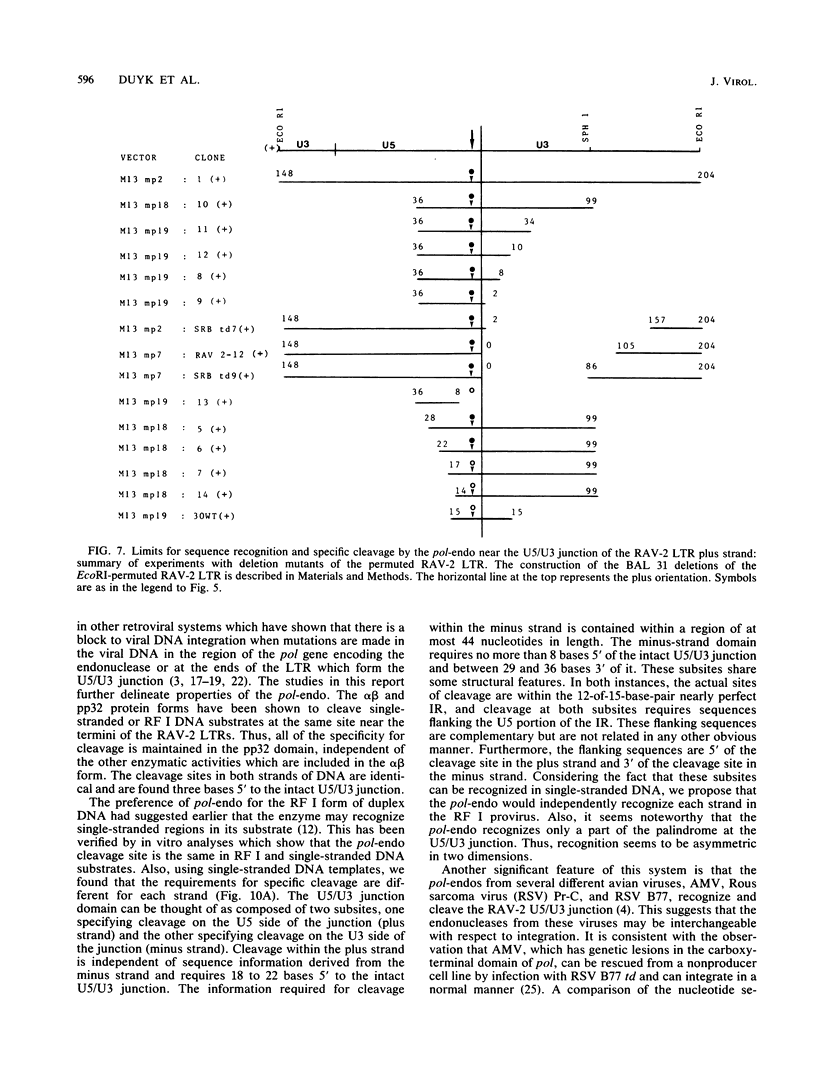
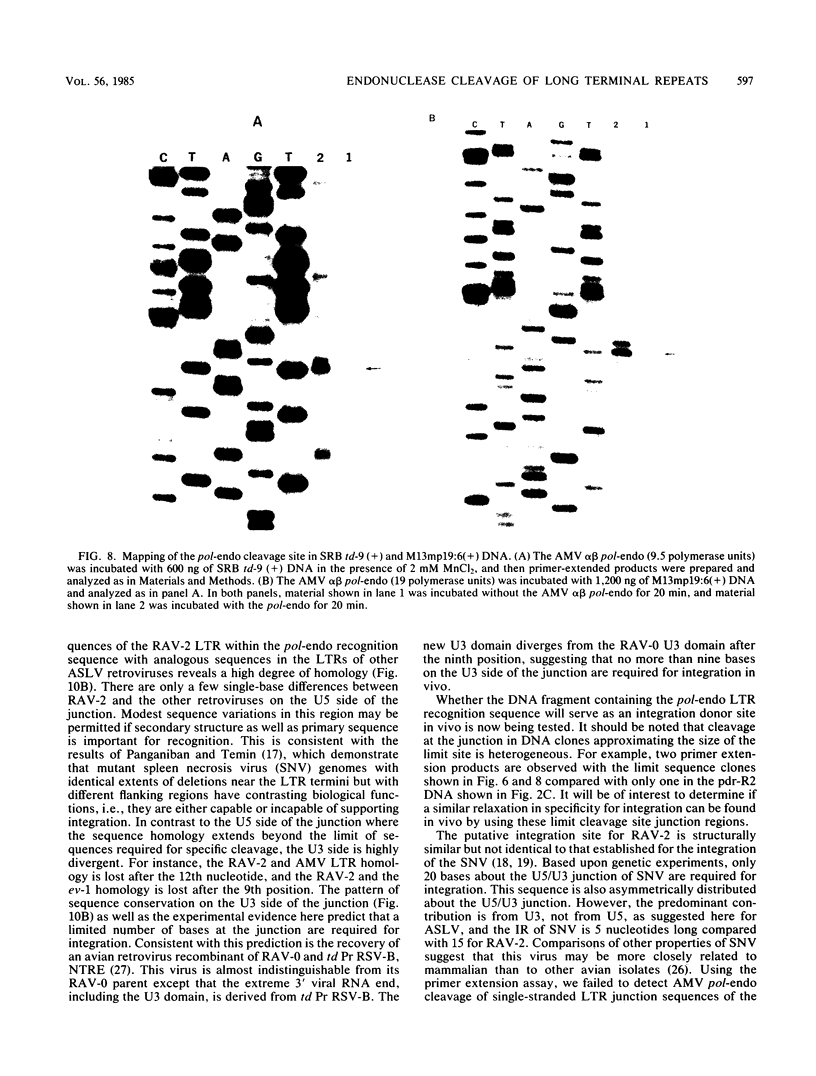
The avian retroviral pol gene-encoded DNA endonuclease (pol-endo) has been shown to selectively cleave the viral long terminal repeat sequences (LTRs) in single-stranded DNA substrates in a region known to be joined to host DNA during integration (G. Duyk, J. Leis, M. Longiaru, and A.M. Skalka, Proc. Natl. Acad. Sci. USA 80:6745-6749, 1983). The preferred sites of cleavage were mapped to the unique U5/U3 junctions found only in covalently closed circular DNA molecules containing two tandem LTRs. The cuts occurred three nucleotides 5' to the axis of symmetry of the 12-of-15-base-pair nearly perfect inverted repeat which marks the LTR junction. Experiments with double-stranded supercoiled DNA substrates revealed a similar specificity for nicking. Also, the endonuclease associated with the pol cleavage product, pp32, has the same specificity as the alpha beta form. The limits of sequence required for site-selective cleavage near the U5/U3 junction were established with single-stranded DNA substrates. A domain no larger than 44 base pairs allowed site-selective cleavage in each strand in vitro. Recognition of either strand appeared to be independent of the other, and in each case, the critical sequence was asymmetrically distributed with respect to the U5/U3 junction. The predominant contribution was from the U5 domain; this is consistent with its conservation in the LTR sequences of a number of avian sarcoma and leukosis viruses.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Caruthers M. H., Beaucage S. L., Efcavitch J. W., Fisher E. F., Goldman R. A., deHaseth P. L., Mandecki W., Matteucci M. D., Rosendahl M. S., Stabinsky Y. Chemical synthesis and biological studies on mutated gene-control regions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):411–418. doi: 10.1101/sqb.1983.047.01.048. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Leis J., Longiaru M., Skalka A. M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Verma I. M. Studies on the reverse transcriptase of RNA tumor viruses. Structural relatedness of two subunits of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4991–4994. doi: 10.1073/pnas.71.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P., Mason W. Virus-coded DNA endonuclease from avian retrovirus. J Virol. 1981 May;38(2):548–555. doi: 10.1128/jvi.38.2.548-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Knaus R. J., Hippenmeyer P. J. Antibodies against a synthetic peptide of the avian retrovirus pp32 protein and the beta DNA polymerase subunit. Virology. 1983 Oct 15;130(1):257–262. doi: 10.1016/0042-6822(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Griffith J., Kornberg A. Mini M13 bacteriophage: circular fragments of M13 DNA are replicated and packaged during normal infections. Virology. 1974 May;59(1):139–152. doi: 10.1016/0042-6822(74)90211-6. [DOI] [PubMed] [Google Scholar]
- Ju G., Boone L., Skalka A. M. Isolation and characterization of recombinant DNA clones of avian retroviruses: size heterogeneity and instability of the direct repeat. J Virol. 1980 Mar;33(3):1026–1033. doi: 10.1128/jvi.33.3.1026-1033.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Leis J., Duyk G., Johnson S., Longiaru M., Skalka A. Mechanism of action of the endonuclease associated with the alpha beta and beta beta forms of avian RNA tumor virus reverse transcriptase. J Virol. 1983 Feb;45(2):727–739. doi: 10.1128/jvi.45.2.727-739.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Rho H. M., Grandgenett D. P., Green M. Sequence relatedness between the subunits of avian myeloblastosis virus reverse transcriptase. J Biol Chem. 1975 Jul 10;250(13):5278–5280. [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978 Oct;28(1):279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. M., Duyk G., Longiaru M., DeHaseth P., Terry R., Leis J. Integrative recombination--a role for the retroviral reverse transcriptase. Cold Spring Harb Symp Quant Biol. 1984;49:651–659. doi: 10.1101/sqb.1984.049.01.073. [DOI] [PubMed] [Google Scholar]
- Souza L. M., Baluda M. A. Identification of the avian myeloblastosis virus genome. I. Identification of restriction endonuclease fragments associated with acute myeloblastic leukemia. J Virol. 1980 Nov;36(2):317–324. doi: 10.1128/jvi.36.2.317-324.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Donehower L., Hager G., Zeller N., Malavarca R., Astrin S., Skalka A. M. Sequence comparison in the crossover region of an oncogenic avian retrovirus recombinant and its nononcogenic parent: genetic regions that control growth rate and oncogenic potential. Mol Cell Biol. 1982 Nov;2(11):1331–1338. doi: 10.1128/mcb.2.11.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. F., Alianell G. A., Bencen G. H., Gray H. B., Jr Isolation and comparison of two molecular species of the BAL 31 nuclease from Alteromonas espejiana with distinct kinetic properties. J Biol Chem. 1983 Nov 25;258(22):13506–13512. [PubMed] [Google Scholar]
- Zasloff M., Ginder G. D., Felsenfeld G. A new method for the purification and identification of covalently closed circular DNA molcules. Nucleic Acids Res. 1978 Apr;5(4):1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Goldman R. A., Cech C. L., Caruthers M. H. Chemical synthesis and biochemical reactivity of bacteriophage lambda PR promoter. Nucleic Acids Res. 1983 Feb 11;11(3):773–787. doi: 10.1093/nar/11.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]