Abstract
Aspergillus fumigatus is a common mould whose spores are a component of the normal airborne flora. Immune dysfunction permits developmental growth of inhaled spores in the human lung causing aspergillosis, a significant threat to human health in the form of allergic, and life-threatening invasive infections. The success of A. fumigatus as a pathogen is unique among close phylogenetic relatives and is poorly characterised at the molecular level. Recent genome sequencing of several Aspergillus species provides an exceptional opportunity to analyse fungal virulence attributes within a genomic and evolutionary context. To identify genes preferentially expressed during adaptation to the mammalian host niche, we generated multiple gene expression profiles from minute samplings of A. fumigatus germlings during initiation of murine infection. They reveal a highly co-ordinated A. fumigatus gene expression programme, governing metabolic and physiological adaptation, which allows the organism to prosper within the mammalian niche. As functions of phylogenetic conservation and genetic locus, 28% and 30%, respectively, of the A. fumigatus subtelomeric and lineage-specific gene repertoires are induced relative to laboratory culture, and physically clustered genes including loci directing pseurotin, gliotoxin and siderophore biosyntheses are a prominent feature. Locationally biased A. fumigatus gene expression is not prompted by in vitro iron limitation, acid, alkaline, anaerobic or oxidative stress. However, subtelomeric gene expression is favoured following ex vivo neutrophil exposure and in comparative analyses of richly and poorly nourished laboratory cultured germlings. We found remarkable concordance between the A. fumigatus host-adaptation transcriptome and those resulting from in vitro iron depletion, alkaline shift, nitrogen starvation and loss of the methyltransferase LaeA. This first transcriptional snapshot of a fungal genome during initiation of mammalian infection provides the global perspective required to direct much-needed diagnostic and therapeutic strategies and reveals genome organisation and subtelomeric diversity as potential driving forces in the evolution of pathogenicity in the genus Aspergillus.
Author Summary
Airborne spores of the fungus Aspergillus fumigatus are present in significant quantities worldwide and are responsible for a range of illnesses from allergy to deadly invasive lung infection. A number of fungal properties are likely required for germination and growth of the fungus in the host, and now that the genome sequence of A. fumigatus is available it is possible to address which genes become important during initiation of infection. Understanding this might lead to new therapeutics and diagnostic tools. We have compared A. fumigatus gene activation during infection in a murine model to that in a laboratory culture to identify fungal attributes preferentially employed during disease. Our analysis entailed measurement of activity from most of the >9000 A. fumigatus genes, identifying iron limitation, alkaline stress, and nitrogen starvation as prominent stresses imposed by the host environment. We also found that genes preferentially employed for infection occur in clusters and are more likely to reside near the end of chromosomes, otherwise known as telomeres.
Introduction
A small fraction of the estimated 1.5 million fungal species on Earth can colonise and infect human beings. Among them, the ascomycete Aspergillus fumigatus is the leading cause of mould-related death, most of which results from invasive lung disease in immune-deficient patients[1]. The ascomycetes' ecologically important saprophytism demands metabolic diversity and species-specific inventories of secreted enzymes. These are attributes which may ultimately contribute to the pathogenicity of certain species in plants and humans and have long influenced interpretations of virulence[1],[2]. Despite the importance of appropriate transcriptional control in orchestrating these processes, accurate data from within the host niche has eluded researchers, principally due to difficulties associated with sample recovery.
Most A. fumigatus infections are a direct consequence of the enormous propensity of A. fumigatus spores for airborne dispersal in large quantities, such that the human lung is constantly exposed to them. If infection ensues, its nature and severity is governed by the status of the host, which determines whether the spores are cleared effectively or whether they go on to germinate in, colonise, or even to invade the surrounding lung tissue[3]. Ex vivo and epidemiological analyses place macrophages and neutrophils on the frontline of cell-mediated defense[4],[5] against A. fumigatus infection and have implicated the fungal secondary metabolite gliotoxin (GT), in an immunotoxigenic capacity, as contributing to virulence[6]. This hypothesis was recently substantiated within a physiological context with the finding that the proapoptotic Bcl-2 family member, Bak, is required for GT-induced apoptosis in murine embryonic fibroblasts. Moreover, Bak knockout mice are resistant to A. fumigatus infection with a GT-producing clinical isolate[7]. Deletion of gliotoxin biosynthetic genes can differentially affect virulence, dependent upon immunsuppressive regimen, in murine infection supporting an important role of the host environment in determining pathogenic potential of A. fumigatus. This dichotomy of virulence phenotype renders the clinical significance of gliotoxin uncertain at the current time[8]–[12].
Fungi that sporulate or produce fruiting bodies demonstrate co-ordinate expression of biologically active secondary metabolites and spore-related products during development[13]. Such regulation is mediated at the level of transcription, from clusters of physically linked, co-ordinately regulated genes and is profoundly affected by both the developmental program under execution, and environmental factors such as pH and nutrient availability[14]. Renewed interest in the significance of secondary metabolites in establishing Aspergillus infection has been triggered by the discovery of a global transcriptional regulator of A. fumigatus secondary metabolite biosynthesis, LaeA[15]. laeA deletion does not affect gross changes in growth or sporulation in media in vitro, but it does result in reduced virulence in mice. This impairment in the ability to cause infection can be correlated with a loss of detectable gliotoxin as well as mis-regulation of gene expression within 13 gene clusters[16]. Siderophore-mediated iron uptake and storage, also partially under LaeA control[16] is indispensable for A. fumigatus virulence[17],[18], and adequate nutrient acquisition within the host niche is inextricably linked to pathogenesis of invasive disease being necessary to support sufficient growth to promote infection[19]–[21].
Transcriptional profiling can greatly illuminate the host pathogen interaction but the potential of this approach remains limited due to difficulties associated with obtaining good quality RNA in sufficient quantities from sites of infection. The feasibility of performing microarray analyses on limited material has been tested by a number of researchers employing linear amplification of mRNA, an approach which has recently proven successful in murine candidiasis[22], though a truly global transcriptional signature has yet to be reported for any fungal pathogen initiating mammalian invasive disease. The Eberwine method of mRNA amplification involves reverse transcription of mRNA with an oligo dT primer bearing a T7 RNA polymerase promoter site, to direct in vitro transcription of antisense RNA (aRNA) after double stranded cDNA synthesis and is favoured for linear mRNA amplification from limited quantities of starting material. It provides the basis for the methodology employed in our study, and in the majority of reported instances where mRNA amplification has been applied to samples destined for microarray analysis[23].
To identify fungal attributes preferentially employed during adaptation to the host niche, and thus contributing to the virulence of the saprophytic parasite A. fumigatus, we compared the transcriptomes of developmentally matched A. fumigatus isolates following laboratory culture or initiation of infection in the neutropenic murine lung. We report the development of a highly robust methodology for global profiling of A. fumigatus gene expression in germlings rescued directly from the murine lung, a tool which will empower the analysis of virulence in this pathogen. Our methodology employs facile molecular manipulations which, combined with custom bioinformatic scripts based on the latest annotations of the A. fumigatus Af293 genome, mark a significant advance in understanding orchestration of fungal virulence. Our analyses identify iron limitation, alkaline stress and nutrient deprivation as relevant host-imposed stresses during early-stage A. fumigatus infection and reveal a biased distribution of host-adaptation genes (relative to laboratory culture) in subtelomeric regions of chromosomes. Finally we assess lineage specificity of functions favoured during initiation of infection within varyingly virulent members of the genus Aspergillus.
Results
A. fumigatus RNA extraction and mRNA amplification
Microarray analyses are constrained by the availability of sufficient RNA for fluorophore labelling and hybridisation. One strategy to overcome the requirement for large quantities of material is global amplification of the sample. This approach has been employed for a number of reported microarray expression analyses[23],[24] with favourable results. We chose to analyse an early time point of infection to facilitate separation of fungal cells from those of the host with the advantage of studying a stage at which germinating hyphae have sensed, and are adapting to, their host. Early time points of A. fumigatus infection represent a vulnerable phase of morphogenesis in vivo since epithelial invasion and formation of mycelial mass have yet to occur. They also mark a point at which diagnoses capable of distinguishing infection from carriage of fungal spores would be most desirable, and antifungal therapy most effective.
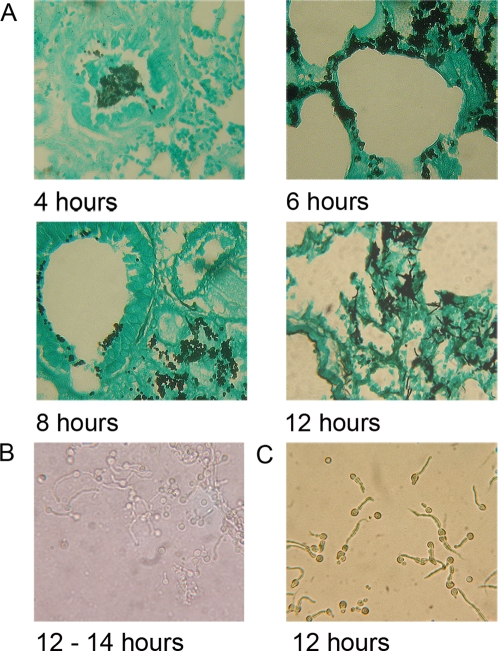
We firstly characterised the time course of hyphal development in the sequenced clinical isolate Af293 by histopathological examination of infected neutropenic murine lung tissues (Figure 1). Lung sections collected and formalin-fixed at 4, 6, 8 and 12 hours post-infection contained numerous A. fumigatus spores in close association with murine epithelium in the bronchioles and alveoli (Figure 1A). At 12–14 hours post-infection >80% of A. fumigatus conidia had undergone germination and primary hyphal production. We therefore performed all BAL extractions for downstream analyses on concurrently infected neutropenic mice within a two hour window of infectious growth corresponding to 12–14 hours post-infection. At this time point recovery of germlings in BAL fluid was routinely achievable in the order of 103 germlings per lavaged lung (Figure 1B).
Figure 1. Comparative time-course of A. fumigatus Af293 germination and hyphal development in the murine lung, and laboratory culture.
(A) Time-course of Af293 germination and hyphal development in the neutropenic murine lung. (B) Microscopic appearance of Af293 germlings recovered from a typical single murine BALF, (harvested at 12–14 hours post-infection). (C) Microscopy of developmentally matched laboratory cultured Af293 germlings, following liquid culture for 12 hours in YPD at 37°C.
To isolate fungal RNA from the site of A. fumigatus infection we inoculated pools of 24 neutropenic CD1 male mice with 108 conidiospores and culled after 12–14 hours. Bronchoalveolar lavage was performed immediately using pre-warmed sterile saline and samples (BALFs) were snap frozen prior to RNA extraction and amplification. Within infection groups BALFs were pooled prior to RNA extraction and mRNA amplification. Total RNA yields from pooled BALFs ranged from 108–800 ng and yielded up to 258 µg aRNA after 2 rounds of linear amplification. Amplification factors therefore ranged from 2.8×103–3.9×105 based upon 2% of the total RNA population being mRNA (Table S1). In vitro reference RNA samples were similarly prepared from developmentally matched A. fumigatus germlings (Figure 1C) which were harvested, and snap-frozen, following 12 hour culture at 37°C in rich medium and subjected to two rounds of mRNA amplification prior to co-hybridisation (Table S1).
Impact of mRNA amplification on preservation of transcript ratios
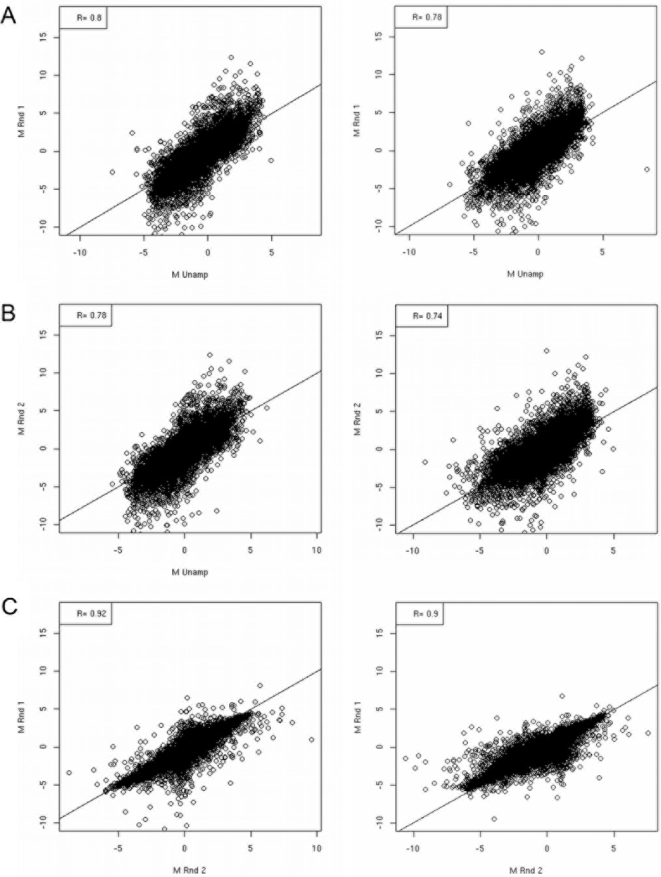
A suitable global mRNA amplification protocol should provide sufficient material for fluorophore cDNA labelling reactions whilst preserving the samples' original relative transcript abundance. To estimate amplification-related error (and distinguish such error from systematic error inherent to microarray methodologies) a mock experiment was devised to quantify the proportion of transcripts having significantly aberrant log2 ratios as a result of RNA amplification. This was achieved by indirectly comparing cDNA samples from different amplification protocols in a statistical linear model and fitting relevant contrast matrices (see Materials and Methods and Figure S1). For the mock experiment total RNA (totRNA) was isolated from two A. fumigatus cell populations T0 and T60, and subjected to either one (aRNAr1) or two (aRNAr2) rounds of mRNA amplification prior to cDNA fluorophore labelling and microarray hybridisation (Figure 2). To evaluate whether ratios are preserved between amplification protocols, we adopted the approach of Nygaard et. al. [23] who used corrected gene-wise t-tests to identify differences in the mean log intensity between aRNA2 and total RNA populations. In preparing the data for multiple t-testing, we excluded spots that were flagged by the the TIGR spotfinder software, or where the intensity was lower than twice the background intensity in either the Cy5 or Cy3 channel. The proportion of excluded spots was used as an indication of the hybridisation quality of the slide as a whole. Slides hybridised with aRNA had fewer spots (13.49–18.22%) removed by the filtering process than did slides hybridised with cDNA (44.56–48.95%), see Table S2. This apparent amplification-related improvement in hybridisation quality has been previously reported[24]. In our study, this can be attributed to a greater signal to noise ratio, and more specifically to increased foreground intensity on these slides (data not shown). We identified 8.49% of retained spots showing evidence of amplification protocol dependent differences in the log2 ratios. Thus our estimated measures of confidence from the above quality-control (QC) exercises fall within a range conducive for deriving biologically useful information, based upon reports of analytical studies published to date[23],[24] where Pearson correlation coefficients range from 0.75 to 0.99 (Figure 2) and rejected gene sets approximate 10% of spots included in QC analysis.
Figure 2. Correlation of log2 ratios resulting from comparative transcriptional analysis of the laboratory cultured A. fumigatus cell populations T0 and T60 under varying mRNA amplification protocols.
Correlation of technically duplicated log2 ratios between competitive hybridisations using single (aRNA1), double (aRNA2) and unamplified (totRNA) RNA samples. (A and B) Correlation between log2 ratios obtained using cDNA derived from amplified and total RNA (totRNA v aRNAr1 r = 0.74–0.80, totRNA v aRNAr2 r = 0.74–0.80) (C) Cross-protocol pairings revealed highest correlations between slides using cDNA derived from amplified RNA (aRNAr1 v aRNAr2 r = 0.88–0.91) Surprisingly, technical replicates of slides using cDNA derived from total RNA (totRNA r = 0.80) were comparable to cross-protocol pairings (data not shown).
A. fumigatus transcript profile during initiation of mammalian infection
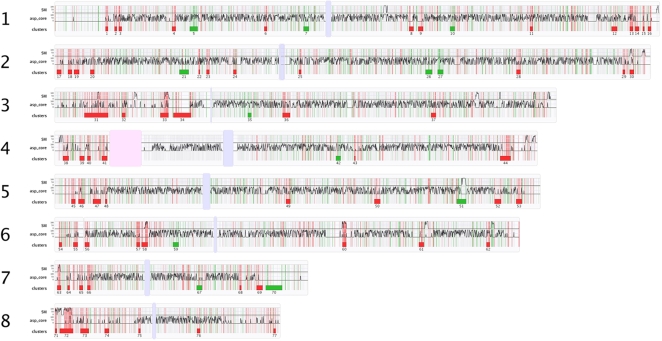
A common reference design was adopted for the microarray experiment from which data were analysed and processed as described in the Materials and Methods section. We performed the infection experiment 5 times in total generating pooled (n = 24) BALFs from five independent Af293 infections (samples A to E, Table S1). These samples were co-hybridised with similarly amplified mRNA prepared from developmentally matched laboratory cultured Af293 germlings (Materials and Methods and Table S1). Fluorescent signals from 9075 out of a possible 9516 represented ORFs were detectable from these hybridisation analyses (Dataset S1 and Array Express (http://www.ebi.ac.uk/microarray-as/aer/#ae-main0 Accession number E-TABM-327). Of 2180 genes (22.6% of the whole genome) having a fold-change in log intensity ratio of 2 or greater, 1281 were up-regulated and 897 were down-regulated. The entire expression dataset is graphically represented in Figure 3 which plots log2 ratios against chromosomal locus. Initial interpretations of the dataset were performed by the Expression Analysis Systematic Explorer[25] (EASE) to infer function by homology to Saccharomyces cerevisiae and identify over-represented Gene Ontology (GO) terms among differentially expressed genes. The results of these analyses are partially listed in Table 1 (full listings in Table S3). Distinct trends among favoured cellular processes are evident from these analyses which reveal a marked investment, by the host-adapting A. fumigatus cell, into transport of metal ion, cation, carbohydrate and siderophore iron. Given the importance of iron acquisition for microbial pathogenesis in general, and the absolute requirement for siderophore biosynthesis during murine A. fumigatus pathogenesis in this model of infection [17],[18] we expected transcripts from genes involved in iron mobilization and transport to be differentially abundant in this analysis, relative to iron-rich laboratory culture media. Accordingly we could identify a minimum of eleven siderophore biosynthesis/transport genes as important during growth in the murine lung (Table 2) including two ferric-chelate reductases (Afu1g17270 and Afu6g13750). Thirteen amino acid permease genes were more abundantly represented during host-adaptation than growth in YPD (Table 2) including 4 GABA (Afu8g01450, Afu7g0040, Afu5g14660 and Afu5g00710), and three proline permeases (Afu2g11220, Afu8g02200 and Afu7g01090) as well as the general amino acid permease, Gap1 (Afu7g04290). Nine genes annotated as maltose permeases or transporters in the current Af293 annotation were also more abundantly represented during initiation of murine infection (Table 2). Extracellular proteases have been implicated as virulence factors in invasive aspergillosis, as well as antigens causing inflammatory irregularity during allergic A. fumigatus disease[1]. Our analysis identified increased abundance of transcripts from the elastinolytic metalloprotease (Mep) (Afu8g07080), an aorsin-like serine protease (Afu6g6g10250) and three dipeptidylpeptidases (Afu4g09320, Afu2g09030 and AfuAfu3g07850). Thus transcription of this subset of A fumigatus proteases is significantly higher in the murine lung relative to rich laboratory culture. Functional categories of ergosterol biosynthesis, heme biosynthesis and aerobic respiration were significant among genes underrepresented during infection, relative to laboratory culture (Table 1) as well as multiple functional categories representing ribosome biogenesis and assembly, and protein biosynthesis and processing. This may reflect the poor nutritional value of murine lung relative to YPD and/or reduced growth (due to any number of stresses) during host-adaptation compared to broth culture. This trend is evidenced on multiple levels within our dataset, comprising repression of genes directing ribosomal protein synthesis, rRNA synthesis, RNA polymerase I and II activity, translation initiation and elongation, tRNA processing and synthesis, intracellular trafficking, secretion and vesicular trafficking (Table 1 and Table S3). While such metabolic dampening is often observed in microbial systems under stress, the observed A. fumigatus regulatory signature mimics that of rapamycin-mediated TOR kinase[26] inhibition and typifies fungal starvation. The S. cerevisiae TOR proteins TOR1 and TOR2 are phosphatidylinositol kinase homologues, first identified as the targets of the immunophilin-immunosuppressant complex FKBP-rapamycin[27], combined deletion of which causes yeast cells to arrest growth, undergo a reduction in protein synthesis, accumulate the storage carbohydrate glycogen and acquire thermotolerance. Comparison of our dataset to that obtained following rapamycin-induced TOR inhibition in S. cerevisiae [26] reveals extensive overlap in induced (n = 35) and repressed (n = 90) homologous genes between the two datasets (Table S4). Thus a clear TOR repression-like starvation signature, relative to laboratory culture, is observable during early-stage infection, which may derive from the relative nutritional status of the tested conditions and/or slower growth within the context of our experiment. The indicated cellular down-turn in metabolism observed is strongly countered by up-regulation of genes encoding functions associated with amino acid and carbohydrate catabolism (Tables 1 and S3).
Figure 3. A genome-wide transcriptional snapshot of A. fumigatus Af293 during intiation of murine infection.
Red and green vertical lines correspond to individual up- and down-regulated genes, respectively. Thin light gray vertical lines indicate the positions of all other genes. (SM) and (asp_core) are density graphs of secondary metabolite and Aspergillus-core genes, respectively, expressed as a percentage of the total bases contained per gene type, per non-overlapping 2 kb of chromosomal sequence. Induced and repressed gene clusters, are depicted by red and green rectangles, respectively, below each chromosome. A complete listing of genes housed in these co-regulated clusters can be found in Table S6. Light blue/gray vertical bars represent putative centromeres and the pink vertical bar in chromosome 4 represents a region of ribosomal DNA.
Table 1. Over-represented Gene Ontology terms among differentially expressed genes.
| Over represented biological processes among genes induced in vivo | ||||||
| GO ID | GO term | List hits | List size | Pop.hits | Pop. Size | Fisher's Exact |
| GO:0006810 | transport | 133 | 448 | 761 | 4219 | 1.08E-10 |
| GO:0008643 | carbohydrate transport | 19 | 448 | 40 | 4219 | 3.41E-09 |
| GO:0009063 | amino acid catabolism | 14 | 448 | 35 | 4219 | 5.37E-06 |
| GO:0044242 | cellular lipid catabolism | 10 | 448 | 19 | 4219 | 6.30E-06 |
| GO:0044270 | nitrogen compound catabolism | 15 | 448 | 42 | 4219 | 1.28E-05 |
| GO:0030001 | metal ion transport | 19 | 448 | 74 | 4219 | 1.76E-04 |
| GO:0015892 | siderophore-iron transport | 5 | 448 | 8 | 4219 | 5.63E-04 |
| GO:0006812 | cation transport | 21 | 448 | 94 | 4219 | 6.45E-04 |
| GO:0019541 | propionate metabolism | 3 | 448 | 4 | 4219 | 4.38E-03 |
| GO:0006830 | high-affinity zinc ion transport | 2 | 448 | 2 | 4219 | 1.13E-02 |
| GO:0006631 | fatty acid metabolism | 13 | 448 | 62 | 4219 | 1.17E-02 |
Table lists selected biological processes significantly over-represented among differentially expressed genes, with respect to their occurrence in the A. fumigatus Af293 genome. To identify over-represented Gene Ontology terms, loci having significantly different expression were analyzed by the Expression Analysis Systematic Explorer (EASE) (PMID:14519205), which is implemented in MEV within the TIGR TM4 microarray data analysis suite (http://TM4.org). Numbers of genes in the indicated Gene Ontology categories were subjected to statistical analysis by EASE[25] to identify categories overrepresented compared with the whole genome data set. Only categories with Fisher's exact test probabilities below 5.00E-02 were included. Full results of the analysis can be found in Table S3.
Table 2. Functional classification of selected genes having altered transcript abundance, relative to laboratory culture, in the murine lung.
| Ergosterol and Heme Biosynthesis | Iron acquisition | ||||
| Log2 Ratio | ORF Number | Gene annotation | Log2 Ratio | ORF number | Gene annotation |
| −2.072748506 | Afu1g05720 | c-14 sterol reductase | 2.3932159 | Afu7g06060 | siderochrome-iron transporter (Sit1) |
| −2.135790025 | Afu1g03950 | cytochrome P450 sterol C-22 desaturase | 2.5525794 | Afu7g04730 | siderochrome-iron transporter |
| −2.1829312 | Afu8g07210 | hydroxymethylglutaryl-CoA synthase | 3.9717973 | Afu6g13750 | ferric-chelate reductase |
| −2.297904455 | Afu5g02450 | farnesyl-pyrophosphate synthetase | 6.105128 | Afu3g03440 | MFS family siderophore transporter |
| −2.346162536 | Afu7g03740 | 14-alpha sterol demethylase Cyp51B | 3.4018665 | Afu4g14640 | low affinity iron transporter |
| −2.895486437 | Afu1g07140 | c-24(28) sterol reductase | 2.6380832 | Afu3g03650 | sidG |
| −2.923648336 | Afu1g03150 | c-14 sterol reductase | 6.4549049 | Afu3g03640 | siderochrome-iron transporter (MirB) |
| −2.970089164 | Afu2g00320 | sterol delta 5 | 6.1431709 | Afu3g03420 | sidD |
| −3.376200503 | Afu6g05140 | sterol delta 5 | 6.4520669 | Afu3g03400 | siderophore biosynthesis acetylase AceI (sidF) |
| −3.422246472 | Afu5g14350 | c-24(28) sterol reductase | 3.4125222 | Afu3g03390 | siderophore biosynthesis lipase/esterase |
| −3.931510834 | Afu4g06890 | 14-alpha sterol demethylase Cyp51A | 4.8541282 | Afu3g03350 | nonribosomal peptide synthase (sidE) |
| −4.401905784 | Afu4g09190 | S-adenosyl-methionine-sterol-C- methyltransferase | 4.0089025 | Afu3g01360 | siderochrome-iron transporter |
| −4.300123651 | Afu1g07480 | coproporphyrinogen III oxidase | 5.2921431 | Afu1g17270 | ferric-chelate reductase (Fre2) |
| −2.435558859 | Afu5g06270 | 5-aminolevulinic acid synthase | 2.6644565 | Afu1g17200 | nonribosomal peptide synthase (sidC) |
| −2.128197751 | Afu5g07750 | ferrochelatase precursor | 2.6144266 | Afu8g01310 | metalloreductase (FRE1) |
| −3.016960611 | Afu6g07670 | cytochrome c oxidase assembly protein cox15 | Carbohydrate transport | ||
| Nitrate assimilation | 5.7013458 | Afu7g06390 | maltose permease | ||
| −4.050567754 | Afu1g12840 | nitrite reductase | 5.8795989 | Afu7g05190 | maltose permease |
| −0.297138231 | Afu1g12850 | nitrate transporter (nitrate permease) | 4.6926397 | Afu6g11920 | maltose permease |
| −2.332483829 | Afu1g12830 | nitrate reductase NiaD | 2.722422 | Afu5g00500 | maltose permease |
| 0.471365637 | Afu5g10420 | nitrate reductase | 4.7493339 | Afu3g01700 | maltose permease |
| 7.059699432 | Afu1g17470 | high affinity nitrate transporter NrtB | 2.917548 | Afu2g10910 | maltose permease |
| 0.55716766 | Afu6g13230 | Nit protein 2 | 2.6084555 | Afu1g03280 | maltose permease |
| Secreted Proteins | 3.2031907 | Afu3g03380 | maltose O-acetyltransferase | ||
| 7.40411592 | Afu5g14190 | beta-glucanase | 2.5614034 | Afu8g07070 | maltase |
| 5.898216645 | Afu1g17510 | lipase/esterase | 4.678081 | Afu7g06380 | maltase |
| 5.644563577 | Afu2g09380 | cutinase | 3.4802952 | Afu4g00150 | MFS maltose transporter |
| 5.496020587 | Afu8g07090 | extracellular proline-serine rich protein | 2.2425193 | Afu8g07240 | MFS maltose permease |
| 5.069338693 | Afu2g05150 | cell wall galactomannoprotein Mp2 | 3.0811489 | Afu6g01860 | MFS lactose permease |
| 5.009338142 | Afu5g00540 | extracellular signaling protein FacC | 2.0826028 | Afu1g17310 | MFS lactose permease |
| 4.974193046 | Afu7g01180 | extracellular lipase | 4.3502528 | Afu3g01670 | MFS hexose transporter |
| 4.926922531 | Afu1g16250 | alpha-glucosidase B | 4.6946836 | Afu2g08120 | MFS monosaccharide transporter (Hxt8) |
| 4.701754948 | Afu3g14030 | alkaline phosphatase | 3.3131505 | Afu5g14540 | MFS monosaccharide transporter |
| 4.683929957 | Afu2g00490 | glycosyl hydrolase | 4.8648019 | Afu4g00800 | MFS monosaccharide transporter |
| 4.678081027 | Afu7g06380 | maltase | 5.0594343 | Afu7g00780 | MFS monocarboxylate transporter |
| 4.659742223 | Afu8g01050 | lipase/esterase | 2.7697017 | Afu3g03320 | MFS monocarboxylate transporter |
| 4.62947329 | Afu3g14910 | extracellular signalling protein (factor C) | 3.0355716 | Afu3g03240 | MFS monocarboxylate transporter |
| 4.628176816 | Afu8g01130 | alpha-galactosidase C | 2.8084929 | Afu6g03060 | monosaccharide transporter |
| 4.576610248 | Afu4g01070 | acid phosphatase | 3.4709519 | Afu5g01160 | monosaccharide transporter |
| 4.53846138 | Afu7g05610 | glucanase | 3.7652613 | Afu4g13080 | monosaccharide transporter |
| 4.316438927 | Afu4g00870 | antigenic cell wall galactomannoprotein | 2.8100298 | Afu7g05830 | MFS sugar transporter |
| 4.069715654 | Afu6g02980 | extracellular exo-polygalacturonase | 6.9692415 | Afu6g14500 | MFS sugar transporter |
| 3.883566446 | Afu8g04710 | xylosidase | 2.0479063 | Afu5g06720 | MFS sugar transporter |
| 2.176380223 | Afu3g07850 | dipeptidyl aminopeptidase Ste13 | 4.4698085 | Afu1g11050 | MFS sugar transporter |
| 3.497276152 | Afu2g09030 | secreted dipeptidyl peptidase | 2.4191786 | Afu3g12010 | high-affinity hexose transporter |
| 3.821050502 | Afu6g11500 | dipeptidase | 2.8865489 | Afu3g00430 | high-affinity glucose transporter |
| Antigens | 3.2509899 | Afu8g04480 | hexose transporter protein | ||
| 5.642295426 | Afu4g09320 | antigenic dipeptidyl-peptidase Dpp4 | 3.6472387 | Afu6g06730 | l-fucose permease |
| 4.316438927 | Afu4g00870 | antigenic cell wall galactomannoprotein | Amino acid transport | ||
| 7.128239041 | Afu4g09580 | major allergen Asp F2 | 4.7164066 | Afu7g04290 | amino acid permease (Gap1) |
| 6.439814589 | Afu8g07080 | elastinolytic metalloproteinase Mep | 4.0522077 | Afu2g08800 | amino acid permease (Dip5) |
| 3.218425558 | Afu6g10250 | alkaline serine protease AorO | 3.7333962 | Afu8g06090 | amino acid permease |
| Carbohydrate and/or protein glycosylation | 5.6472247 | Afu5g09440 | amino acid permease | ||
| 2.540391388 | Afu8g02020 | glycosyl transferase | 2.9622857 | Afu2g10560 | amino acid permease |
| 6.098633033 | Afu4g14070 | glycosyl transferase | 3.2688804 | Afu1g09120 | amino acid permease |
| 2.229215652 | Afu5g00670 | glycosyl hydrolase family 35 | 2.9580145 | Afu8g02200 | proline permease |
| 2.266930252 | Afu6g11910 | glycosyl hydrolase family 3 | 5.7069031 | Afu7g01090 | proline permease |
| 3.177447564 | Afu4g00390 | glycosyl hydrolase | 3.3818247 | Afu2g11220 | proline permease |
| 2.065677868 | Afu2g03270 | glycosyl hydrolase | 4.1850538 | Afu8g01450 | GABA permease |
| 4.683929957 | Afu2g00490 | glycosyl hydrolase | 2.3940135 | Afu7g00440 | GABA permease |
| 2.14112188 | Afu2g03120 | cell wall glucanase (Utr2) | 2.9136458 | Afu5g14660 | GABA permease |
| 3.080401687 | Afu8g05610 | cell wall glucanase (Scw11) | 6.28253 | Afu5g00710 | GABA permease |
| 5.069338693 | Afu2g05150 | cell wall galactomannoprotein Mp2 | 6.7336819 | Afu1g14700 | allantoate transporter |
| Carbohydrate catabolism | Metal ion transport/homeostasis | ||||
| 2.832926255 | Afu6g14490 | beta-glucosidase | 4.9722279 | Afu5g09360 | calcineurin A |
| 2.109492985 | Afu3g00230 | beta-glucosidase | 5.7140897 | Afu7g01030 | Calcium-transporting ATPase 1 (PMC1) |
| 7.40411592 | Afu5g14190 | beta-glucanase | 3.30078 | Afu3g10690 | calcium-translocating P-type ATPase(PMCA-type) |
| 3.165725818 | Afu5g14550 | beta-galactosidase | 2.2461463 | Afu3g08540 | Ca2+ binding modulator protein (Alg2) |
| 2.183493763 | Afu1g14170 | beta-galactosidase | 2.7693449 | Afu6g00470 | plasma membrane zinc ion transporter |
| 7.164853239 | Afu7g06140 | beta-D-glucoside glucohydrolase | 4.2562039 | Afu5g03550 | plasma membrane H(+)ATPase |
| 3.586205167 | Afu6g08700 | beta glucosidase | 3.2581249 | Afu1g02480 | plasma membrane ATPase |
| 3.25484732 | Afu1g16700 | beta galactosidase | 4.1291169 | Afu1g01550 | high affinity zinc ion transporter |
| 4.926922531 | Afu1g16250 | alpha-glucosidase B | 3.103044 | Afu8g01890 | Na+/H+ exchanger family protein |
| 3.825416885 | Afu4g10150 | alpha-glucosidase | 3.3213401 | Afu7g04570 | Na/K ATPase alpha 1 subunit |
| 4.628176816 | Afu8g01130 | alpha-galactosidase C | Oxidative stress resistance | ||
| 6.513162955 | Afu1g01200 | alpha-galactosidase | 4.2704386 | Afu1g14550 | Mn superoxide dismutase MnSOD |
| 2.86708433 | Afu8g07300 | alpha/beta hydrolase | −0.7096154 | Afu4g11580 | Mn superoxide dismutase (SodB) |
| 4.46285962 | Afu8g00570 | alpha/beta hydrolase | 2.3390424 | Afu8g01670 | bifunctional catalase-peroxidase Cat2 |
| 2.441133251 | Afu8g00530 | alpha/beta hydrolase | |||
| 2.041648909 | Afu7g00830 | alpha/beta hydrolase | |||
| 5.206583195 | Afu3g01280 | alpha/beta hydrolase | |||
Differentially regulated transcripts were defined as having log2(Cy5 – Cy3) greater than the arbitrary thresholds of plus and minus two.
Lineage specificity and locational analyses of differentially expressed genes
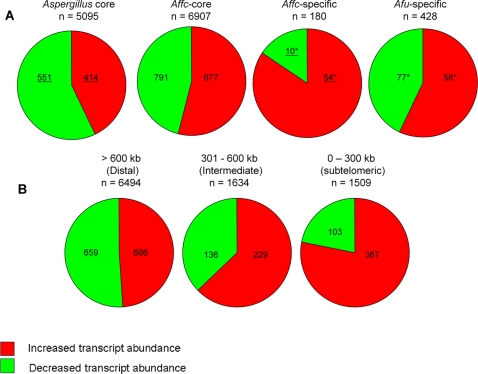
From an evolutionary perspective, relative proportions of genes being over- and underrepresented in the analysis differed significantly within lineage-specific gene cohorts (Figure 4A and Table 3). Genes having increased transcript abundance during infection are significantly enriched (p<0.0001 by chi-square analysis, see Table 3) among differentially expressed genes (n = 64) unique to the A. fumigatus lineage. Thus 93.6% of A. fumigatus genes having orthologues restricted to two very closely related, but differentially virulent, species Neosartorya fischeri (anamorph of Aspergillus fischerianus) (AAKE00000000) and Aspergillus clavatus (AAKD00000000) are more abundantly represented during the initiation of infection (Table 3). In contrast only 8% of genes having orthologues in all six Aspergillus species sequenced to date (i.e. the Aspergillus ‘core’ genome) are more abundantly represented under these conditions (n = 5095). This invariable ‘core’ genome encodes many functions associated with information processing, central metabolism and cell growth, retention of which is most likely to be essential for cellular survival[28]. Narrowing the phylogenetic sampling to include only A. fumigatus and its relatives N. fischeri and A. clavatus distinguishes several subtelomeric ‘genomic islands’ upon which phenotypic variation between species, including differing pathogenicity, might depend[28]. Accordingly we found differentially expressed genes to be unevenly distributed amongst A. fumigatus chromosomes (Figure 4B and Table 3). Induced genes form a significantly increased proportion of differentially regulated functions in intermediate (p<0.001) and subtelomeric (p<0.001) regions of the chromosomes (Figure 4B and Table 3). While only 16% of the predicted A. fumigatus gene repertoire is housed within 300 kb of telomeres (classed as the subtelomeric gene repertoire in our analyses), 29% of transcripts having increased abundance, relative to laboratory culture, in the murine lung are located to such subtelomeric areas, compared to just 11% of down-regulated transcripts. Moreover, 28% of the entire subtelomeric gene repertoire is represented in the induced dataset compared to only 8% of subtelomeric genes represented among down-regulated functions (Table 3).
Figure 4. Distribution of lineage specific and telomere-proximal genes among differentially expressed host adaptation dataset.
(A) Lineage specificity of A. fumigatus genes having altered transcript abundances, relative to laboratory culture, in the murine lung. The Aspergillus-core (Asp-core) set contains A. fumigatus Af293 proteins that have orthologues in A. clavatus (AAKD00000000), N. fischeri (AAKE00000000), Aspergillus terreus NIH2624 (AAJN01000000), Aspergillus oryzae RIB40[70], A. nidulans FGSC A4[71] and Aspergillus niger CBS 513.55[72] The Affc-core set were defined as A. fumigatus Af293 proteins that have ortholouges in N. fischeri and A. clavatus. The Affc-unique set is a sub-set of Affc-core proteins that do not have ortholouges in A. terreus, A. oryzae, A. nidulans or A. niger. Asterisks indicate gene sets which are listed in Table S5. Underlined values significantly deviate from the null hypothesis that an equal number of induced and repressed genes will occur in each cohort, as estimated by Chi-square analysis (Table 3). (B) Chromosomal distribution of A. fumigatus genes having altered transcript abundances, relative to laboratory culture, in the murine lung. Distances from telomeres (kb) are noted above pie charts. Asterisked gene sets are listed in Supplementary Table S5. Underlined values significantly deviate from the null hypothesis that an equal number of induced and repressed genes will occur in each cohort, as estimated by Chi-square analysis (Table 3).
Table 3. Distribution of induced and repressed genes among lineage specificity cohorts and chromosomal locations.
| Number of genes | Differentially expressed | UP' (observed) | DOWN' (observed) | UP' (expected) | DOWN' (expected) | χ2 | p | |
| Aspergillus core | 5095 | 965 | 414 | 551 | 482.5 | 482.5 | 19.45 | <0.0001 |
| Affc-core | 6907 | 1468 | 791 | 677 | 734 | 734 | 8.853 | 0.0029 |
| Affc-specific | 180 | 64* | 54 | 10 | 32 | 32 | 30.25 | <0.0001 |
| Afu-specific | 428 | 135* | 77 | 58 | 67.5 | 67.5 | 2.674 | 0.102 |
Chi-square analysis (with one degree of freedom) was used to test the distribution of induced and repressed genes with respect to lineage specificity and sub-genomic locations, based upon the null hypothesis that equal numbers of induced and repressed genes occur in each cohort. Asterisked gene populations are listed in Supplementary Table S5.
Clustering of induced genes
Regarding our A. fumigatus gene expression dataset as a function of chromosomal locus (Figure 3) we identified that many induced A. fumigatus genes are found in contiguous clusters. To investigate this further we generated a custom script to automate cluster identification which identified numerous genomic loci within which co-ordinate regulation of a minimum of 5 closely neighbouring genes can be observed (Figure 3 and Table S6). Co-ordinate expression of physically clustered genes is a prominent feature of the induced, but not repressed, gene set and we observe a large proportion (40%) of up-regulated physically clustered genes to reside within 300 kb of chromosome ends (Figure 3 and Table 4). The clusters are comprised of up to 34 co-ordinately expressed genes and include loci directing biosynthesis of siderophores (cluster 33) and two known secondary metabolites, pseurotin and gliotoxin (clusters 72 and 60, respectively). The gliotoxin biosynthetic cluster is not subtelomerically located being 700 kb from the telomere (as annotated by Perrin et. al. [16]). The pseurotin biosynthetic cluster, however, (as annotated by Maiya et. al. [29]) is contained within the fumitremorgen cluster (Afu8g00100–8g00720) at 100 kb from the telomere[16]. Pseurotin[29]–[31] is a neuritogenic, nematicidal quinone and gliotoxin an immunotoxin which supports A. fumigatus virulence in some murine models of invasive pulmonary aspergillosis[8]–[12]. We observed four other postulated, but uncharacterised, secondary metabolite gene clusters induced during early stage A. fumigatus infection, including a large proportion of genes on the left arm of chromosome 8, predicted to encode a fumitremorgen biosynthesis supercluster[32]. Thus, it would seem that selective expression of a subset of secondary metabolite loci facilitates initiation of mammalian infection. While gliotoxin biosynthesis is dispensable for virulence in some murine models, our analysis demonstrates that this host environment is nonetheless conducive to immunotoxin production, and further insights on virulence mechanisms relevant to neutropenia and/or corticosteroid therapy await comparative analyses of fungal gene expression in each of these strikingly different host settings, an analysis which is currently underway in our laboratory. A fully annotated table of clusters, indicating between-species synteny within cluster loci, and all accession numbers, can be viewed in Dataset S2.
Table 4. Comparative analysis of genetic distribution among genes having differential transcript abundances in microarray analyses using A. fumigatus RNA following exposure to in vitro or murine-adaptive stress, or laeA gene deletion.
| Experimental condition | Total genes in dataset | Proximal | % | Intermediate | % | Distal | % | A. fumigatus-specific | % |
| Genome stats | 9632 | 1509 | 16 | 1634 | 17 | 6494 | 67 | 2201 | 23 |
| Clustered in vivo up | 658 | 227 | 34 | 129 | 20 | 302 | 46 | 218 | 33 |
| ΔlaeA up | 415 | 102 | 32 | 59 | 14 | 254 | 61 | 122 | 29 |
| in vivo up | 1282 | 367 | 29 | 229 | 18 | 686 | 54 | 374 | 30 |
| Nitrogen starvation up | 626 | 180 | 29 | 115 | 18 | 331 | 53 | 33 | 33 |
| in vivo up (no sec mets) | 1196 | 327 | 27 | 223 | 19 | 646 | 54 | 355 | 30 |
| Neutrophils 60 minutes up | 312 | 83 | 27 | 81 | 26 | 148 | 47 | 80 | 26 |
| Alkaline adaptation up | 211 | 53 | 25 | 39 | 18 | 113 | 54 | 6 | 6 |
| ΔlaeA up (no sec mets) | 318 | 70 | 22 | 58 | 18 | 190 | 60 | 50 | 16 |
| Acid stress up | 83 | 18 | 22 | 21 | 25 | 44 | 53 | 43 | 52 |
| Iron limitation up | 28 | 4 | 14 | 4 | 15 | 20 | 71 | 8 | 30 |
| ΔlaeA down | 528 | 93 | 18 | 99 | 19 | 336 | 64 | 108 | 20 |
| Oxidative stress up | 54 | 9 | 17 | 9 | 17 | 36 | 67 | 22 | 54 |
| in vivo down | 898 | 103 | 11 | 136 | 15 | 659 | 73 | 126 | 14 |
Gene density in telomere-proximal (o-300 kb from telomeres), intermediate (300–600 kb from telomeres) and telomere-distal (>600 kb from telomeres) regions of the A. fumigatus genome are indicated (Genome stats). (no sec mets) indicates omission of secondary metabolite biosynthetic genes (identified as detailed in the Materials and Methods section) from the tested dataset.
Comparative analyses of murine adaptation- and in vitro gene expression datasets
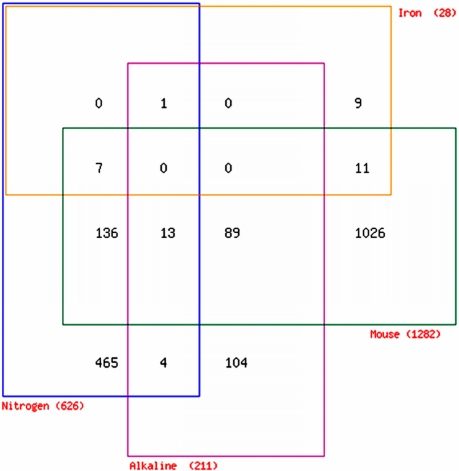
The complexity of the transcriptional signature derived from host adaptation analyses is likely to originate from convergence of multiple environmental cues, coupled with metabolic and morphologic effects. Although functional categorisation of differentially expressed functions (Tables 1 and S3) could identify metabolic and physiological trends during initiation of infection, environment-related signatures were less easy to discern. To ascertain physiologically relevant features of the host environment we compared the transcriptome of host-adapting germlings to those of in vitro A. fumigatus cultures exposed to iron limitation, nutrient limitation, alkaline stress, acid stress, neutrophils, oxidative stress or anaerobic stress. The resulting transcriptomic responses varied in magnitude (Table 4) but assignment of cut-off log2 ratio values of +2 and −2 across all of the analyses enabled us to distinguish several important aspects of the A. fumigatus host-adaptive response (Figure 5). The transcriptional signatures of paramount importance among those examined were alkaline adaptation, iron deprivation and nutrient starvation, which are remarkably prominent in the infection dataset. Using the aforementioned log2 cut-offs, 24 of 43 iron-regulated genes were identified as differentially expressed during host adaptation, of these 18 were more abundantly represented (Figure 5 and Dataset S3), and 6 less abundantly represented. Relaxing the cut-off criterion to encompass all differentially regulated genes in the host adaptation dataset allowed complete capture of the iron regulon (Figure 6). Among these genes are the siderophore biosynthetic genes sidA (Afu2g07680), sidD (Afu3g03420) and sidC (Afu1g17200), the latter two discerning biosynthesis of both intra- and extracellular siderophores during infection as substantiated by previous findings[18], and four siderochrome/siderophore transporter proteins Afu7g06060, Afu7g04730, Afu3g03440 and afu3g03640. Preferential gene expression following a 60 minute shift from acid to alkaline medium could also be strongly correlated with that observed during infection (Figure 5 and Dataset S3). Alkaline adaptive capability, previously found to be essential for A. nidulans virulence in neutropenic mice[33], is likely to be important for growth of A. fumigatus spores at physiological pH. Accordingly we identified 102 genes preferentially expressed during both murine infection and in vitro alkaline adaptation (Figure 5 and Dataset S3). Among them are 36 genes having unknown function, two sodium ATPases (Afu6g03690 and Afu4g09440), the plasma membrane zinc ion transporter (Afu6g00470) and an alkaline phosphatase (Afu3g14030). Interestingly, we found no concordance between iron starvation and alkaline adaptation (Figure 5). Since acidification of the macrophage phagolysosome is an essential step in ROS-mediated A. fumigatus killing we also assessed the transcriptome of Af293 germlings upon shift from pH7 to pH3, using a rich medium and hydrochloric acid. No concordance between the resulting dataset and that of host adaptation was evident, indeed (despite the magnitude of the murine infection dataset) most functions upregulated in response to an in vitro acid shift were less abundantly represented in our infection analyses (n = 18, Figure S2) thus we can confidently conclude that acid stress, at least within the context tested in this analysis (which can only approximate conditions encountered in the host) is not relevant during host adaptation. This agrees with our observation that alkaline adaptation is a physiological cue of primary importance in this murine model of infection (Figure 5).
Figure 5. Overlap between murine adaptation and in vitro stress datasets.
Venn diagrammatic representation of overlap between murine adaptation dataset and those of nitrogen starvation, iron starvation and alkaline shift. Genes are listed in Dataset S3.
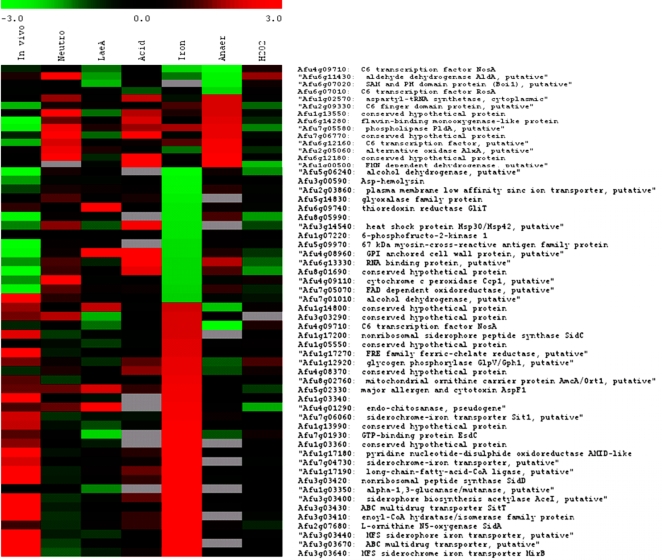
Figure 6. Comparative analysis of A. fumigatus gene expression datsets.
A pan-experimental comparison of A. fumigatus gene expression aligning log2 ratios obtained during host adaptation (mice); exposure to neutrophils (neut), increased expression in parental strain versus ΔlaeA mutant, acid shift (acid), iron starvation (iron), oxygen depletion (anaer) and oxidative stress (H2O2) for various genes. The colour bar indicates the range of log2 expression ratios, grey bars indicate genes from which signals were undetectable for technical reasons. Experimental conditions are described in Materials and Methods. LaeA dataset is taken from Perrin et. al. [16]. Comparative analyses were implemented in TM4 http://www.jcvi.org/cms/research/software/.
An essential component of phagocyte defense against A. fumigatus conidia and hyphae is the NADPH oxidase-mediated respiratory burst which generates reactive oxygen species (ROS) required for fungal killing. To assess the A. fumigatus transcriptional response to oxidative stress, conidia were grown in rich medium at 37°C prior to shift into similar medium containing 17 mM hydrogen peroxide. Comparison of the resulting dataset to that obtained from host-adapting germlings revealed some concordance, this time revealing a subset of genes having decreased transcript abundance both in vitro and during murine infection (Figure S2). A common theme among this group of genes is ergosterol and heme biosynthesis, evidenced by common behaviour of both 14-alpha sterol demethylase Cyp51A-encoding genes (Afu4g06890 and Afu7g03740) and a coproporphyrinogen III oxidase homologue (Afu1g07480). Oxygen depletion was achieved by transfer of A. fumigatus hyphae, following 16 hour growth in rich medium, to anaerobic chambers containing two palladium catalysts. We were unable to correlate gene expression under these anaerobic stress conditions with gene expression during infection (Figure 6). Finally, we assessed germlings grown in 20 ml of RPMI1640 with L-glutamine, 25 mM HEPES and 5% fetal bovine serum for 7 hours at 37°C following exposure to human neutrophils at a multiplicity of infection of 1∶1 for 60 minutes, the reference sample for this analysis being germlings incubated in the absence of neutrophils. Of 57 A. fumigatus genes upregulated in response to neutrophil exposure in vitro, 18 (60%) were also more abundantly represented during initiation of murine infection (Figures 6 and S2). Interestingly these included the two major A. fumigatus antioxidant enzymes, Mn superoxide dismutase (Afu1g14550) and the bifunctional catalase-peroxidase Cat2 (Afu8g01670). Whether representation of these transcipts among those differentially expressed in both murine and laboratory culture is indeed neutrophil-specific remains to be determined.
Nutrient limitation is a relevant physiological cue during A. fumigatus initiation of murine infection
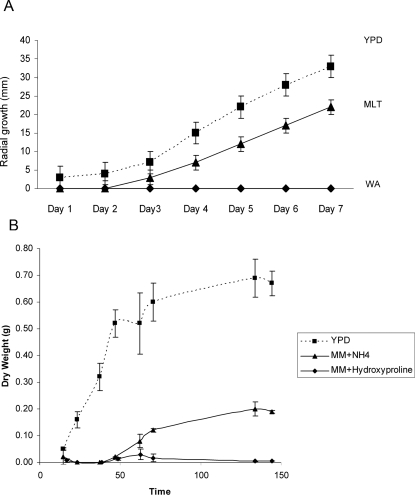
To investigate the relevance of nutrient starvation during host adaptation we made several growth and gene expression analyses. Hypothesising that YPD is nutritionally more robust than murine lung tissue we compared radial growth of A. fumigatus, in triplicate from an inoculum of 100 spores grown at an agar/air interface on Petri dishes, on YPD and on a synthesized murine lung tissue medium (MLT) composed of homogenised murine lung tissue (80%) and water/agar (20%), overlaid upon a water/agar baseplate. Growth of A. fumigatus was completely unsupported by water/agar base with no evidence of conidial germination after 7 days. Growth on YPD produced conidiating colonies averaging 66 mm (n = 3) in diameter after 7 days at 37°C, whereas MLT as growth medium supported significantly less growth, reaching a maximum colony diameter of 44 mm (Figure 7A). Any concern that reduced radial growth observed on MLT medium originates from iron deprivation can be allayed by reference to radial growth analysis of two independent wild type A. fumigatus isolates, CEA10 and ATTC46645[17], where equivalent growth is observed in the presence and absence of iron, and on blood agar medium. Thus, from this solid growth analysis, we conclude that MLT supports slower A. fumigatus colony growth than YPD under laboratory conditions. Our MLT analysis could not support the volume of liquid culture required to perform growth curve analyses, moreover, the viscosity of the medium would have hindered dry weight measurements. To support our conclusions on nutritive status of the host environment, within the context of the experimentation performed during our infection analyses, we assessed log2 ratios obtained from competitive microarray hybridisation, using doubly amplified A. fumigatus mRNA extracted from nitrogen starved germlings, and the same YPD reference sample used for the initial host adaptation analysis (samples F and G, respectively, Table S1). Nitrogen starvation was exerted in shaken liquid culture using minimal medium, with hydroxyproline as nitrogen source, for a period of five hours. Hydroxyproline is a rational candidate nitrogen source during initiation of mammalian pulmonary infection, being a widely used surrogate marker of lung injury whose concentration in bronchoalveolar lavage fluid permits quantitative assessment of collagen breakdown[34],[35]. Transcript levels of the proline permeases afu2g11220, Afu8g02200 and Afu7g01090 suggest induction of proline uptake during initiation of infection, and laboratory culture on solid medium confirms that hydroxyproline can support aconidial filamentous growth of A. fumigatus on minimal medium in the presence of a repressing carbon source such as glucose (data not shown). To confirm the inferiority of hydroxyproline as a nitrogen source (relative to YPD) we performed dry weight growth curve analyses (Figure 7B) including a widely used Aspergillus minimal medium (MM) for comparison.
Figure 7. Characterisation of A. fumigatus growth, relative to YPD.
(A) Comparative analysis of Af293 radial growth on YPD and synthetic murine lung tissue medium (MLT). Triplicated, spot-inoculated plates containing single 100 spore inocula were incubated at 37°C. (B) Growth curve analysis of Af293, performed in triplicate using liquid YPD, or AMM containing 1% glucose and either 5 mM ammonium tartrate or 5 mM hyroxyproline as nitrogen source. Cultures were inoculated to a final concentration of 5×106 spores/ml and incubated under aerobic conditions at 37°C with shaking at 150 rpm. At selected timepoints mycelia were harvested on Miracloth, encased in Whatmann paper and dried at 37°C for 48 hours before weighing.
Nitrogen starvation rendered 1047 genes subject to differential expression, relative to the doubly amplified YPD reference. Several notable features of the resultant dataset (Dataset S4) support our conclusion that, relative to YPD laboratory culture, initiation of murine infection occurs under nutrient stress. As with our analyses of host adaptation, over-represented Gene Ontology (GO) terms among differentially expressed genes identified ribosome biogenesis and assembly, and protein biosynthesis and processing as the most significantly down-regulated functional categories (Table S7). We also identified significant over-representation of cell cycle-related functions among genes preferentially expressed during growth in hydroxyproline, including cell cycle regulation, mitosis, nuclear migration, chromosome segregation and karyogamy (Table S7). This is a particularly satisfying finding since, supported by our growth curve analyses (Figure 7B), a clear distinction between A. fumigatus growth phase in the compared media is discernable. This was not a feature of the murine-YPD comparison which, importantly, lessens the probability that differences associated with cell cycle stage or growth rate preside over environmental cues within the host adaptation dataset. Direct comparisons of the murine and nitrogen starvation datasets identified an overlap of 280 differentially expressed genes common to both, of these 24 genes are also common to the TOR kinase nutrient limitation geneset, and are indicated in Table S4. 156 genes preferentially expressed under nitrogen starvation conditions (relative to YPD laboratory culture) were similarly favoured during host adaptation (Dataset S3). Assessing locational bias among the hydroxyproline dataset, 29% of the geneset was found to reside subtelomerically (Table 4) with 180 out of 634 preferentially expressed genes housed within 300 kb of telomeres. This is comparable to the level of subtelomeric gene expression identified during initiation of murine infection (Table 4) and indicates that, relative to growth in a nutrient-rich laboratory culture, adaptation to growth in a nutritionally challenging environment prompts expression of a subtelomeric gene repertoire. A number of physically linked coregulated genes came to prominence in the hydroxyproline starvation dataset (Table S8). 16 clusters conforming to the previously applied cluster algorithm were identified, 8 of which were subtelomeric and 3 of which encompass genes in secondary metabolite loci, as defined by bioinformatic analyses. Of the three latter loci one cluster (Afu6g03390–6g03490) subject to regulation by the LaeA methyltransferase[16], the product of which is currently unknown, is also expressed during murine infection. Beyond this, correlation between clustered gene regulation in murine and nitrogen starved growth was modest.
The LaeA regulon is represented among genes preferentially expressed during intitation of murine infection
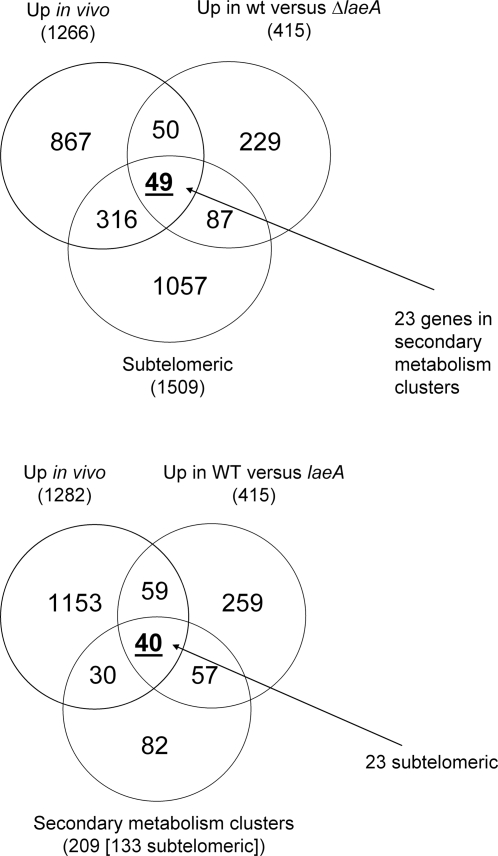
Given the predominance of clustered and subtelomeric gene loci among host adaptation genes, we compared our dataset to that produced by Perrin et. al. [16] during study of laeA gene deletion. An A. fumigatus ΔlaeA (1g14660) mutant, which has decreased virulence in both neutropenic and hydrocortisone treated mice[32],[36], demonstrates significantly lower expression of genes in 13 secondary metabolite biosynthetic clusters including that of gliotoxin. LaeA was found to influence expression of a subset of lineage- and species-specific genes therefore we tested the overlap between functions down-regulated in the absence of LaeA and functions having greater transcript abundance during murine infection, hoping to decipher a link between genes under LaeA control, and those active during initiation of murine infection. Out of 415 genes down-regulated in the absence of LaeA we identified 99 genes having increased abundance during initiation of murine infection (Figure 8). Functional categorisation of shared genes revealed that 40% (n = 40) were involved in secondary metabolite biosynthesis, among these we could identify three complete secondary metabolite clusters, those directing gliotoxin and pseurotin biosynthesis as well as the genetic locus mentioned above (Afu6g03390–6g03490) whose biosynthetic product is unknown. Perrin et. al. also found 54% of the LaeA-regulated gene clusters showing differential expression under laboratory conditions were located within 300 kb of the telomeres. We therefore extended our analysis still further, determining the proportions of subtelomeric and secondary metabolism cluster genes shared between the two datasets. This identified 49 and 40 genes, having subtelomeric locations and secondary metabolite biosynthetic functions respectively (Figure 8).
Figure 8. Expression of LaeA-regulated genes during initiation of murine infection.
Venn diagram representation of overlap between genes repressed in ΔLaeA[16] and those having increased transcript abundance during murine infection, according to proportions having subtelomeric locations, and secondary metabolism functionality (on the basis of annotation).
Finally we analysed all of the in vitro and in vivo datasets comparatively to assess the occurrence of subtelomeric bias among differentially expressed genes, aiming to determine whether the induction of genes at the telomeric extremities of chromosomes was a standard feature of adaptation to environmental alterations rather than a host adaptation phenomenon (Table 4). The analysis identified murine adaptation, laeA deletion, neutrophil exposure and nitrogen starvation as the conditions most enriching for transcript abundance among subtelomeric genes, where 29%, 32%, 27% and 29% of the respective cohorts were located. Interestingly, removal of secondary metabolism genes from the analysis dramatically reduced the LaeA regulated subtelomeric gene cohort to 22% while only minimally impacting representation of murine adaptation genes (Table 4).
Taken together these analyses indicate that a significant component of the LaeA regulon, comprised mainly of secondary metabolism genes, is represented among transcripts more abundant during infection. Furthermore the subtelomeric bias observed among differentially expressed murine adaptation genes extends beyond secondary metabolite biosynthesis and does not appear to be a general feature of adaptation to environmental change.
Discussion
We present a methodology for A. fumigatus transcript profiling during initiation of murine infection and a comparative analysis of global transcriptional programming, in laboratory culture and the mammalian lung. We were able to optimize a technical and statistical framework sufficiently robust to reproducibly quantify relative transcript abundances using minute samplings of A. fumigatus germlings. The statistical analysis, though seemingly complex, utilizes standard statistical methods which are well documented in biological analyses, particularly in microarrays[37]. We chose to co-hybridize mock samples from the same amplification protocol (thereby determining the ‘true’ biological effect in a benchmark sample, in this case total RNA) and then, by comparison to cohybridisations using unamplified samples, indirectly estimate the systematic effect of the amplification in our analysis. The t-testing framework[23] then allowed us to identify genes where the log ratios are significantly different due to one factor i.e. the amplification protocol. This enabled an estimation of the proportion of genes showing amplification-dependent bias which we carried into the murine experiment, where comparison of unamplified versus amplified material is impossible. Clearly, we assume that this estimate can be reliably applied to similar experiments, in doing so we also assume that the amplification process depends only on the protocol adopted, and not the underlying gene expression dynamics. A further technical consideration in planning experiments comparing murine and laboratory samples is differential treatments. Given the nature of our analysis, differential treatment of the germling samples was necessary to avoid osmotic shock in either instance. Water lavage would impose an osmotic shift on germlings rescued from the lung and saline treatment would osmotically shock laboratory cultured germlings. Thus the technical limitations imposed by such comparative analysis must be accepted, however, in terms of maximally preserving transcript abundances within the context of the experiment, we believe our treatments of the samples to be appropriate.
To identify factors governing adaptation to the host niche we compared laboratory and murine lung samples aiming to find fungal attributes preferentially employed during infection. Therefore our findings document genes having increased transcript abundance in the A. fumigatus sample of murine origin, relative to that derived from laboratory cultured fungus. We performed our comparative analysis using doubly amplified mRNA from developmentally matched A. fumigatus germlings following laboratory culture, or growth in neutropenic murine lungs. In total we identified 2164 genes having altered transcript abundance. Functional analysis of the dataset flagged certain putatively relevant physiological cues which we then pursued by additional in vitro analyses.
Careful interpretation of the dataset with respect to specific nutrient acquisition mechanisms, and within the context of the comparison performed here, can lend powerful insight into accessible nutrients in the mammalian niche. Nitrate assimilation (which is strongly inducible by nitrate in the absence of preferred nitrogen sources ammonium and glutamine)[38] is not preferentially employed in the murine host as evidenced by co-ordinate down-regulation of crnA (Afu1g12850), niaD (Afu12830) and niiA (Afu1g12840). This may be due to a) equivalent nutritional status of YPD and murine lung or b) to the absence of nitrate as a nitrogen source during initiation of infection, or both. We were able to confirm, by radial growth analyses in vitro using YPD and a synthetic lung tissue medium MLT, that YPD is nutritionally superior to MLT based upon the rate of radial growth supported (Figure 7A). Coupled with the observation that the nitrogen metabolite repression gene product AreA is required for full virulence[19] a likely explanation for slowed growth and repression of nitrogen assimilation, is the utilisation of alternative non-preferred nitrogen sources during establishment of disease, such as amino acids. Strong support for this conclusion is provided by high level induction of the areA-dependent nitrogen-scavenging enzyme L-amino acid oxidase LaoA (Afu7g06810) which enables Aspergilli to catabolise a broad spectrum of amino acids in nitrogen starvation conditions[39]. Notably a by-product of such catabolism is ammonium. However the likliehood that sufficient ammonium is produced by these reactions to prevent starvation is diminished by the observed general starvation response in our dataset. Catabolism of amino acids during initiation of infection is also evidenced by induction of the methyl citrate synthase enzyme (Afu6g03590) which acts to detoxify the intermediates of propionyl-coA generating carbon sources[40], such as cysteine, isoleucine and methionine. An essential role for methylcitrate synthase in murine aspergillosis has recently been demonstrated[41] thereby demonstrating the value of our approach in generating physiological information on virulence mechanisms within the context of murine infection.
Strong themes among genes having lowered transcript abundance during murine infection are ribosome biogenesis and assembly, and protein bisoynthesis and folding (Table 1). Such signatures are commonly observed among microbes under stress, and in this instance might indicate a slowing of growth in the murine lung, relative to laboratory culture. We reasoned that a mechanism possibly linking such transcriptional profiles to nutrient starvation is TOR mediated ribosomal gene regulation, which tightly couples protein synthesis and cellular growth to availability of nutrients and physiological status to balance the opposing forces of protein synthesis and degradation. This is pivotal for cellular fate determination in many organisms[42],[43], propelling cells towards either proliferation (through the cell cycle) or vegetative growth (increase in size). Comparison of our dataset to that generated following rapamycin-mediated TOR kinase inhibition in S. cerevisiae revealed a very marked overlap (n = 125) in differentially regulated homologous genes (Table S4). This is in keeping with defined roles for TOR kinase function in S. cerevisiae, which includes the regulation of transcription in response to nutrients[26]. A strong correlation between developmental programming and microbial secondary metabolite biosynthesis has been well-documented[14] and while plasticity of nitrogen metabolism demonstrably supports A. fumigatus virulence[20] the role of the single, and likely essential, A. fumigatus TOR kinase homologue, TorA, (Afu2g10270) in this process remains untested. Intriguingly, however, a link between TOR kinase function and secondary metabolite production, partially through AreA, has recently been established in the rice pathogen Fusarium fujikuroi [44] where, in addition to the target genes in common with yeast and other eukaryotes, the AreA-regulated giberellin and bikaverin biosyntheis genes are also under the control of TOR. This raises the possibility of a relatively limited investment in secondary metabolism, within the context of our analysis, at the tested timepoint of murine infection.
Genes expressed during nitrogen starvation are also expressed during virulence in Magneporthe grisea [45], where two wide-range regulators of nitrogen catabolism genes, NPR1 and NPR2 [46] are required for virulence. Contrary to our analysis nitrate and nitrite reductase activities were found to be relevant for M. grisea during rice infection[45], however, a number of amino acid transporting proteins predominated among induced functions during both in vitro nitrogen starvation and infection. Common to that analysis, and ours, was increased abundance of proline oxidase (Afu3g02300) and proline permease (Afu2g11220, Afu7g10190, Afu8g02200) proteins, which, in the context of our experiment might have special relevance given the high hydroxyproline of collagen tissue, and the notable proline requirement of an attenuated A. fumigatus deletion mutant lacking the Ras-related protein RhbA[47]. To test the effect of nitrogen limitation on gene expression, in the context of the host adaptation study performed, we returned to the YPD reference sample employed for the initial analysis (sample F, Table S1), this time performing competitive hybridisations with doubly-amplified RNA obtained from a nitrogen-starved laboratory culture (sample G, Table S1). Given the predominance of hydroxyproline among collagen amino acids content, and its release into bronchoalveolar lavage fluid upon tissue injury[34] we adopted hydroxyproline as sole source of nitrogen for the starvation experiment. Hydroxyproline supported markedly slowed growth of A. fumigatus in liquid culture (Figure 7B) relative to YPD. We identified a significant overlap between the two datasets amounting to 280 genes. Functional anlaysis of the resulting datasets revealed that, in keeping with the murine adaptation dataset, ribosome biogenesis and protein biosynthesis were markedly down regulated, moreover, functions associated with mitosis and cell cycle were more abundantly represented among the categories favoured under nitrogen starvation, relative to rich laboratory growth. Given the differences between the tested media in terms of ability to support A. fumigatus growth (Figure 7B) it is pleasing to see that such a theme was not apparent from the murine adaptation dataset where the degree of nutrient limitation is unlikely to be as severe as that imposed in the nitrogen starvation conditions we used here. Interestingly 29% of the genes preferentially expressed during nitrogen starvation were subtelomerically located, and a degree of clustered gene expression was observable. Thus induction of the subtelomeric gene repertoire becomes important during nutrient deprivation, a trend which was not observable in response to any of the other in vitro stresses, other than neutrophil exposure the physiological relevance of which requires further investigation.
The uptake and catabolism of other amino acids released by proteolytic digestion of murine lung parenchyma might provide a source of nitrogen during A. fumigatus infection, which is supported in our analyses by increased transcript abundance of various secreted proteases (Table 2). Extracellular proteases are implicated as virulence actors in invasive aspergillosis[1]. An elastolytic protease, produced when A. fumigatus is cultured on the insoluble matrix from bovine lung is also produced during spore germination in infected lungs of neutropenic mice, as judged by immunogold cytochemical localization[48], and mutationally-derived mutants unable to produce this protease were deficient for virulence in the murine model. An elastinolytic metalloprotease, characterized by the same group was similarly visualized during murine lung infection[49]. Many species of human pathogenic fungi secrete proteases in vitro or during infection. Full virulence associated with A. fumigatus protease mutants [1],[50] is presumed to be due to redundancy among the many enzymes produced by this organism which may degrade the lung parenchyma to release utilisable carbon and nitrogen during infection. A directed analysis of the role of such proteins in virulence might now be possible on the strength of our data. Importantly, however, any conclusions reached on the basis of this study reflect the comparative nature of the anlaysis, thus transcripts equally abundant under both conditions tested will not have been be identified. Time course analyses of A. fumigatus growth during murine infection will reveal stage-specific gene expression in the absence of confounding sample treatments. These analyses are underway in our laboratories.
Fungal oxygen-sensing mechanisms have been linked to cell membrane sterol levels in Schizosaccharomyces pombe and Cryptococcus neoformans where homologues of the mammalian Sterol Regulatory Element Binding Protein (SREBP) transcription factors, in complex with SREBP cleavage-activating protein (SCAP) partners, undergo cellular translocation (from the endoplasmic reticulum to the golgi) prior to proteolytic SREBP activation[51],[52]. The C. neoformans SREBP homologue, Sre1p, plays an important role in low oxygen adaptation and infection[52]. Biosynthesis of sterols and unsaturated fatty acids is an aerobic process in Saccharomyces cerevisiae and in C. neoformans, the hypoxia-mimicking agent cobalt chloride and oxygen limitation target sterol biosynthetic gene expression. This was intriguing to us given the broadly observed reduction in transcript abundance among ergosterol biosynthetic genes, relative to laboratory culture (Table 2). Our initial hypothesis attributed this effect to oxygen limitation in the murine lung environment since airway obstruction, intraalveolar exudates and inflammation, or damage to alveolar capillaries (all observed in our murine modelling of pulmonary aspergillosis) pose a significant barrier to proper oxygenation in human lungs[53]. Oxygen deprivation in mammals leads to a transcriptional induction of genes for adaptation to hypoxia[54] and efforts to characterise the transcription profile of murine immune responses to A. fumigatus infection indicate that hypoxia is relevant in the neutropenic murine lung at 24 hours post-infection (Turnbull, personal communication). Co-hybridisation of murine lung cDNAs, derived from infected immunocompetent and immunocompromised mice with a murine immunology array set identified upregulation of murine ARNT (log2 ratio = 2.09) the obligate heterodimeric binding partner for the hypoxia induced factor HIF-1α[54]. However, on comparing the A. fumigatus murine adaptation gene expression signature to that observed following exposure to anaerobic stress in vitro (Figure 5A) no support for this hypothesis could be gleaned. Rather, repression of the two A. fumigatus 14-alpha sterol demethylase genes (Afu4g06890 and Afu Afu03740), representing a critical step in ergosterol biosynthesis, was observed following hydrogen peroxide-mediated oxygen stress. Therefore, it would seem that this component of the H2O2-mediated oxidative stress response is relevant in vivo. This was found to be distinct from antioxidative action of the A. fumigatus Mn superoxide dismutase (Afu1g14550) and the bifunctional catalase-peroxidase Cat2 (Afu8g01670), both of which were more abundant following murine lung (Table 2) or neutrophil exposure (Figures 6 and S2), suggesting multiple modes of oxidative stress encountered during murine infection. Interestingly H2O2 gradients are detectable across the Saccharomyces cerevisiae plasma membrane upon H2O2 exposure, suggesting a mechanism other than diffusion for H2O2 entry into cells. This, coupled with the observation that S. cerevisiae mutants erg3Δ and erg6Δ, having increased ergosterol biosynthesis, show increased permeability to H2O2 might suggest down-regulation of ergosterol biosynthesis as a protective response against oxidative stress[55].
All of the previously characterised components of the A. fumigatus siderophore biosynthetic pathway[17],[18] were more abundantly represented at transcript level during murine infection. Comparison of the murine dataset with that generated during in vitro iron limitation confirmed the importance of this environmental deficit during murine adaptation (Figure 6). With respect to extracellular iron mobilization fungal siderophores bind ferric iron with a high affinity, delivering the ferric chelate to specific receptors at the cell surface for translocation into the cytoplasm. Existing evidence supports an essential role for this uptake mechanism during infection[17],[18]. Reductive iron assimilation (RIA) is dispensable for murine virulence[17] but may assist in iron acquisition during infection since RIA inhibition in the absence of extracellular siderophore biosynthesis prevents growth in vitro [18]. This hypothesis is supported by our finding that numerous RIA components are induced during initiation of infection (Table 2) in addition to siderophore biosynthetic genes. We were also able to correlate differential gene expression following in vitro alkaline shift to the host adaptation transcriptome (Figure 5). This was to be expected given the broad requirement for fungal pH adaptation during mammalian pathogenesis[56] but is nonetheless pleasing to observe particularly given that the in vitro acid stress transcriptome showed an opposite trend (Figure S2) and since pleiotropic activity of the virulence-directing family of PacC/Rim101 pH sensing transcription factors[56] does not permit virulence defects to be soley, or thus far absolutely, correlated with pH growth phenotypes.
Fungal secondary metabolite biosynthesis is mediated at the level of transcription, from clusters of physically linked, co-ordinately regulated genes and is profoundly affected by environmental factors such as pH and nutrient availability[14]. For the most part, defined cues prompting gene expression from such clusters remain unknown, as do the functions of the molecules produced by them in the natural environment, but popular theory regards microbial secondary metabolites as chemical determinants of selective advantage[57]. Noting that clusters of physically linked, co-ordinately regulated genes were a prominent feature of our dataset we applied a custom script to systematically identify them. To limit the identification of false negatives from our analysis (due, for example, to poor spot quality or absence on array) we allowed for ‘gaps’ of up to a maximum of 4 non-up- or down- regulated genes permissable per cluster. Gap size variation influenced the architecture of clusters detectable in the dataset, further widening the extremities of pre-existing clusters rather than creating new ones (a complete breakdown of the cluster anlaysis is available as Dataset S2). Thus a gap size of zero identified 14 upregulated gene clusters and a gap size of one identified 38. The maximum gap size applied (gap size = 4) identified 65 groups of physically linked genes having increased transcript abundance during murine infection (Figure 3). A large proportion (34%) of up-regulated, physically clustered, genes was found to reside within 300 kb of chromosome ends (Figure 3 and Table 4). The clusters are comprised of up to 34 co-ordinately expressed genes and include loci directing biosynthesis of siderophores (cluster 33) and two known secondary metabolites, pseurotin and gliotoxin (clusters 70 and 59, respectively). The importance of fungal gene clusters in virulence has recently been characterised in the fungal plant pathogen, Ustilago maydis where co-induction of physically linked, secreted protein-encoding genes is seen during infection[58]. We were unable to find evidence of clustered secreted protein genes from our analyses of A. fumigatus (data not shown) however, from an evolutionary perspective, clusters 8, 40, 53, 68 (down-regulated), 69 and 70 (Table S6) deviate significantly from the least virulent sequenced species considered here, N. fischeri, and therefore merit further analysis. N. fischeri is extremely rarely identified as a human pathogen[59]–[61], while prolonged exposure to A. clavatus spores can cause extrinsic allergic alveolitis known as malt worker's lung[62]. From a functional perspective some relevant deductions regarding the clusters identifiedby our analyses might be possible using present genome annotations. However, closer scrutinization of clusters is required to address the likelihood that A. fumigatus genes located in them encode functions required for virulence. We anticipate that the inference of shared functionality among neighbouring co-regulated genes in our dataset will empower the functional annotation of the A. fumigatus genome; moreover, it significantly raises the profile of such gene-regulatory paradigms within the context of fungal pathogenicity.
Our observations of biased localization among genes overrepresented during growth in the murine lung (Figures 3 and 4, Table 3) prompted comparison of our dataset to that obtained by Perrin et. al. [16] who identified genes under control of the global seconday metabolism regulator, LaeA. Significant overlap is observable, notably among secondary metabolism genes, most specifically between genes in the gliotoxin and pseurotin biosynthetic gene clusters. Hypovirulence of a ΔlaeA deletion mutant cannot be explained solely on the basis of a lack of gliotoxin biosynthesis, and no investigation of the role of other secondary metabolite clusters, with respect to virulence, has been undertaken in A. fumigatus, therefore at this time it is not possible to reach any conclusions on the contribution made to virulence by the molecules whose synthesis is directed by these loci. With regard to our comparison of functions under LaeA control and those important during murine infection, differing experimental conditions (Perrin study[16] performed at 25°C in liquid shaking culture and glucose minimal medium[63] for 60 hours) and methodologies pose a contentious issue. A truly illuminating analysis of LaeA activity during murine pathogenesis might be forthcoming from time course analyses in neutropenic mice, which we are currently attempting. DNA sequences in subtelomeric regions undergo ectopic recombination at a much higher rate than expected for homologous recombination[64] allowing the expansion and diversification of gene families located at chromosome ends. For some organisms gene expression is governed by subtelomeric localisation, for example two A. nidulans secondary metabolism gene clusters, directing penicillin and sterigmatocystin biosynthesis, are activated following deletion of the hdaA histone deacetylase gene[65]. In other organisms subtelomeres provide an ideal setting for genes involved in antigenic variation, such as in the parasites Plasmodium falciparum and Trypanosoma brucei [66] and in cytoadhesion, such as the EPA family of Candida glabrata adhesins[67]. Thus the importance of sub-telomeric chromosomal regions within the context of eukaryotic pathogenesis is gaining significance in this post-genomic era of microbial studies.
These analyses of transcript abundance, drawn from minute quantities of fungal material convey a programme of A. fumigatus cellular regulation directly from the site of pulmonary infection. From a fungal physiological perspective the mammalian host restricts iron, and likely various nutrients, as well as exerting multiple degrees of oxidative stress. Regarded as a function of the genomic landscape the observed transcriptional changes reveal genome organisation and subtelomeric diversity as effectors of the remarkable versatility of A. fumigatus with respect to the niches it successfully inhabits, one of which is the neutropenic human lung.
Materials and Methods
Full details of the methods used are available through Array Express (http://www.ebi.ac.uk/microarray-as/aer/#ae-main0) Accession number E-TABM-327. The sequenced A. fumigatus isolate Af293 has been previously described[68] and was used for all analysis reported in this study.
A. fumigatus strains and growth conditions
A. fumigatus Af293 in vitro isolates for murine infection experiments were grown in shaken liquid culture at 37°C in YPD medium. For oxidative stress Af293 conidia were inoculated into Aspergillus complete medium (CM)[69] and grown for 16 hours with shaking at 37°C. The resultant hyphal culture was transferred to CM containing 17 mM H2O2 and aliquots were harvested for RNA isolation upon transfer (T0) and at 60 minutes after the initiation of H2O2 exposure. For acid stress Af293 conidia were incubated in liquid CM medium with shaking for 6 hours at 37°C. At that time the culture was split, one portion of the culture being adjusted to pH3 using HCl (time T0). Aliquots of each were taken at time 60 mins for RNA purification and microarray expression analysis. pH 3 was verified at T0 and at the end of the time course. For alkaline stress A. fumigatus germlings were cultured for 16 hours in shaken liquid AMM containing 100 mM glycolic acid pH5.0, 10 ml/L vitamin solution[7], 5 mM ammonium tartrate and 1% w/v glucose. Germlings were filtered using Miracloth and shifted to similar, prewarmed medium containing 100 mM Tris-HCl pH8.0. After 1 hour incubation at 37°C, mycelia were washed with cold AMM pH5.0 and RNA extraction was performed immediately. For anaerobic stress hyphae were grown in CM (107 conidia in 20 ml in 100 mm Petri dishes without agitation) for 16 hours at 37°C. Three plates were placed in anaerobic chambers and anaerobic conditions were established using 2 palladium catalysts (GasPak Plus, Becton Dickinson). A control plate was placed in a chamber without palladium catalysts and all plates were returned to a 37°C incubator for the duration of the experiment. Plates placed in anaerobic chambers were removed at 60 minutes, the mycelium rapidly harvested on Miracloth, rinsed with ice-cold water and rapidly frozen in liquid nitrogen. The RNA was extracted and used in competitive hybridization with the 16 hour control RNA sample. For neutrophil exposure 107 germlings grown in 20 ml of RPMI1640 with L-glutamine, 25 mM Hepes, and 5% (v/v) fetal bovine serum for 7 hours at 37°C in 100 mm Petri dishes were exposed to 107 human neutrophils. At 60 minutes of exposure to neutrophils, RNA was prepared. For iron limitation A. fumigatus isolate ATCC46645 was grown for 15 hours at 37°C in –Fe Aspergillus minimal medium (AMM, iron depleted conditions) according to Pontecorvo[63] containing 1% (wt/vol) glucose as carbon source, 20 mM glutamine as nitrogen source. After ths time, 10 µM FeSO4 was added to the medium and germlings harvested after a further 60 minutes. Growth curve analyses were performed in triplicate using liquid YPD, or AMM containing 1% glucose and either 5 mM ammonium tartrate or 5 mM hyroxyproline as nitrogen source. Cultures were inoculated to a final concentration of 5×106 spores/ml and incubated under aerobic conditions at 37°C with shaking at 150 rpm. At selected timepoints mycelia were harvested on Miracloth, encased in Whatmann paper and dried at 37°C for 48 hours before weighing.
Murine infections
Groups of 24 outbred male mice (strain CD1, 18–22 g, Harlan Ortech) were housed in individually vented cages and allowed free access to food and water. Mice received cyclophosphamide (150 mg/kg, ENDOXANA, Asta Medica) by intraperitoneal injection on days −3 and −1. A single dose of hydrocortisone acetate (112.5 mg/kg, HYDROCORTISTAB, Sovereign Medical) was administered subcutaneously on day −1. All mice received tetracycline hydrochloride 1 mg/l and ciprofloxacin 64 mg/l in drinking water as prophylaxis against bacterial infection. Aspergillus spores for inoculations were grown on solid ACM, containing 5 mM ammonium (+)-tartrate and 1% (w/v) Oxoid Agar Number 3 for 5 days prior to infection. Conidia were freshly harvested using sterile saline (Baxter Healthcare Ltd. England) and filtered through MIRACLOTH (Calbiochem). Conidial suspensions were spun for 5 minutes at 3000 g, washed twice with sterile saline, counted using a hemocytometer and re-suspended at a concentration of 2.5×1010 colony forming units (c.f.u)/ml. Viable counts from administered inocula were determined following serial dilution by plating on Aspergillus complete medium and growth at 37 °C. Mice were anesthetized by halothane inhalation and infected by intranasal instillation of 108 conidia in 40 µl of saline. Groups of infected mice were culled and processed collectively during a 2 hour window corresponding to a time point 12–14 hours post-infection. Bronchoalveolar lavage was performed immediately after culling using three 0.5 ml aliquots of pre-warmed sterile saline. BALFs were snap frozen immediately following harvest using liquid nitrogen.
RNA extraction and amplification
Prior to RNA extraction snap-frozen BALF samples were centrifuged, washed with 1 ml of ice-cold sterile water to lyse contaminating host cells and pooled. Following a further cycle of snap freezing pelleted pooled product was homogenized using a pestle and mortar and liquid nitrogen. RNA extraction was performed immediately using the ‘filamentous fungi’ protocol of the RNeasy mini kit (Qiagen). RNA concentration and integrity was measured by Nanodrop. Reference RNA was prepared from snap frozen ground mycelium. RNA from reference samples was prepared from snap-frozen homogenized germlings or young mycelium.
Estimation of fidelity of transcript abundance following mRNA amplification
The corrected, multiple t-testing methodology of Nygaard[23] formed the basis of our investigation. However, for computational ease it was modified to use the limma linear modelling framework for the R statistical environment.
Spots were filtered as previously described and systematic spatial effects were normalized within arrays using the print-tip lowess method (limma package for R); no between-array normalization was performed in order to preserve the amplification-protocol variation existent between arrays. Least squares regression was used to fit the average log ratio, and error term of each transcript in each of the amplification protocols. The resulting coefficient vectors contained the mean relative abundance of each transcript (T0 v T60) in one of the three amplification protocols: aRNAr1, aRNAr2 or totRNA. The amplification protocol effects and their associated Benjamini-Hochberg corrected p-values (contrast1, contrast2 and contrast3 in Figure S1) were extracted by fitting a contrast matrix to the original linear model. We defined the null hypothesis H0 as no difference in log ratios due to the amplification protocol, and an alternative hypothesis H1 as loss of log ratio fidelity due to the amplification protocol. Spots were partitioned into the three sets based on the Benjamini-Hochberg adjusted p-values associated with each of the fitted contrasts. The rejected, undetermined and conservative sets were populated by genes whose adjusted p-values were 0<p<0.01, 0.01<p<0.1 and ≫0.1<p<1 respectively. This approach rejected 2325 (8.49%) of the spots in the aRNA1 - totRNA comparison, i.e. the genes whose log ratios are significantly altered as a result of 1 round of mRNA amplification. The analogous rejected sets for the aRNA2 – totRNA and aRNA1 – aRNA2 comparisons contained 2604 (9.52%) and 73 (0.27%) rejected spots respectively. Comparison of the rejection sets showed that 80.69% of the spots in the aRNA1- totRNA rejection set were also present in the aRNA2- totRNA rejection set. This finding, in addition to the observed small number of genes in the aRNA1 – aRNA2 rejection set, suggests that most of the amplification specific noise is introduced in the first round.
Microarray hybridisations
All experiments used Af293 DNA amplicon microarrays[68]. Labelling reactions with RNA and hybridisations were conducted as described in the TIGR standard operating procedures found at http://atarrays.tigr.org. Independent verification of selected log2 ratios was achieved using quantitative RT-PCR (See Figure S3 and Table S9).
Hybridised slides were scanned using the Axon GenePix 4000B microarray scanner and the TIFF images generated were analyzed using TIGR Spotfinder (http://www.tigr.org/software/) to identify poor quality spots. mRNA underwent two rounds of linear amplification by T7 promoter-directed transcription. Data processing and analysis was performed in the R statistical framework, using the limma, multcomp and fdrtool packages (http://www.bioconductor.org). The intensity data retained after filtering (91.5% of the total spots) were normalized within (print-tip lowess) and between (scale) arrays. Least squares regression (lmFit) was then used to estimate a vector of regression coefficients (representing the mean log2 (Cy5/Cy3) across the 5 biological replicates for the in vivo effect), the associated residuals and unadjusted p-values. Differentially regulated transcripts (Benjamini-Hochberg corrected p-value<0.05) were further categorized by the magnitude and direction of the fitted log ratios. Up-regulated transcripts were defined as having log2 (Cy5/Cy3) greater than an arbitrary threshold of plus, or minus, two. Accuracy of microarray data was independently verified by quantitative RT-PCR on selected transcripts (See Figure S3). Oligonucleotides used for this analysis are detailed in Table S9).
For exposure to in vitro stresses microarray experiments were performed as previously described[68].
Automated biological theme determinations
To identify over-represented Gene Ontology terms, loci showing significantly different expression were further analyzed by the Expression Analysis Systematic Explorer (EASE) (PMID:14519205), which is implemented in MEV within the TIGR TM4 microarray data analysis suite (http://TM4.org). Numbers of genes in the indicated Gene Ontology categories were subjected to statistical analysis by EASE[25] to identify categories overrepresented compared with the whole genome data set. Only categories with Fisher's exact test p-values<0.05 were included.
Core and lineage-specific gene sets
Orthologous proteins in the genomes were identified using a reciprocal-best-BLAST-hit (RBH) approach with a cut-off of 1e-05. The orthologous clusters, as well as synteny visualization and comparative analysis tools can be also found in the Aspergillus Comparative Database at http://www.tigr.org/sybil/asp.
Gene cluster identifications
Clusters of co-regulated neighbouring genes were defined using a simple, two parameter algorithm. The first parameter, minimum_block_size (mbs), specifies the minimum number of up- or down- regulated genes that a cluster must contain. The second parameter, maximum_gap_size (mgs), specifies the maximum number of adjacent non-up- or down- regulated genes permissable per cluster. Four analyses were performed corresponding to mgs values of n = 0, 1, 2, 3 and 4. Figure 3 shows data from mgs = 4. Assigned cluster numbers are based on chromosomal position. More details on the clustering algorithm are available at http://sybil.sourceforge.net/documentation.html#algorithms. Secondary metabolite biosynthesis genes were identified using SMURF, a web-based software tool (http://www.tigr.org/software/genefinding.shtml - temporarily hosted at http://binf.gmu.edu/fseifudd/smurf/index1.html). Density of the SM genes was estimated as the exon length per kb.
Supporting Information
Schematic representation of hybridisations for assessing maintenance of relative transcript abundance during A. fumigatus mRNA amplification. Boxes represent the biological source material from which the RNA was isolated. The open and shaded arrows depict dye-swapped hybridisations for all pairs of samples. For all comparisons Af293-derived cDNA sampled at media shift (t0) was cohybridised with Af-derived cDNA sampled at 60 minutes post-shift (t60). Horizontally aligned boxes depict cohybridisation of samples subjected to one (aRNA1), two (aRNA2) or zero (totRNA) rounds of linear mRNA amplification. Amplification protocols were compared indirectly by fitting a contrast matrix (contrast1, contrast2, contrast3).
(0.03 MB JPG)
Comparative analysis of A. fumigatus gene expression datsets. A pan-experimental comparison of A. fumigatus gene expression aligning log2 ratios obtained during host adaptation (mice); exposure to neutrophils (neut), increased expression in parental strain versus ΔlaeA mutant, acid shift (acid), iron starvation (iron), oxygen depletion (anaer) and oxidative stress (H2O2) for various genes. The colour bar indicates the range of log2 expression ratios, grey bars indicate genes from which signals were undetectable for technical reasons. Experimental conditions are described in Materials and Methods. LaeA dataset is taken from Perrin et. al. [16]. Comparative analyses were implemented in TM4 http://www.jcvi.org/cms/research/software/
(70.39 MB TIF)
Concordance between estimates of relative abundance from triplicate qPCR measurements (yellow) and microarray experiments (grey) for selected A. fumigatus ORFs. Oligo sequences are given in Table S9.
(1.00 MB TIF)
RNA samples
(0.05 MB DOC)
Filtered genes
(0.03 MB DOC)
GO analysis
(0.85 MB DOC)
TOR analysis
(0.37 MB DOC)
Asterisked gene lists
(0.34 MB DOC)
Cluster overview
(1.33 MB DOC)
Nitrogen starvation GO
(0.40 MB XLS)
Nitrogen starvation gene clusters
(0.03 MB XLS)
Oligo sequences for RT-PCR
(0.04 MB DOC)
(0.58 MB XLS)
Gene clusters fully annotated
(0.19 MB XLS)
Lists of genes common to several analyses
(0.04 MB XLS)
(0.93 MB XLS)
Acknowledgments
We thank H. N. Arst Jnr., Sven Krappman and Nora Khaldi for critical discussion and reading of the manuscript and Jimena Gonzalez Gonzalez, Tatiana Munera and C. Kim Nguyen for technical support.
Footnotes
The authors have declared that no competing interests exist.
We acknowledge the support of the Medical Research Council, award numbers G0501164 (E. B.) and G0501397 (for K. H. and A. M.) awarded to Imperial College, National Institute for Allergy and Infectious Diseases (NIAID) NIH, award numbers NIAID U01AI48830 and R21 AI052236 (W. C. N.), The Austrian Science Foundation, award number FWP P-18606-B11 (H. H.) and The National Institutes of Health, award numbers AI051144, AI052236 and N01-AI-30041 (G. M.) and Cancer Support Center, grant number CA16672 (G. M.). The Wellcome Trust (K. H. and D. A-J.) and the Biotechnology and Biological Sciences Research Council (K. H.). G. G. is supported by FAPESP and CNPq, Brazil.
References
- 1.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tekaia F, Latge JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8:385–392. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Zmeili OS, Soubani AO. Pulmonary aspergillosis: a clinical update. QJM. 2007;100:317–334. doi: 10.1093/qjmed/hcm035. [DOI] [PubMed] [Google Scholar]
- 4.Schaffner A, Douglas H, Braude A. Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus. Observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. J Clin Invest. 1982;69:617–631. doi: 10.1172/JCI110489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol. 2006;176:3717–3724. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- 6.Mullbacher A, Eichner RD. Immunosuppression in vitro by a metabolite of a human pathogenic fungus. Proc Natl Acad Sci U S A. 1984;81:3835–3837. doi: 10.1073/pnas.81.12.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardo J, Urban C, Galvez EM, Ekert PG, Muller U, et al. The mitochondrial protein Bak is pivotal for gliotoxin-induced apoptosis and a critical host factor of Aspergillus fumigatus virulence in mice. J Cell Biol. 2006;174:509–519. doi: 10.1083/jcb.200604044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bok JW, Chung D, Balajee SA, Marr KA, Andes D, et al. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun. 2006;74:6761–6768. doi: 10.1128/IAI.00780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramer RA, Jr, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, et al. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell. 2006;5:972–980. doi: 10.1128/EC.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupfahl C, Heinekamp T, Geginat G, Ruppert T, Hartl A, et al. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol Microbiol. 2006;62:292–302. doi: 10.1111/j.1365-2958.2006.05373.x. [DOI] [PubMed] [Google Scholar]
- 11.Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, et al. Gliotoxin Production in Aspergillus fumigatus Contributes to Host-Specific Differences in Virulence. J Infect Dis. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- 12.Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, et al. Gliotoxin Is a Virulence Factor of Aspergillus fumigatus: gliP Deletion Attenuates Virulence in Mice Immunosuppressed with Hydrocortisone. Eukaryot Cell. 2007;6:1562–1569. doi: 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–59, table. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 15.Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, et al. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. doi:10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200:1213–1219. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, et al. Distinct Roles for Intra- and Extracellular Siderophores during Aspergillus fumigatus Infection. PLoS Pathog. 2007;3:e128. doi: 10.1371/journal.ppat.0030128. doi:10.1371/journal.ppat.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel M, Arst HN, Jr, Aufauvre-Brown A, Holden DW. The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol Gen Genet. 1998;258:553–557. doi: 10.1007/s004380050767. [DOI] [PubMed] [Google Scholar]
- 20.Krappmann S, Braus GH. Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med Mycol. 2005;43(Suppl 1):S31–S40. doi: 10.1080/13693780400024271. [DOI] [PubMed] [Google Scholar]
- 21.Krappmann S, Bignell EM, Reichard U, Rogers T, Haynes K, et al. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol Microbiol. 2004;52:785–799. doi: 10.1111/j.1365-2958.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- 22.Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, et al. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- 23.Nygaard V, Loland A, Holden M, Langaas M, Rue H, et al. Effects of mRNA amplification on gene expression ratios in cDNA experiments estimated by analysis of variance. BMC Genomics. 2003;4:11. doi: 10.1186/1471-2164-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 28.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a Hybrid PKS/NRPS Required for Pseurotin A Biosynthesis in the Human Pathogen Aspergillus fumigatus. Chembiochem. 2007;8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi A, Fujioka S, Nukina M, Kawano T, Shimada A, et al. Fumiquinones A and B, nematicidal quinones produced by Aspergillus fumigatus. Biosci Biotechnol Biochem. 2007;71:1697–1702. doi: 10.1271/bbb.70110. [DOI] [PubMed] [Google Scholar]
- 31.Komagata D, Fujita S, Yamashita N, Saito S, Morino T. Novel neuritogenic activities of pseurotin A and penicillic acid. J Antibiot (Tokyo) 1996;49:958–959. doi: 10.7164/antibiotics.49.958. [DOI] [PubMed] [Google Scholar]
- 32.Keller N, Bok J, Chung D, Perrin RM, Keats SE. LaeA, a global regulator of Aspergillus toxins. Med Mycol. 2006;44(Suppl):83–85. doi: 10.1080/13693780600835773. [DOI] [PubMed] [Google Scholar]
- 33.Bignell E, Negrete-Urtasun S, Calcagno AM, Haynes K, Arst HN, Jr, et al. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol Microbiol. 2005;55:1072–1084. doi: 10.1111/j.1365-2958.2004.04472.x. [DOI] [PubMed] [Google Scholar]
- 34.Adamson IY, King GM, Bowden DH. Collagen breakdown during acute lung injury. Thorax. 1988;43:562–568. doi: 10.1136/thx.43.7.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robin J. Methods for measuring hydroxyproline and estimating in vivo rates of collagen biosynthesis and degradation. Methods in Molecular Medicine. 2005. pp. 189–207. [DOI] [PubMed]
- 36.Sugui JA, Pardo J, Chang YC, Mullbacher A, Zarember KA, et al. Role of laeA in the Regulation of alb1, gliP, Conidial Morphology, and Virulence in Aspergillus fumigatus. Eukaryot Cell. 2007;6:1552–1561. doi: 10.1128/EC.00140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 38.Amaar YG, Moore MM. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Genet. 1998;33:206–215. doi: 10.1007/s002940050328. [DOI] [PubMed] [Google Scholar]
- 39.Davis MA, Askin MC, Hynes MJ. Amino acid catabolism by an areA-regulated gene encoding an L-amino acid oxidase with broad substrate specificity in Aspergillus nidulans. Appl Environ Microbiol. 2005;71:3551–3555. doi: 10.1128/AEM.71.7.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maerker C, Rohde M, Brakhage AA, Brock M. Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J. 2005;272:3615–3630. doi: 10.1111/j.1742-4658.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 41.Ibrahim-Granet O, Dubourdeau M, Latge JP, Ave P, Huerre M, et al. Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cellular Microbiology Epub ahead of print. 7 A.D. doi: 10.1111/j.1462-5822.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- 42.Hansen IA, Attardo GM, Roy SG, Raikhel AS. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J Biol Chem. 2005;280:20565–20572. doi: 10.1074/jbc.M500712200. [DOI] [PubMed] [Google Scholar]
- 43.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teichert S, Wottawa M, Schonig B, Tudzynski B. Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot Cell. 2006;5:1807–1819. doi: 10.1128/EC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donofrio NM, Oh Y, Lundy R, Pan H, Brown DE, et al. Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet Biol. 2006;43:605–617. doi: 10.1016/j.fgb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Lau G, Hamer JE. Regulatory Genes Controlling MPG1 Expression and Pathogenicity in the Rice Blast Fungus Magnaporthe grisea. Plant Cell. 1996;8:771–781. doi: 10.1105/tpc.8.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panepinto JC, Oliver BG, Fortwendel JR, Smith DL, Askew DS, et al. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of Invasive pulmonary aspergillosis. Infect Immun. 2003;71:2819–2826. doi: 10.1128/IAI.71.5.2819-2826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolattukudy PE, Lee JD, Rogers LM, Zimmerman P, Ceselski S, et al. Evidence for possible involvement of an elastolytic serine protease in aspergillosis. Infect Immun. 1993;61:2357–2368. doi: 10.1128/iai.61.6.2357-2368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markaryan A, Morozova I, Yu H, Kolattukudy PE. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect Immun. 1994;62:2149–2157. doi: 10.1128/iai.62.6.2149-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaton-Ogay K, Paris S, Huerre M, Quadroni M, Falchetto R, et al. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol Microbiol. 1994;14:917–928. doi: 10.1111/j.1365-2958.1994.tb01327.x. [DOI] [PubMed] [Google Scholar]
- 51.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 53.Tuder RM, Yun JH, Bhunia A, Fijalkowska I. Hypoxia and chronic lung disease. J Mol Med. 2007;85:1317–1324. doi: 10.1007/s00109-007-0280-4. [DOI] [PubMed] [Google Scholar]
- 54.Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell Mol Life Sci. 2003;60:1376–1393. doi: 10.1007/s00018-003-2370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branco MR, Marinho HS, Cyrne L, Antunes F. Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279:6501–6506. doi: 10.1074/jbc.M311818200. [DOI] [PubMed] [Google Scholar]
- 56.Penalva MA, Tilburn J, Bignell E, Arst HN., Jr Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008 doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 57.Rohlfs M, Albert M, Keller NP, Kempken F. Secondary chemicals protect mould from fungivory. Biol Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howlett BJ, Idnurm A, Heitman J. Fungal pathogenesis: gene clusters unveiled as secrets within the Ustilago maydis code. Curr Biol. 2007;17:R87–R90. doi: 10.1016/j.cub.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 59.Chim CS, Ho PL, Yuen KY. Simultaneous Aspergillus fischeri and Herpes simplex pneumonia in a patient with multiple myeloma. Scand J Infect Dis. 1998;30:190–191. doi: 10.1080/003655498750003627. [DOI] [PubMed] [Google Scholar]
- 60.Lonial S, Williams L, Carrum G, Ostrowski M, McCarthy P., Jr Neosartorya fischeri: an invasive fungal pathogen in an allogeneic bone marrow transplant patient. Bone Marrow Transplant. 1997;19:753–755. doi: 10.1038/sj.bmt.1700715. [DOI] [PubMed] [Google Scholar]
- 61.Summerbell RC, de RL, Chartrand C, St GG. Graft-related endocarditis caused by Neosartorya fischeri var. spinosa. J Clin Microbiol. 1992;30:1580–1582. doi: 10.1128/jcm.30.6.1580-1582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blyth W, Grant IW, Blackadder ES, Greenberg M. Fungal antigens as a source of sensitization and respiratory disease in Scottish maltworkers. Clin Allergy. 1977;7:549–562. doi: 10.1111/j.1365-2222.1977.tb01485.x. [DOI] [PubMed] [Google Scholar]
- 63.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 64.Farman ML. Telomeres in the rice blast fungus Magnaporthe oryzae: the world of the end as we know it. FEMS Microbiol Lett. 2007;273:125–132. doi: 10.1111/j.1574-6968.2007.00812.x. [DOI] [PubMed] [Google Scholar]
- 65.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barry JD, Ginger ML, Burton P, McCulloch R. Why are parasite contingency genes often associated with telomeres? Int J Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 67.De Las PA, Pan SJ, Castano I, Alder J, Cregg R, et al. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to. Genes Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 69.Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 70.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 71.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 72.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of hybridisations for assessing maintenance of relative transcript abundance during A. fumigatus mRNA amplification. Boxes represent the biological source material from which the RNA was isolated. The open and shaded arrows depict dye-swapped hybridisations for all pairs of samples. For all comparisons Af293-derived cDNA sampled at media shift (t0) was cohybridised with Af-derived cDNA sampled at 60 minutes post-shift (t60). Horizontally aligned boxes depict cohybridisation of samples subjected to one (aRNA1), two (aRNA2) or zero (totRNA) rounds of linear mRNA amplification. Amplification protocols were compared indirectly by fitting a contrast matrix (contrast1, contrast2, contrast3).
(0.03 MB JPG)
Comparative analysis of A. fumigatus gene expression datsets. A pan-experimental comparison of A. fumigatus gene expression aligning log2 ratios obtained during host adaptation (mice); exposure to neutrophils (neut), increased expression in parental strain versus ΔlaeA mutant, acid shift (acid), iron starvation (iron), oxygen depletion (anaer) and oxidative stress (H2O2) for various genes. The colour bar indicates the range of log2 expression ratios, grey bars indicate genes from which signals were undetectable for technical reasons. Experimental conditions are described in Materials and Methods. LaeA dataset is taken from Perrin et. al. [16]. Comparative analyses were implemented in TM4 http://www.jcvi.org/cms/research/software/
(70.39 MB TIF)
Concordance between estimates of relative abundance from triplicate qPCR measurements (yellow) and microarray experiments (grey) for selected A. fumigatus ORFs. Oligo sequences are given in Table S9.
(1.00 MB TIF)
RNA samples
(0.05 MB DOC)
Filtered genes
(0.03 MB DOC)
GO analysis
(0.85 MB DOC)
TOR analysis
(0.37 MB DOC)
Asterisked gene lists
(0.34 MB DOC)
Cluster overview
(1.33 MB DOC)
Nitrogen starvation GO
(0.40 MB XLS)
Nitrogen starvation gene clusters
(0.03 MB XLS)
Oligo sequences for RT-PCR
(0.04 MB DOC)
(0.58 MB XLS)
Gene clusters fully annotated
(0.19 MB XLS)
Lists of genes common to several analyses
(0.04 MB XLS)
(0.93 MB XLS)