Abstract
Immunoglobulin (Ig) isotype switching is a recombination event that changes the constant domain of antibody genes and is catalyzed by activation-induced cytidine deaminase (AID). Upon recruitment to Ig genes, AID deaminates cytidines at switch (S) recombination sites, leading to the formation of DNA breaks. In addition to their role in isotype switching, AID-induced lesions promote Igh-cMyc chromosomal translocations and tumor development. However, cMyc translocations are also present in lymphocytes from healthy humans and mice, and thus, it remains unclear whether AID directly contributes to the dynamics of B cell transformation. Using a plasmacytoma mouse model, we show that AID+/− mice have reduced AID expression levels and display haploinsufficiency both in the context of isotype switching and plasmacytomagenesis. At the Ig loci, AID+/− lymphocytes show impaired intra- and inter-switch recombination, and a substantial decrease in the frequency of S mutations and chromosomal breaks. In AID+/− mice, these defects correlate with a marked decrease in the accumulation of B cell clones carrying Igh-cMyc translocations during tumor latency. These results thus provide a causality link between the extent of AID enzymatic activity, the number of emerging Igh-cMyc–translocated cells, and the incidence of B cell transformation.
Upon antigen encounter, activated B cells often replace their heavy chain constant domain (Cμ) with one of a set of downstream CH exons (Cγ, Cε, or Cα) in a process known as class switch recombination (CSR). This intrachromosomal recombination event occurs between highly repetitive switch (S) sequences located upstream of each CH gene, with the exception of Cδ. CSR is targeted to specific S regions by 5′ I promoters that mediate germline or sterile S-CH transcription (1), and by 5′ and 3′ heavy chain enhancers that promote S-S synapses (2). These mechanisms cooperate to render S domains accessible to activation-induced cytidine deaminase (AID) (3, 4), a B cell–specific enzyme encoded by the Aicda gene that deaminates cytidine residues to uracils on both strands of S region DNA, resulting in U:G mismatches (5).
Uracils are recognized and removed by uracil DNA glycosylase and mismatch repair enzymes (6, 7), leading ultimately to the formation of DNA double-strand breaks (6, 8, 9). These DNA lesions are processed by the nonhomologous end-joining proteins and other repair mechanisms that ensure efficient recombination (10). If unrepaired, however, AID-mediated DNA breaks can become substrates for chromosomal translocations that often juxtapose protooncogenes to the Ig loci. Canonical Igh-cMyc translocations, for instance, are the hallmark of both Burkitt's lymphomas in humans (T(8;14)) and plasmacytomas in mice (T(12;15)) (11).
The role of AID in the etiology of Igh-cMyc chromosomal translocations was implicated by genetic experiments using AID−/− mice carrying IL-6 or Bcl-xL transgenes (12, 13). In both cases, deletion of AID resulted in the absence of canonical cMyc translocations. In H2AX−/− B cells, AID was also required for CSR-mediated translocations (14). In addition, several lines of evidence indicate that AID somatic hypermutation (SHM) activity may also promote tumor development by targeting non-Ig genes (15–17). Using a plasmacytoma mouse model, we now demonstrate that the extent of AID activity influences the incidence of B cell tumor development by directly determining the number of lymphocytes undergoing Igh-cMyc chromosomal translocations during tumor latency.
RESULTS AND DISCUSSION
Delayed plasma cell tumor development in AID heterozygous mice
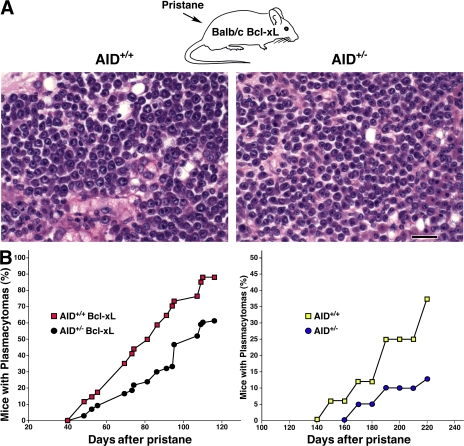
Pristane injection of BALB/c mice expressing Bcl-2 or Bcl-xL transgenes leads to the rapid induction of plasmacytomas carrying canonical Igh-cMyc translocations (13, 18). In the absence of AID, BALB/c-Bcl-xL mice display a reduced incidence of plasmacytomas carrying translocations that are nonreciprocal and do not involve S regions (13). To investigate whether AID gene dosage contributes to tumor susceptibility, we induced plasmacytomas in groups of BALB/c-Bcl-xL mice carrying one (AID+/−) or two copies of AID (AID+/+). After pristane injection, the presence of plasma cell tumors in peritoneal oil granulomas was diagnosed by the appearance of tumor cells in the ascites. We found that AID+/− mice developed tumors phenotypically indistinguishable from AID+/+ as judged by histological features, the expression of mature plasma cell markers, or tumor recovery in transplantation assays (Fig. 1 A and not depicted). However, there was a clear difference in tumor incidence between the two groups of mice: the median tumor latency in AID+/− animals (n = 96) was 102 d, whereas that of AID+/+ mice (n = 41) was 82 d (P = 0.01; Fig. 1 B, left). In the absence of Bcl-xL, plasmacytomagenesis was, as expected, markedly delayed in both groups of mice. Still, AID heterozygous mice showed decreased tumor incidence relative to AID wild-type counterparts (Fig. 1 B, right). These results thus reveal a straight correlation between AID gene dosage and the incidence of plasma cell tumor development.
Figure 1.
Happloinsufficiency in plasma cell tumor development in AID+/− mice. (A) Photomicrographs of plasma cell tumors arising in BALB/c-Bcl-xL AID+/+ and BALB/c-Bcl-xL AID+/− mice show no significant variations in plasma cell morphology. Bar, 20 μm. (B, left) Incidence of plasma cell tumors in the presence of Bcl-xL as determined by the histological appearance of foci, with each containing 50 or more atypical plasma cells (red squares, plasma cell tumors in 36 out of 41 AID+/+ mice injected with pristane; black circles, plasma cell tumors in 58 out of 96 AID+/− mice). (right) Plasmacytoma development in AID+/+ (n = 16) and AID+/− (n = 39) mice in the absence of Bcl-xL.
Reduced CSR in stimulated AID+/− B cells
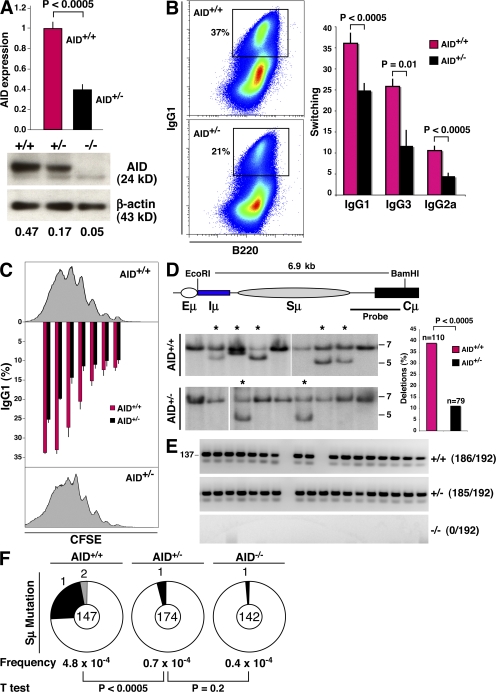
The results described in the previous paragraph imply that AID activity is compromised in AID+/− mice. To confirm this, we stimulated AID+/− and AID+/+ splenic B cells with LPS and IL-4, which induce AID transcription and CSR primarily to IgG1. Real-time PCR and Western blot analyses showed lower AID mRNA and protein in activated AID+/− lymphocytes compared with controls (Fig. 2 A). This reduction in AID expression levels was correlated with impaired switching, as indicated by a 40–50% decrease in surface IgG1 expression in heterozygous cells relative to wild-type (P < 0.0005; Fig. 2 B). B cell activation with LPS and α-δ-dextran or IFN-γ also showed a statistically significant decrease in γ3 and γ2a recombination between AID+/+ and AID+/− lymphocytes (P = 0.01 and P < 0.0005 for γ3 and γ2, respectively; n = 5; Fig. 2 B). CSR levels have been shown to be closely associated with cell division (19, 20). We examined cell proliferation in AID+/− and wild-type cultures by CFSE labeling, and IgG1 switching was simultaneously determined by flow cytometry. We found no obvious differences in cell division between the two groups of B cells (Fig. 2 C, histograms). However, γ1 switching in AID+/− lymphocytes lagged AID+/+ by approximately two cell cycles (Fig. 2 C, graph), thus confirming that CSR levels are compromised in AID+/− cells.
Figure 2.
Reduced AID activity in AID+/− B cells. (A) AID mRNA (graph, AID+/+ and AID+/−; n = 5) and protein (AID+/+, AID+/−, and AID−/−), as measured by real-time PCR and Western blotting. Error bars represent the mean ± SD. (B, left) IgG1 switching (pseudocolor plot) profiles in wild-type and AID+/− B cells activated ex vivo, as determined by flow cytometry. (right) Mean switching to IgG1, IgG3, and IgG2a. Values represent the mean ± SD (n = 5). Antibodies were B220-PercP-Cy5.5 and IgG1-PE. Dead cells were gated out using DAPI. (C) Flow cytometric measurement of cell proliferation and IgG1 switching on CFSE-labeled B cells activated for 96 h with LPS + IL-4. The graph represents percentages of γ1 switching at each cell division (n = 2). Error bars represent the mean ± SD. (D) The percentage of intra-switch deletions as determined by Southern blotting on hybridomas generated from AID+/+ and AID+/− lymphocytes activated for 96 h in the presence of LPS + IL-4. Intact Sμ domains result in a 6.9-kb band, whereas smaller bands (stars) represent intra-switch deletions. (E) Analysis of AID gene expression by single-cell RT-PCR (137 bp) from sorted LPS + IL-4–activated B cells. (F) Sμ mutation frequency in AID+/+, AID+/−, and AID−/− activated B cells. Pie chart segments are proportional to the number of sequences carrying an equal number of mutations (indicated on the periphery). The total number of sequences is portrayed in the middle of each chart, and mutation frequency (calculated as the total mutations per total base pairs sequenced) and p-values are shown below the charts.
Lymphocytes activated for CSR undergo frequent AID-dependent S deletions, predominantly at Sμ (21, 22). To measure the incidence of intra-Sμ deletions, we generated IgM+ hybridoma clones from AID+/+ and AID+/− B cells stimulated for 96 h in the presence of LPS and IL-4. Southern blotting showed a threefold reduction in intra-Sμ deletions in B cells carrying a single copy of Aicda, as only 9 out of 79 AID+/− clones (11%) displayed structural alterations in Sμ, whereas S deletions were present in 43 out of 110 AID+/+ clones analyzed (39%; P < 0.0005; Fig. 2 D).
Reduced inter- and intra-switch recombination in AID+/− cells could conceivably result from monoallelic expression of Aicda. Under this scenario, up to 50% of AID+/− B cells would transcribe the neomycin-deleted allele, leading to a complete absence of AID expression and activity in 50% of cultured cells. To directly evaluate this idea, we monitored AID gene transcription by single-cell RT-PCR (23). By this assay, only the untargeted Aicda allele is amplified using PCR primers specific for AID exons 2 and 3, which are deleted by neomycin insertion in the targeted allele (3). We detected AID in 185 out of 192 single AID+/− B cells assayed (Fig. 2 E), a result that clearly demonstrates Aicda transcription to be biallelic. We conclude that CSR defects in AID+/− cultures result from a reduction in overall AID expression levels.
AID deamination leads to nucleotide substitutions at S domains (24, 25), primarily as a result of DNA replication over uracils and error-prone repair activity (6). To measure S mutation frequency, we cloned and sequenced the 5′ end of Sμ from AID+/− and wild-type lymphocytes stimulated with LPS and IL-4 for 96 h. Sequencing analysis showed a striking reduction in Sμ mutations in heterozygous cells (P < 0.0005; Fig. 2 F). The calculated mutation frequency in AID+/− cells was in fact not significantly higher than the PCR error rate determined using AID−/− controls (P = 0.2; Fig. 2 F). Collectively, our data reveal that the frequency of S mutation and recombination in stimulated B cells is determined, at least in part, by AID gene dosage.
Reduced AID activity in AID+/− mice during the immune response
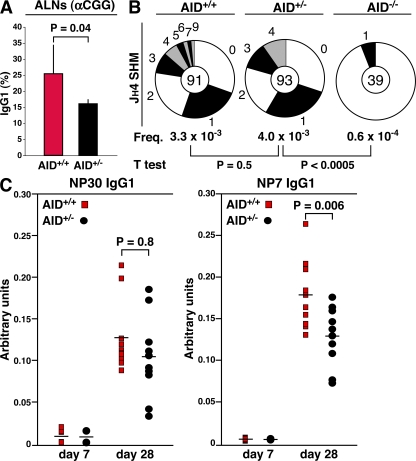
We next examined AID activity during the humoral immune response. AID+/− and control BALB/c littermates were immunized in the front footpads by subcutaneous injection of 50 μg of chicken γ globulin (CGG) in the presence of complete adjuvant. CSR levels were then monitored in germinal centers (GCs) from axillary and brachial lymph nodes 7 d after immunization. In agreement with the B cell culture results, we found fewer IgG1 B220+CD95high GC cells in AID+/− mice relative to controls (P = 0.04; Fig. 3 A). Notably, the disparity in CSR levels between the two strains of mice was not as striking as that seen in ex vivo cultures. This might be explained by the combined facts that switched cells accumulate over time as a function of the cell cycle (Fig. 2 C), and that B cell recruitment to and proliferation in GCs is extensive and asynchronized (26).
Figure 3.
Reduced AID activity in AID+/− mice during the immune response. (A) Measurement of IgG1 levels in GCs from AID+/+ and AID+/− mice. Values represent the mean ± SD (n = 5). (B) Somatic hypermutation as determined by sequencing the JH4 intron from AID+/+, AID+/−, and AID−/− GC cells. (C) ELISA analysis of serum from five AID+/+ and AID+/− mice immunized with NP-CGG. IgG1 antibodies displaying low affinity for NP were captured in this assay using NP30-BSA, whereas high affinity antibodies were monitored with NP7-BSA, as described in Results and discussion. Horizontal bars represent the mean.
To evaluate whether reduced AID expression levels affected V gene hypermutation, we next cloned and sequenced the 5′ end of the JH-Eμ intron (downstream of rearranged VHDJH4 genes) from sorted B220+CD95high GC cells. This analysis showed a comparable hypermutation frequency between the two groups of mice (P = 0.5). Notably, AID+/− GCs harbored fewer clones carrying multiple mutations (Fig. 3 B), suggesting a reduction in the accumulation of point mutations in heterozygous cells. To directly evaluate whether affinity maturation was affected as a function of AID expression, AID+/− and control mice were immunized intraperitoneally with hapten (4-hydroxy-3-nitrophenyl)acetyl (NP) conjugated to CGG (NP25-CGG) adsorbed to alum. The affinity of serum antibodies for NP was then monitored 7 and 28 d after immunization by ELISA. In this assay, low affinity antibodies were captured using a high density NP-substituted BSA (NP30-BSA), whereas high affinity antibodies were specifically monitored with NP7-BSA. Low affinity IgG1 serum levels were not different between the two strains of mice (P = 0.8; Fig. 3 C, left). Nonetheless, we found a statistically significant reduction of high affinity anti-NP antibodies in AID+/− mice compared with wild-type counterparts (P = 0.006; Fig. 3 C, right). Thus, it seems likely that under conditions of limited AID activity, fewer AID+/− B cell clones develop high affinity antibodies during the immune response. Based both on the CSR and SHM data, we conclude that the extent of AID enzymatic activity during the immune response is moderately affected in AID+/− mice.
Diminished chromosome 12 breaks and Igh-cMyc translocation–bearing cells in AID+/− mice
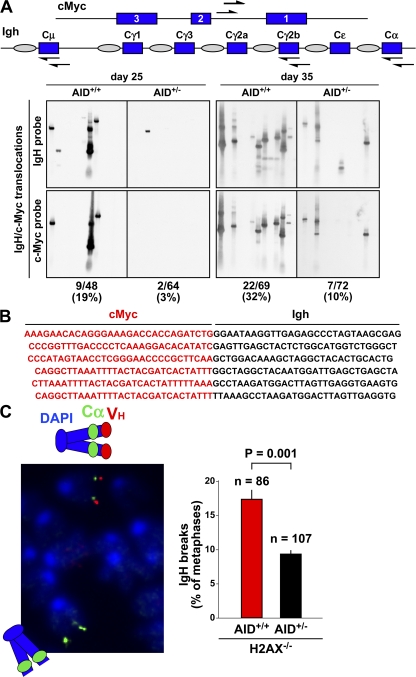
Plasmacytomagenesis in pristane-treated mice is a multistep process initiated by the deregulation of cMyc upon translocation to the Igh loci. Fully malignant monoclonal tumors are thought to arise from a polyclonal pool of premalignant B cells bearing cMyc translocations (27). We reasoned that delayed tumor development in AID+/− mice might result from a reduction in the number of clones carrying cMyc translocations. To investigate this, Bcl-xL transgenic AID+/+ and AID+/− mice were injected with pristane, and the presence of translocation-bearing cells before the onset of malignancy (approximately day 45; Fig. 1 B) was determined by long-distance PCR using nested primers specific for Sμ, Sγ2a, Sα, and cMyc intron 1 (Fig. 4 A, diagram). At days 25 and 35 after pristane treatment, canonical translocations were amplified from DNA samples representing 5 × 105 mononuclear cells isolated from Peyer's patches, mesenteric lymph nodes, or oil granuloma. DNA from three (day 35) or five (day 25) AID+/+ or AID+/− mice was analyzed. The specificity of the PCR was evaluated by Southern blotting using both Igh- and cMyc-specific probes, and unique T(12;15)-positive clones were distinguished by their size (from 1–4 kb; Fig. 4 A) and sequencing analysis (Fig. 4 B; and Table S1, available at http://www.jem.org/cgi/content/full/jem.20081007/DC1). At day 25, an analysis of 57 PCR samples from AID+/+ mice revealed nine distinct translocations (16%, or 1 T(12;15)/105 cells; Fig. 4 A). In contrast, only 2 out of 67 AID+/− samples (3%, or 0.2 T(12;15)/105 cells) showed bona fide Igh-cMyc translocations (P = 0.01; Fig. 4 A and Table S1). Similarly, 22 out of 69 (30%) AID+/+ samples showed translocations at day 35 after pristane treatment, whereas we detected only 7 translocations in 72 (10%) AID+/− samples assayed (2 vs. 0.6 T(12;15)/105 cells, respectively; P = 0.001; Fig. 4 A). In both groups of mice, the majority of the translocations were mapped to Sα (∼70%; Table S1), a feature that suggestively reflects the prevalence of IgA CSR in gut-associated lymphoid tissues (23). Thus, compared with wild-type controls, AID+/− mice display a substantial reduction in the number of translocation-bearing cells emerging upon tumor induction.
Figure 4.
Paucity of translocation-bearing clones and CSR-induced DNA breaks in AID+/− mice. (A) Igh-cMyc chromosomal translocations in BALB/c-Bcl-xL AID+/+ and BALB/c-Bcl-xL AID+/− mice (three mice per time point) were amplified by PCR 25 or 35 d after pristane injection using nested primers (arrows) specific for cMyc intron 1 and Igh Cμ, Cγ2b, and Cα constant domains. PCR products were analyzed by Southern blotting using cMyc- and Igh-specific probes. (B) PCR products appearing multiple times were sequenced and counted once if they represented a single clone. (C) Igh FISH in metaphase spreads from AID+/+H2AX−/− and AID+/−H2AX−/− B cells activated for CSR. (left) The loss of the red VH signal with an intact green Cα signal (micrograph) denotes a chromosome 12 break. (right) Total number of breaks observed for each strain (metaphases analyzed are indicated on top of each bar).
S recombination and Igh-cMyc translocations originate from DNA double-strand breaks targeting S domains in activated B cells (14, 28). To evaluate whether the frequency of S DNA breaks was affected in AID heterozygous mice, we bred the AID-deleted allele onto the H2AX−/− background, where a large number of unrepaired CSR lesions progress into chromosome breaks (14, 28, 29). These lesions, detected in metaphase spreads by fluorescence in situ hybridization (FISH) using VH- and Cα-specific probes, are good indicators of the relative frequency of S DNA breaks occurring in G1 (14, 28, 29). Analysis of metaphase spreads from AID+/+H2AX−/− and AID+/−H2AX−/− B cells stimulated with LPS and IL-4 for 96 h showed a direct correlation between the frequency of CSR breaks and AID gene dosage. Although 17.5% of all AID+/+H2AX−/− metaphases displayed split VH/Cα signals, only 9% of all AID+/−H2AX−/− metaphases showed evidence of chromosome 12 breaks (Fig. 4 C). We conclude that S domain DNA breaks are diminished in AID heterozygous B cells.
We have shown that the absence of a single copy of Aicda affects the rate of intrachromosomal recombination and interchromosomal translocations involving the Igh loci. This haploinsufficiency for AID is likely explained by the marked reduction in the frequency of formation and availability of S-DNA lesions in AID+/− cells. The strong inference is that AID protein content falls below a critical threshold in lymphocytes carrying a single copy of Aicda. Such a threshold might be imposed by mechanisms controlling nuclear AID (30, 31), which represents a minor fraction of total AID protein in activated B cells (32). Alternatively, error-free repair pathways, which have been shown to inhibit both CSR and SHM (17, 33), may compromise AID activity to a greater extent under conditions of limited AID transcription. Regardless of the mechanism at work, our findings demonstrate that AID expression levels directly determine the extent of AID-dependent DNA remodeling events, including chromosomal translocations. Thus, AID expression levels appear to determine the incidence of plasmacytomagenesis by specifying the number of cMyc translocation–bearing clones emerging during tumor induction. Overt transformation of such clones may subsequently rely on a mistargeted SHM, which as we have shown is also dependent on the amount of AID.
Our previous work revealed that noncanonical T(12;15) translocations can occur in the absence of AID (13). On the other hand, canonical translocations targeting S region DNA and cMyc intron 1 absolutely require AID enzymatic activity (12, 13). We now extend these results by showing that AID expression levels directly determine the rate of canonical translocations emerging during tumor induction in vivo. Ramiro et al. have similarly shown that cMyc translocations can be induced in an AID-dependent manner in vitro (28). In that study, translocations were rarely present under physiological AID levels in LPS- and IL-4–stimulated B cells, whereas AID overexpression dramatically increased their frequency (28). In like manner, Igh-cMyc translocations are produced to a disproportionately high degree in stimulated B lymphocytes deficient for microRNA 155 (34), which normally suppresses the steady state of Aicda mRNA and AID protein content (34, 35). We find it noteworthy that although the frequency of off-target translocations is highly sensitive to AID expression levels, in vivo on-target SHM and CSR are only mildly affected by AID overexpression (34) or underexpression (this report). It seems likely that AID enzymatic activity is differentially constrained at on and off targets. The unraveling of such regulatory mechanisms might help clarify how S DNA breaks promote physiological recombination or pathological translocations in the B cell compartment.
MATERIALS AND METHODS
Tumor induction.
SV40-Eμ-Bcl-xL and AID−/− (a gift from T. Honjo, Kyoto University, Kyoto, Japan) mice were backcrossed for 11 generations onto BALB/c mice in a conventional facility. Bcl-xL-AID+/+ and Bcl-xL-AID+/− mice were given 0.5 ml of pristane oil intraperitoneally at 8 wk of age, followed by a second injection of pristane 60 d later. BALB/c-Bcl-xL AID+/+ and AID+/− mice were intraperitoneally inoculated with 0.4 or 0.5 ml pristane (Aldrich Chemicals) at 2 mo of age. Using Wright-Giemsa–stained cytofuge preparations of ascites, plasma cell tumors were diagnosed by the presence of 10 or more tumor cells per field. All animal experiments were performed according to the NIH guidelines for laboratory animals and were approved by the Scientific Committee of the NIAMS and NCI Animal Facilities.
B cell culture.
To induce S recombination ex vivo, B lymphocytes were isolated from spleens by immunomagnetic depletion using CD43 MACS beads (Miltenyi Biotec). Purified cells were then activated in B cell media (RPMI 1640, 10% FCS, 1× antibiotic-antimycotic, 1% glutamine, 1× nonessential amino acids, 1% sodium pyruvate, 14.2 M 2-β-mercaptoethanol, and 10 mM Hepes) for 72 or 96 h in the presence of 25 μg/ml LPS (Escherichia coli 0111:B4; Sigma-Aldrich) and 5 ng/ml IL-4 (Invitrogen) to induce switching to IgG1, 2.5 ng/ml α-δ-dextran (Fina Biosolutions) for IgG3 CSR, or 2 ng/ml IFN-γ (PeproTech) for IgG2a switching. Cell proliferation was monitored by CFSE staining according to the manufacturer's instructions (Invitrogen).
Flow cytometry.
Unless otherwise stated, the following antibodies were obtained from BD Biosciences: B220-PercP-Cy5.5, Fab′2-IgM-Cy5 (Jackson ImmunoResearch Laboratories), IgG1-PE, IgG3-biotin, streptavidin-PE, and IgG2a-FITC. Apoptotic cells were gated out using TO-PRO-3 (Invitrogen) or DAPI (Sigma-Aldrich). Flow cytometers used were the FACSCalibur (BD Biosciences) and the Cyan (Dako), and a MoFlo instrument (Dako) was used for cell sorting. Flow cytometry data were analyzed with FlowJo software (Tree Star, Inc.).
qRT-PCR.
RNA was extracted with RNAqueous-Micro (Ambion) and treated with DNaseI. cDNA was synthesized, and the level of endogenous AID transcripts was monitored by real-time PCR with the following primers: PrimerBank-mAID5′ (5′-gccaccttcgcaacaagtct-3′) and PrimerBank-mAID3′ (5′-ccgggcacagtcatagcac-3′). cDNA was normalized based on Ku80 mRNA with the following primers: QRT-mKu80-5′ (5′-CTTCCTGGACGCCCTGATTG-3′) and QRT-mKu80-3′ (5′-GCTGCGGAGGTCAGTGAAC-3′).
Western blotting.
To detect AID protein, cultured B cells in the presence of LPS and IL-4 were lysed with NP-40 lysis buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 1 mM DTT, and protease inhibitor cocktail). 50 μg per well of lysates was separated using NuPAGE 4–12% Bis-Tris gels (Invitrogen). Gels were transferred to polyvinylidene fluoride membranes (Invitrogen) and analyzed by immunoblotting AID antisera (a gift from J. Stavnezer, University of Massachusetts, Worcester, MA) and anti–β-actin antibody (Millipore). For secondary antibodies, goat anti–mouse IgG (Millipore) and goat anti–rabbit IgG conjugated with horseradish peroxidase (HRP; Abcam) were used. Blots were visualized using the ECL-PLUS kit (GE Healthcare).
Sμ mutations.
Mutations at Sμ were determined by amplifying genomic DNA from LPS + IL-4–activated (96 h) B cells with primers Sμ(B) (5′-gtaaggagggacccaggctaag-3′) and Sμ(D) (5′-cagtccagtgtaggcagtaga-3′) at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s (product = 650 bp). The final product was ligated into ZeroBlunt vector (Invitrogen), and positives clones were sequenced with M13 primers.
FISH analysis.
At 72 h, cultured B cells were arrested at mitosis with 100 ng/ml colcemid (Roche), swollen in prewarmed 0.075 M KCl for 15 min at 37°C and fixed by methanol/acetic acid treatment, and dropped in microscope slides using a humidity chamber (Thermotron). Slides were denatured in 75°C 70% formamide for 7 min; hybridized with following probes overnight at 37°C; washed, dehydrated, and stained with DAPI; and mounted with Vectashield (Vector Laboratories). BAC probes containing the IgH Cα and Vh regions were labeled by digoxigenin and a biotin nick translation kit (Roche), and were detected by antidigoxigenin FITC antibody (Roche), and Alexa Fluor 568–conjugated streptavidin (Invitrogen). FISH images were captured using an epifluorescence microscope (DMRB Leitz; Leica) and a charge-coupled device camera (model C4742-95; Hamamatsu Photonics).
Hybridoma analysis.
Analysis of CD43− MACS-isolated B cells were stimulated with LPS + IL-4 for 96 h and fused to the SP2/0Ag-14 myeloma cell line. IgM-secreting clones, as determined by ELISA, were expanded in culture for further analysis. Genomic DNA was prepared and Southern blot analysis was performed using standard techniques.
Detection of Igh-cMyc translocations and plasma cell tumors.
Tissue samples were harvested 25 or 35 d after pristane injection. DNA samples from three (day 35) and five (day 25) mice were prepared as followed: Peyer's patches (three samples per mouse), mesenteric lymph nodes (two samples per mouse), and oil granuloma (two samples per mouse). Tissues were collected in separate tubes and DNA was isolated individually. The sensitivity of the translocation PCR was measured using serial dilution of positive control cells with wild-type mouse liver cells and calculated as >6.25 T(12;15) positive cells/105 negative cells. PCR products were separated on 1% agarose gels and denatured for 15 min in the presence of 0.4 M NaOH before being transferred to nylon membranes. The Southern blot probe was amplified and labeled by the ECL Direct Labeling and Detection System (GE Healthcare). Probe primer sequences were as follows: c-myc forward, 5′-gctagcgcagtgaggagaag-3′; c-myc reverse, 5′-gacgccactgcaccagaga-3′; Cμ forward, 5′-agccagtgtctcagaggga-3′; Cμ reverse, 5′-cttaaggcaggcagggcta-3′; Cγ2β forward, 5′-aagtcagaccgtcacctgca-3′; Cγ2β reverse, 5′-cagaggatcatgtccttgagtg-3′; Cα forward, 5′-tgctggctgatcttcagtctc-3′; and Cα reverse, 5′-ttgaacatgacctgcaggtagtc-3′.
ELISA.
Five AID+/+ and AID+/− mice were immunized intraperitoneally with 100 μg NP25-CGG precipitated in Alum (Thermo Fisher Scientific). Sera were collected before and at 7 or 28 d after immunization. Serum anti-NP levels were measured by standard ELISA. Plates were coated with 5 μg/ml NP30-BSA or NP7-BSA in PBS. Serum samples were diluted, and bound antibody was detected using HRP-conjugated anti–mouse IgG (Bethyl) followed by 3,3′,5,5′-tetramethylbenzidine treatment.
Online supplemental material.
Table S1 shows the translocation PCR/Southern blot results from AID+/+, AID+/−, and AID−/− mice carrying Bcl-xL. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081007/DC1.
Supplementary Material
Acknowledgments
We thank Janet Stavnezer for anti-AID antisera, Tasuku Honjo for AID−/− mice, Siegfried Janz and Beverly Mock for helpful comments, and Jim Simone for technical assistance.
This research was supported in part by the Intramural Research Program of NIAMS and NCI of the NIH.
The authors declare no competing financial interests.
References
- 1.Stavnezer, J., J.E. Guikema, and C.E. Schrader. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wuerffel, R., L. Wang, F. Grigera, J. Manis, E. Selsing, T. Perlot, F.W. Alt, M. Cogne, E. Pinaud, and A.L. Kenter. 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 27:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 4.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 5.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 6.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 7.Bardwell, P.D., C.J. Woo, K. Wei, Z. Li, A. Martin, S.Z. Sack, T. Parris, W. Edelmann, and M.D. Scharff. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5:224–229. [DOI] [PubMed] [Google Scholar]
- 8.Guikema, J.E., E.K. Linehan, D. Tsuchimoto, Y. Nakabeppu, P.R. Strauss, J. Stavnezer, and C.E. Schrader. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 204:3017–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson, E.D., W.J. Cummings, D.W. Bednarski, and N. Maizels. 2005. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol. Cell. 20:367–375. [DOI] [PubMed] [Google Scholar]
- 10.Yan, C.T., C. Boboila, E.K. Souza, S. Franco, T.R. Hickernell, M. Murphy, S. Gumaste, M. Geyer, A.A. Zarrin, J.P. Manis, et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482. [DOI] [PubMed] [Google Scholar]
- 11.Küppers, R. 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 5:251–262. [DOI] [PubMed] [Google Scholar]
- 12.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 13.Kovalchuk, A.L., W. duBois, E. Mushinski, N.E. McNeil, C. Hirt, C.F. Qi, Z. Li, S. Janz, T. Honjo, M. Muramatsu, et al. 2007. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J. Exp. Med. 204:2989–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco, S., M. Gostissa, S. Zha, D.B. Lombard, M.M. Murphy, A.A. Zarrin, C. Yan, S. Tepsuporn, J.C. Morales, M.M. Adams, et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualucci, L., G. Bhagat, M. Jankovic, M. Compagno, P. Smith, M. Muramatsu, T. Honjo, H.C. Morse III, M.C. Nussenzweig, and R. Dalla-Favera. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112. [DOI] [PubMed] [Google Scholar]
- 16.Okazaki, I.M., H. Hiai, N. Kakazu, S. Yamada, M. Muramatsu, K. Kinoshita, and T. Honjo. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, M., J.L. Duke, D.J. Richter, C.G. Vinuesa, C.C. Goodnow, S.H. Kleinstein, and D.G. Schatz. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. [DOI] [PubMed] [Google Scholar]
- 18.Silva, S., A.L. Kovalchuk, J.S. Kim, G. Klein, and S. Janz. 2003. BCL2 accelerates inflammation-induced BALB/c plasmacytomas and promotes novel tumors with coexisting T(12;15) and T(6;15) translocations. Cancer Res. 63:8656–8663. [PubMed] [Google Scholar]
- 19.Hasbold, J., L.M. Corcoran, D.M. Tarlinton, S.G. Tangye, and P.D. Hodgkin. 2004. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat. Immunol. 5:55–63. [DOI] [PubMed] [Google Scholar]
- 20.Rush, J.S., M. Liu, V.H. Odegard, S. Unniraman, and D.G. Schatz. 2005. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. USA. 102:13242–13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reina-San-Martin, B., S. Difilippantonio, L. Hanitsch, R.F. Masilamani, A. Nussenzweig, and M.C. Nussenzweig. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudley, D.D., J.P. Manis, A.A. Zarrin, L. Kaylor, M. Tian, and F.W. Alt. 2002. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA. 99:9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crouch, E.E., Z. Li, M. Takizawa, S. Fichtner-Feigl, P. Gourzi, C. Montano, L. Feigenbaum, P. Wilson, S. Janz, F.N. Papavasiliou, and R. Casellas. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen, S., R. Casellas, B. Reina-San-Martin, H.T. Chen, M.J. Difilippantonio, P.C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D.R. Pilch, et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaoka, H., M. Muramatsu, N. Yamamura, K. Kinoshita, and T. Honjo. 2002. Activation-induced deaminase (AID)–directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med. 195:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwickert, T.A., R.L. Lindquist, G. Shakhar, G. Livshits, D. Skokos, M.H. Kosco-Vilbois, M.L. Dustin, and M.C. Nussenzweig. 2007. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 446:83–87. [DOI] [PubMed] [Google Scholar]
- 27.Potter, M. 2003. Neoplastic development in plasma cells. Immunol. Rev. 194:177–195. [DOI] [PubMed] [Google Scholar]
- 28.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callen, E., M. Jankovic, S. Difilippantonio, J.A. Daniel, H.T. Chen, A. Celeste, M. Pellegrini, K. McBride, D. Wangsa, A.L. Bredemeyer, et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75. [DOI] [PubMed] [Google Scholar]
- 30.McBride, K.M., V. Barreto, A.R. Ramiro, P. Stavropoulos, and M.C. Nussenzweig. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brar, S.S., M. Watson, and M. Diaz. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401. [DOI] [PubMed] [Google Scholar]
- 32.Rada, C., J.M. Jarvis, and C. Milstein. 2002. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. USA. 99:7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, X., and J. Stavnezer. 2007. DNA polymerase β is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 204:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorsett, Y., K.M. McBride, M. Jankovic, A. Gazumyan, T. Thai, D.F. Robbiani, M. Di Virgilio, B. Reina San-Martin, G. Heidkamp, T.A. Schwickert, et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 28:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng, G., P. Hakimpour, P. Landgraf, A. Rice, T. Tusch, R. Casellas, and F.N. Papavasiliou. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.