Abstract
MicroRNAs (miRNAs) are implicated in the differentiation and function of many cell types. We provide genetic and in vivo evidence that the two RNaseIII enzymes, Drosha and Dicer, do indeed function in the same pathway. These have previously been shown to mediate the stepwise maturation of miRNAs (Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V.N. Kim. 2003. Nature. 425:415–419), and genetic ablation of either within the T cell compartment, or specifically within Foxp3+ regulatory T (T reg) cells, results in identical phenotypes. We found that miRNA biogenesis is indispensable for the function of T reg cells. Specific deletion of either Drosha or Dicer phenocopies mice lacking a functional Foxp3 gene or Foxp3+ cells, whereas deletion throughout the T cell compartment also results in spontaneous inflammatory disease, but later in life. Thus, miRNA-dependent regulation is critical for preventing spontaneous inflammation and autoimmunity.
Appropriate regulation of immune homeostasis is critical for ensuring adequate immunity toward harmful pathogens without uncontrolled inflammation that could lead to tissue damage. T reg cells have a critical role in homeostasis by suppressing the activation of autoreactive T cells. Either mutations in the gene encoding Foxp3, a transcription factor required for T reg cell differentiation, or ablation of T reg cells results in devastating autoimmune lymphoproliferative disease (1–4). Chronic inflammation and autoimmunity can also result from defective activation of apoptosis that is normally required for the contraction of effector T cells after an immune response (5–7). Dysregulated cytokine signaling can also lead to spontaneous T cell activation and inflammatory disease, independently of antigen stimulation (8–10).
A hallmark of T reg cells is the expression of the forkhead transcription factor Foxp3. “Natural” Foxp3-expressing T cells arise in the thymus from a subset of thymocytes selected on class II MHC (11). Expression of Foxp3 is also induced in naive CD4+ T cells in response to antigen stimulation in the presence of TGF-β. These inducible T reg cells have also been shown to have suppressive activities (12).
The decision of precursor cells to differentiate into phenotypically and functionally distinct lineages, such as occurs when immature CD4+8+ double-positive (DP) thymocytes differentiate into cytotoxic CD8+, helper CD4+, or regulatory Foxp3+ T cells, is ultimately governed by the regulation of gene expression in response to intrinsic or extrinsic signals. Gene expression is regulated at multiple levels, from chromatin organization to transcription to posttranscriptional controls. It is now clear that not only proteins but also small regulatory RNAs play critical roles at each of these levels of regulation.
MicroRNAs (miRNAs) are short (∼22 nt) noncoding RNAs, expressed from endogenous genes, that act on target protein–encoding mRNAs through the RNA-induced silencing complex, targeting them for translational repression or degradation (13). The biogenesis of miRNAs involves two processing steps that have been largely defined in cell-based and biochemical studies. Primary miRNA (pri-miRNA) transcripts are first cleaved by the nuclear “microprocessor” complex containing the RNaseIII enzyme Drosha and its double-stranded RNA (dsRNA) binding partner DGCR8/Pasha, leaving short hairpin pre-miRNAs (14, 15). These pre-miRNAs are then exported to the cytoplasm, where they are further processed by another RNaseIII enzyme, Dicer, and its dsRNA binding partner, TRBP, to liberate the mature miRNAs for loading into the RNA-induced silencing complex (16).
Genetic studies have clearly demonstrated a critical requirement for Dicer in vivo. Dicer deficiency in the nematode Caenorhabditis elegans causes sterility (17), whereas deficiency in mice results in early embryonic lethality (18). Dicer has also been shown to have critical functions in the differentiation of specific tissues, such as skeletal muscle (19), pancreatic islets (20), and B lymphocytes (21). The function of Dicer, however, is not limited to miRNA biogenesis. Dicer is also required for the generation of small inhibitory RNAs (siRNAs) derived from endogenous dsRNA transcripts or exogenous sources of dsRNAs, including viruses and experimental siRNAs. siRNAs derived from endogenous dsRNAs may also play important roles in gene regulation. siRNAs generated from pseudogene-derived dsRNAs may regulate complementary protein–coding transcripts, analogous to miRNAs, whereas siRNAs from retrotranspon-derived dsRNAs appear to be important for genome stability (22, 23). Thus, although the loss of miRNA-dependent regulation is implicated in the phenotypes caused by Dicer deficiency, it is not known whether they are a consequence solely of the loss of miRNAs or if the loss of other Dicer functions also contributes.
Unlike Dicer, Drosha is thought to be required for the biogenesis of miRNAs but not in the generation of other siRNAs. However, an early report suggested that this RNaseIII enzyme is required for processing of the preribosomal RNA (pre-rRNA) (24). To investigate specifically the function of miRNA-dependent regulation, we generated mice with a targeted conditional LoxP allele of the Drosha gene and compared these with mice with a conditional LoxP allele of Dicer. By specifically inactivating these genes in T lymphocytes, we found that Drosha is indeed required for the biogenesis of miRNAs but not for the expression of rRNA subunits. Both Drosha- and Dicer-dependent pathways were critical for the induction of Foxp3 and function of the T reg lineage. Consequently, the loss of Drosha throughout the T cell compartment resulted in spontaneous T cell activation, inflammatory disease, and premature lethality. The absence of either Drosha or Dicer specifically in Foxp3+ T reg cells was even more devastating, resulting in early onset lymphoproliferative disease, analogous to Foxp3 or T reg deficiency. Thus, we provide the first genetic and in vivo evidence that Drosha and Dicer function in the same pathway, and that this pathway, in the biogenesis of miRNAs, is critical for immune regulation and preventing lethal inflammatory disease.
RESULTS
Specific miRNA deficiency in mice with Drosha deficiency in the T cell compartment
The mouse Drosha gene (also known as Rnasen or Etohi2) consists of 35 exons spanning 110 kb on chromosome 15 (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). The full-length Drosha transcript was predicted to be 4.5 kb and to encode a 1,373-residue polypeptide (Fig. S1 B). However, an initial survey of expressed sequence tags that mapped to the Drosha locus suggested that multiple splice isoforms might exist. By using sequential primer pairs to amplify cDNA reverse transcribed from various mouse tissues, two alternately spliced isoforms were identified in addition to the full-length transcript. These were products of splicing between exons 2 and 7 and exons 4 and 7 of the primary transcript (Fig. S1, C and D). Although the full-length transcript was always the predominant species, both alternately spliced isoforms were predicted to be in frame. We therefore used a targeting strategy that would disrupt all three potential Drosha transcripts. By gene targeting in embryonic stem cells, a conditional Drosha allele (F) was generated by flanking exon 9 with LoxP sites (Fig. S2 A), which if deleted was predicted to cause a frame shift and the appearance of multiple stop codons in exon 11. Mice were generated from these cells, and subsequent recombination of the conditional allele in the germ line yielded the null allele (Δ; Fig. S2 B).
Heterozygous DroshaF/Δ mice, which harbor only one functional Drosha allele, were viable. These were bred with transgenic mice expressing cre under regulation of the CD4 promoter/enhancer/silencer (25). Mice were maintained as DroshaF/Δ heterozygotes to ensure efficient deletion whenever cre was expressed. CD4-cre efficiently recombined the conditional allele at the DNA and transcript levels at the DP thymocyte stage. Deletion of the conditional allele was similarly observed in mature peripheral T cells, indicating that there was no preferential selection of cells that had failed to undergo recombination at the Drosha locus (Fig. 1 A and Fig. S2 C). Expression of the full-length Drosha protein, which was the only isoform detected in wild-type lymphocytes by Western blotting, was markedly reduced in DP thymocytes and was completely absent from mature T cells in DroshaF/Δ CD4-cre mice. This resulted in the loss of pre-miRNAs and mature miRNAs specifically in T cells (Fig. 1 C). However, the expression of some mature miRNAs, such as miR-29a, was only partially down-regulated in the absence of Drosha, suggesting that different miRNA species may have different stabilities. Consistent with a function for the processing of pri-miRNA, an accumulation of pri–miR-150 transcript was detected in Drosha-deficient T cells (Fig. 1 D).
Figure 1.
Drosha deficiency abrogates miRNA but not rRNA processing. (A) CD4-cre efficiently deletes exon 9 from the conditional Drosha transcript from the DP thymocyte stage onwards. cDNA from the indicated populations was analyzed for expression of the Drosha exons 5/6 or exon 9/10 junction by quantitative RT-PCR. DN4, CD90+TCRβloCD4/8/44/25−; DP, CD4+8+; CD4SP, TCRβhiCD24lo4+8−; CD8SP, TCRβhiCD24lo4−8+. (B) Drosha protein expression in DP thymocytes, and peripheral TCRβ+ T cells and B220+ B cells from DroshaF/Δ CD4-cre and control DroshaF/+ CD4-cre mice. Molecular weights are shown. (C) Total RNA from DP thymocytes and peripheral T and B cells was resolved on a polyacrylamide gel and Northern blotted for expression of mature miRNAs. Also shown is the ethidium bromide gel for expression of small rRNA and transfer RNA species. (D) Total RNA was resolved on an agarose-formaldehyde gel and Northern blotted for expression of pri-miR-150. Also shown is the ethidium bromide gel for expression of the large rRNA subunits.
It was previously suggested that Drosha might be required for processing of the pre-rRNA transcript into the smaller ribosomal subunits (24). However, we found no difference in the level of 28S, 18S, 5.8S, and 5S rRNAs expressed in Drosha-deficient T cells compared with control cells (Fig. 1, C and D). Therefore, Drosha appears to be specific for the processing of pri-miRNA transcripts.
Drosha deficiency at the DP thymocyte stage results in T lymphopenia
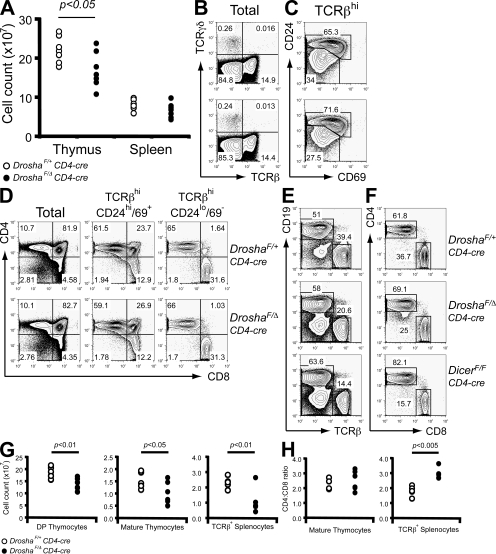
A reduction in the number of total thymocytes was observed in DroshaF/Δ CD4-cre mice at 6 wk of age (Fig. 2 A). An analysis of cell surface markers did not reveal a loss of any particular thymocyte subpopulation. TCRβhi versus TCRγδ+ populations appeared similar to littermate controls, as did expression of CD4 and CD8 at various stages of thymocyte differentiation (Fig. 2, B and D). One subtle difference observed was a reduction in the frequency of TCRβ+ thymocytes that had down-regulated CD24 and CD69 (Fig. 2 C), suggesting a partial perturbation in the ability of Drosha-deficient thymocytes to mature after selection.
Figure 2.
Drosha deficiency at the DP thymocyte stage partially perturbs thymocyte output. (A) Total thymocyte and splenocyte numbers in DroshaF/Δ CD4-cre and DroshaF/+ CD4-cre (control) mice. (B) TCRβ versus TCRγδ expression on total thymocytes. (C) CD24 and CD69 expression on TCRβhi gated thymocytes. (D) CD4 versus CD8 expression on total, postselected (TCRβhiCD24hi/69+) and mature (TCRβhiCD24lo/69−) thymocytes. (E and F) FACS analysis of populations in total splenocytes (E) and TCRβ+ splenocytes (F). Also shown is the effect of Dicer deficiency. Percentages of cells are shown in B–F. (G) Absolute CD4+8+ DP, mature thymocyte, and TCRβ+ splenocyte numbers. (H) CD4/CD8 ratio in the mature thymocyte and TCRβ+ splenocyte compartments.
Although thymocyte subpopulation profiles appeared normal, absolute numbers of DP and mature thymocytes were significantly reduced with Drosha deficiency (Fig. 2 G). This translated into a reduction in the number of mature T cells found in the periphery (Fig. 2, E and G). Furthermore, although the ratio of CD4 to CD8 mature thymocytes was similar between DroshaF/Δ CD4-cre mice and littermate controls, there was a significant reduction in the frequency of mature CD8+ T cells in the periphery (Fig. 2, F and H). Thus, T cell–specific Drosha deficiency results in T lymphopenia, particularly in the CD8+ compartment, and suggests a requirement for miRNAs in the homeostasis of mature T cells.
T cell–specific deficiency results in spontaneous inflammatory disease and premature mortality
To investigate the consequence of losing Drosha-dependent regulatory pathways in T cells, a cohort of DroshaF/Δ CD4-cre mice and various littermate controls was monitored in a long-term survival study. From around 4 mo of age, cachexia was observed in many of the DroshaF/Δ CD4-cre mice. These became moribund soon thereafter, at which point they were killed with littermate controls for analysis. By 6 mo, there was a 50% mortality rate in the DroshaF/Δ CD4-cre group (Fig. 3 A).
Figure 3.
T cell–specific Drosha deficiency results in spontaneous inflammatory disease and premature mortality. (A) Kaplan-Meyer survival plot of DroshaF/Δ CD4-cre and control mice (DroshaF/F, DroshaΔ/+ , or DroshaF/+ CD4-cre). (B) Aberrant activation of CD4+ T cells in lymphoid organs of moribund DroshaF/Δ CD4-cre mice. (C and D) Cytokine expression by CD4+ (C) and CD8+ (D) T cells. (E) Frequent IFN-γ and IL-17A coexpression by CD4+ T cells. (F) Granulocyte (Gr-1+) and γδT cells populations in lymphoid organs. Percentages of cells are shown in B–F. (G) H&E-stained sections of the lung (i and iii) and liver (ii, iv, and v). Bars, 100 μm.
A reduction in muscle mass and a complete loss of visceral adipose tissue was evident in all moribund DroshaF/Δ CD4-cre mice. A substantial increase in the frequency of activated (CD62Llo44hi) CD4+ T cells was observed in the spleen and lymph nodes (Fig. 3 B). Furthermore, very high frequencies of IFN-γ– and IL-17A–secreting cells were observed (Fig. 3 C). IL-17A expression was particularly prominent, as significant levels of IL-17A are normally only observed at mucosal surfaces, such as the small intestine, and in some autoimmune diseases, such as in experimental autoimmune encephalomyelitis (26, 27). Another prominent feature in moribund DroshaF/Δ CD4-cre mice was the presence of a high frequency of IFN-γ/IL-17A double-secreting cells (Fig. 3 E), which has also been observed in autoimmune models such as experimental autoimmune encephalomyelitis. A marginal increase in the frequency of IFN-γ+–or IL-17A–secreting CD4+ T cells could already be detected in DroshaF/Δ CD4-cre mice at 3 mo of age (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1), suggesting a gradual accumulation of inflammatory cells in aging mice. Consistent with aberrant IL-17A expression, there was a significant increase in the frequency of granulocytes in these lymphoid organs (Fig. 3 F). An increase in γδT cells was also observed. Aberrant activation and cytokine secretion by CD8+ T cells was usually not observed in these mice (Fig. 3, B and D).
The liver and lung appeared to be the major organs affected by inflammation (Fig. 3 G; and Table S1, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). Infiltrating cells also accumulated in the lung, primarily around blood vessels, resulting in epithelial thickening of the surrounding tissue. Infiltrating cells accumulated around most vessels in the liver, with additional foci of inflammatory cells evident throughout the sinusoids (Fig. 3 G, v). In addition, small foci of inflammatory cells were more prevalent in the small intestine and colon of DroshaF/Δ CD4-cre mice compared with littermate controls.
Despite aberrant CD4+ T cell activation, inflammatory cytokine secretion, and tissue inflammation, the spleen and lymph nodes of DroshaF/Δ CD4-cre mice were no larger than those in littermate controls (Fig. S4, A and B, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). In fact, the reduced thymic output and T lymphopenia remained a feature of these older mice (Fig. S4, C–E). Thus, T cell–specific Drosha deficiency results in lethal inflammatory disease associated with lymphopenia.
Drosha-dependent pathways are required for the induced Foxp3 expression and suppressive activity of T reg cells
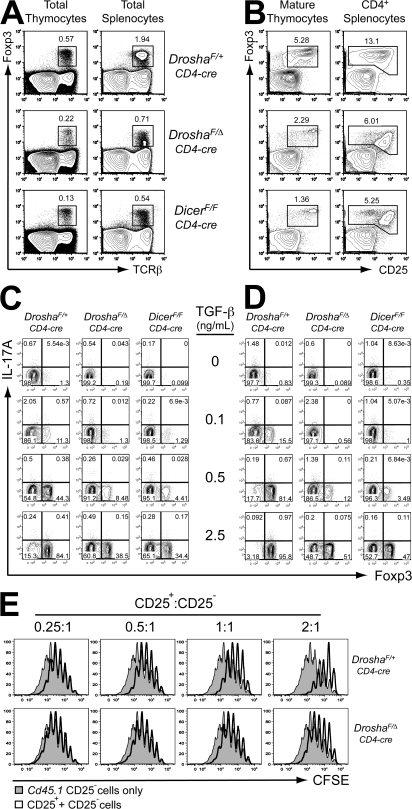
It was previously suggested that Foxp3 contributes to the miRNA signature in T reg cells and that miRNA pathways contribute to the regulation of Foxp3 expression (28). Indeed, a reduction in the frequency of Foxp3+ thymocytes and mature peripheral T reg cells was observed in both DroshaF/Δ CD4-cre and DicerF/F CD4-cre conditional-deficient mice (Fig. 4 A). This reduction was not simply concurrent with the general T lymphopenia, because it was still evident when examining only the CD4+ compartment (Fig. 4 B). With both Drosha and Dicer deficiency, the reduction in the T reg cell population was largely caused by a loss of the Foxp3+CD25lo cells.
Figure 4.
Drosha- and Dicer-dependent pathways are required for efficient induction of Foxp3 expression and function of T reg cells. (A) Reduced TCRβ+Foxp3+ T reg cell populations in the thymus and spleen of DroshaF/Δ CD4-cre and DicerF/F CD4-cre mice. (B) Foxp3 expression in peripheral T reg cell populations in the absence of Drosha or Dicer. (C and D) Drosha and Dicer are required for efficient differentiation of induced T reg cells. Naive CD62Lhi44lo25− CD4+ T cells from DroshaF/Δ CD4-cre, DicerF/F CD4-cre, and control DroshaF/+ CD4-cre mice were activated in vitro with anti-CD3/28 antibodies plus 10 U/ml IL-2, and TGF-β (C) or TGF-β plus 10 nM RA (D) for 3 d. The cells were restimulated with PMA/ionomycin plus GolgiStop for 3 h, and Foxp3 and IL-17A expression was analyzed by intracellular FACS. Percentages of cells are shown in A–D. (E) FACS-purified naive CD4+ cells from Cd45.1 mice were loaded with CFSE and mixed with purified CD4+25+ cells from DroshaF/Δ CD4-cre or DroshaF/+ CD4-cre (Cd45.2) mice at the indicated ratios. These were then incubated with inactivated splenocytes and anti-CD3 antibody. After 4 d, the CD45.1+ cells were analyzed for CFSE dilution.
Foxp3 expression can also be induced in naive CD4+ T cells upon antigen stimulation in the presence of TGF-β, and this expression is enhanced and stabilized by retinoic acid (29, 30). To examine if Drosha- and Dicer-dependent pathways may also be required for inducible T reg cells, naive CD4+ T cells were purified from DroshaF/Δ CD4-cre and DicerF/F CD4-cre mice and were activated in vitro in the presence of TGF-β (Fig. 4 C) or TGF-β and retinoic acid (RA; Fig. 4 D). Induction of Foxp3 was markedly blunted in both Drosha- and Dicer-deficient cells, even in the presence of RA. The magnitude of Foxp3 induction was similar between Drosha- and Dicer-deficient cells. The defect in Drosha- and Dicer-deficient cells was especially noticeable at lower concentrations of TGF-β. This defect in the ability of naive CD4+ T cells from DroshaF/Δ CD4-cre mice to up-regulate Foxp3 expression could be caused by generation of abnormal T cells that had been aberrantly selected in the absence of Drosha. To examine the requirement of Drosha-dependent pathways specifically during inducible T reg cell differentiation, naive CD4+ T cells were purified from DroshaF/Δ Rosa26CreER/+ mice, in which Cre only deletes upon the addition of tamoxifen (Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). Foxp3 induction by TGF-β (Fig. S5 B) or TGF-β and RA (Fig. S5 C) was still blunted when the conditional Drosha allele was deleted in vitro by tamoxifen addition, indicating that the observed defect was not caused by abnormal thymic selection.
Although the induction of Foxp3 expression was dramatically perturbed in DroshaF/Δ CD4-cre mice, Foxp3+CD25hi cells were still present. To examine if these cells were functional, CD25+CD4+ T cells were purified from DroshaF/Δ CD4-cre and control mice, and their ability to suppress the in vitro proliferation of CD25−CD4+ T cells in response to APCs and anti-CD3 antibody stimulation was examined (Fig. 4 E). CD25+CD4+ T cells from DroshaF/Δ CD4-cre mice could partially suppress the proliferation of CD25−CD4+ T cells at high CD25+/CD25− ratios. However, this activity was substantially reduced in comparison with that of CD25+CD4+ T cells from control DroshaF/+ CD4-cre mice. Thus, not only is there a reduction in T reg cell numbers and Foxp3 induction in DroshaF/Δ CD4-cre mice, but the remaining T reg cells have reduced suppressive activity, at least in vitro. This suggests that a defect in the T reg cell compartment may be the cause of or a major contributor to the spontaneous inflammatory disease in DroshaF/Δ CD4-cre mice.
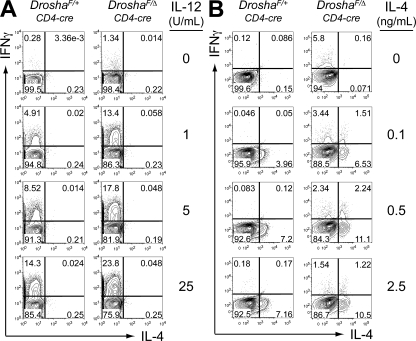
Drosha deficiency does not prevent the differentiation of Th1 or Th2 phenotype cells
It is possible that Drosha-dependent pathways may be specifically required for the differentiation and function of T reg cells. It is also possible that the differentiation of all T cell subsets may be affected by Drosha deficiency. The finding that older DroshaF/Δ CD4-cre mice display very high frequencies of proinflammatory cytokine-secreting cells suggests that the differentiation of other Th cell subsets may not be dependent on Drosha. To examine this further, naive CD4+ T cells were purified from DroshaF/Δ CD4-cre and control mice and were activated in vitro under Th1 (Fig. 5 A) or Th2 (Fig. 5 B) polarizing conditions. Drosha-deficient naive CD4+ T cells were still capable of differentiating into either IFN-γ–secreting Th1 cells or IL-4–secreting Th2 cells. In fact, Drosha-deficient CD4+ T cells consistently expressed more IFN-γ than controls under Th1 polarizing conditions. Again, this was not caused by defective thymocyte differentiation in DroshaF/Δ CD4-cre mice, because naive CD4+ T cells from DroshaF/Δ Rosa26CreER/+ mice also released more IFN-γ when the conditional Drosha allele was deleted in vitro upon the addition of tamoxifen (Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). CD8+ T cell activation and IFN-γ secretion did not appear to contribute to the disease in old DroshaF/Δ CD4-cre mice, and CD8+ T cells from these mice were capable of secreting IFN-γ after activation in vitro (Fig. S7).
Figure 5.
Drosha deficiency does not prevent polarization of naive CD4+ T cells into Th1 or Th2 cells. Naive CD62Lhi44lo25− CD4+ T cells from DroshaF/Δ CD4-cre and control DroshaF/+ CD4-cre mice were activated in vitro with anti-CD3/28 antibodies and 10 U/ml IL-2, plus (A) IL-12 and 1 μg/ml anti–IL-4 antibody or (B) IL-4 and 1 μg/ml anti-IFN-γ antibody for 4 d. The anti-CD3/28 antibodies were removed from the culture for the last 24 h, after which the cells were restimulated with PMA/ionomycin for 3 h in the presence of GolgiStop and analyzed for intracellular IFN-γ and IL-4 expression by FACS. Percentages of cells are shown.
Thus, it appears that the defective T reg cell differentiation and function, together with the normal or enhanced differentiation of conventional CD4+ T cells into proinflammatory subsets, may be the mechanism underlying the spontaneous inflammatory disease and premature mortality in DroshaF/Δ CD4-cre mice.
Drosha- and Dicer-dependent pathways are indispensable within the T reg cell compartment
To determine if defective T reg cells enhanced Th1 cell differentiation, or if both are required for spontaneous inflammatory disease in DroshaF/Δ CD4-cre mice, we examined the effect of inactivating the Drosha gene specifically in T reg cells. Mice with IRES-Cre knocked into the X-linked Foxp3 gene were used for T reg cell–specific deletion (31). Both female mice homozygous for and male mice hemizygous for the Cre insertion were examined, and alleles for both are designated as Foxp3Cre/Cre(Y). Both DroshaF/Δ Foxp3Cre/Cre(Y) and DicerF/F Foxp3Cre/Cre(Y) mice became sick at 2–3 wk of age, with a 100% mortality rate (Fig. 6 A). Sick DroshaF/Δ Foxp3Cre/Cre(Y) mice displayed massive lymphadenopathy and, to a lesser extent, splenomegaly (Fig. 6, B and C). In moribund mice, all leukocyte populations appeared to have expanded equally, except for a consistent increase in the frequency of CD8+ T cells (Fig. 6 D). Increases in the frequencies of CD62Llo44hi CD4+ and CD8+ T cells were observed, suggesting aberrant activation (Fig. 6 E). Hyper–IFN-γ secretion by both CD4+ and CD8+ T cells, and hyper–IL-4 secretion by CD4+ T cells were also observed in these mice (Fig. 6, F and G). Large areas of the liver and lung were infiltrated by inflammatory cells in moribund DroshaF/Δ Foxp3Cre/Cre(Y) mice (Fig. 6 J; and Table S2, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). Cuffing around blood vessels was also observed in other tissues, including the pancreas, kidney, and skeletal muscle. This phenotype is identical to mice lacking Foxp3 or Foxp3-expressing cells (2, 3). This phenotype was also recapitulated in DicerF/F Foxp3Cre/Cre(Y) mice (unpublished data).
Figure 6.
T reg cell–specific Drosha or Dicer deficiency recapitulates the scurfy phenotype. (A) Kaplan-Meyer survival plot of DroshaF/Δ Foxp3Cre/Cre(Y), DicerF/F Foxp3Cre/Cre(Y), and control mice (DroshaF/+ Foxp3Cre/Cre(Y), DroshaF/Δ Foxp3Cre/+, DroshaF/+ Foxp3Cre/+, DicerF/+ Foxp3Cre/Cre(Y), and DicerF/+ Foxp3Cre/+). (B) Massive lymphadenopathy in a female DroshaF/Δ Foxp3Cre/Cre mouse. Shown are the cervical lymph nodes. (C) Cell counts of lymphoid organs from moribund DroshaF/Δ Foxp3Cre/Cre or DroshaF/Δ Foxp3Cre/Y mice and littermate controls. Data represent the mean ± SD of four mice at 2.5–3 wk of age. (D) Only marginal changes in the proportions of B (CD19+), γδT (TCRγδ+), myeloid (CD11b+), and T cells (TCRβ+) in the spleen and lymph nodes of DroshaF/Δ Foxp3Cre/Cre mice. (E) Aberrant T cell activation in the spleen and lymph nodes of moribund DroshaF/Δ Foxp3Cre/Cre mice. (F and G) Cytokine expression by CD8+ (F) and CD4+ (G) T cells from DroshaF/Δ Foxp3Cre/Cre mice. (H and I) Foxp3 expression in total splenocytes (H) and CD4+ T cells (I). Percentages of cells are shown in D–I. (J) H&E-stained sections of the lung (i and iii) and liver (ii and iv). Designation of Foxp3Cre/Cre(Y) alleles indicates that both males and females were included in the experiments. Bars, 100 μm.
This early lymphoproliferative phenotype was not caused by a loss of T reg cells but rather by loss of function, because Foxp3+ cells could still be detected in these mice (Fig. 6, H and I). In fact, the T reg cell phenotype in these mice was similar to that in DroshaF/Δ CD4-cre mice. There was a reduction of the Foxp3+CD25lo population, but Foxp3+CD25hi cells were present.
It has previously been suggested that Foxp3+ T reg cells express a high frequency of high-affinity self-reactive TCRs (32). This hypothesis remains controversial, as a more recent analysis found that TCRs derived from T reg and non–T reg cells recognize foreign peptide–MHC complex with similar frequencies (33). Nevertheless, if a subset of Foxp3+ cells does express self-reactive TCRs, Drosha deficiency potentially may not only abrogate suppressive activity but also allow for activation through such TCRs. Thus, a Foxp3+ population itself may be responsible for this inflammatory phenotype in DroshaF/Δ Foxp3Cre/Cre(Y) mice. However, female DroshaF/Δ Foxp3Cre/+ heterozygous mice, in which only half of the Foxp3+ cells express cre because of random X chromosome inactivation, do not develop inflammatory disease and remain healthy. This suggests that the loss of Drosha-dependent regulatory pathways simply abrogates the function of T reg cells, and in heterozygous DroshaF/Δ Foxp3Cre/+ mice, the remaining 50% of T reg cells that express Drosha are sufficient to prevent this early onset disease. Furthermore, Foxp3+ T cells from either moribund DroshaF/Δ CD4-cre or DroshaF/Δ Foxp3Cre/Cre(Y) mice did not coexpress IFN-γ or IL-17 (Fig. S8, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). The finding that both T reg cell–specific Drosha- and Dicer-deficient mice display a phenotype identical to that of Foxp3-deficient mice demonstrates an absolute requirement for miRNA pathways in the function of T reg cells.
DISCUSSION
In this study, we have shown identical phenotypes when either the Drosha or Dicer gene was inactivated in TCRαβ T cells and in Foxp3+ T reg cells. These results confirm that these two RNaseIII enzymes function in the same biological pathway in miRNA biogenesis. However, it has previously been shown that Dicer also functions in the biogenesis of other siRNAs, in addition to miRNAs (22, 23), and Drosha was suggested to have a role in rRNA maturation (24). Our results indicate that there is no defect in rRNA production in the absence of Drosha, and they are consistent with loss of miRNAs as the primary cause underlying the phenotypes observed. Although it is quite possible that Dicer-deficient cells will also exhibit other more subtle phenotypes, such as the derepression of retrotransposons (22, 23), it appears that it is miRNAs, rather than other Dicer-dependent small RNAs, that are critical for most cells.
Ablation of the miRNA biogenesis pathway late in thymocyte differentiation, either by deleting Drosha or Dicer, did not prevent the development or maintenance of a peripheral mature T cell pool. Similarly, deletion of Dicer in terminally differentiated pancreatic β cells (20) or in dopaminoceptive postmitotic neurons (34) did not result in the loss of these cells. Instead, functional defects were manifest in these cells. In the case of the T cell compartment, a functional defect was particularly prominent in the T reg lineage. Either Drosha or Dicer deficiency led to defective Foxp3 induction and suppressive activity in these cells. Loss of miRNA biogenesis in T reg cells had a devastating effect on the survival of mice when the rest of the immune system was miRNA competent. Even though Foxp3+ cells were still present in DroshaF/Δ Foxp3Cre/Cre(Y) and DicerF/F Foxp3Cre/Cre(Y) mice, the animals succumbed to autoimmune lymphoproliferative disease indistinguishable from that in mice that lack Foxp3+ cells or a functional Foxp3 gene. Even in DroshaF/Δ CD4-cre mice, in which all T cells lacked the ability to generate mature miRNAs, spontaneous lethal inflammatory disease still ensued, albeit much later in life. This indicates that miRNAs are indispensable for the function of T reg cells. This is unlike what has been described for some cell types, such as postmitotic Purkinje neurons, in which the loss of Dicer activity results in cell death (35). Why the loss of miRNA-dependent regulation in some cell types results in cell death instead of functional perturbations is unclear. It may be that neurodegeneration is secondary to the loss of function. Alternatively, components of the apoptotic machinery may be subject to miRNA-dependent regulation (21).
How might miRNAs enable a T reg cell to be immunosuppressive? T reg cells have been postulated to act via a myriad of mechanisms. These include secretion of immunomodulatory cytokines, such as TGF-β and IL-10, modulation of co-stimulatory molecules on antigen-presenting cells, cytotoxic T lymphocyte antigen 4–induced tryptophan catabolism and direct cytotoxicity (36). Both in vitro and in vivo studies have provided compelling evidence that all of these and other mechanisms contribute to T reg cell–mediated immunosuppresion. However, abrogation of any one mechanism does not significantly impair the function of these cells. Even the ablation of IL-10 production by T reg cells results in only mild inflammation at mucosal surfaces (31). It is likely that T reg cells use multiple mechanisms to maintain immune homeostasis. Ablation of miRNA biogenesis is the first example that completely phenocopies the loss of Foxp3+ cells. This suggests that miRNAs may concurrently modulate multiple pathways in T reg cells, and the loss of this modulation may in effect abrogate multiple immunosuppressive mechanisms used by these cells. In addition to conferring some of the immunosuppressive properties of T reg cells (37), Foxp3 itself has been shown to be responsible for a significant component of the miRNA signature in these cells (28). Because miRNA pathways are also required for inducing and maintaining Foxp3 expression, this suggests that there is a positive feedback loop between Foxp3 and miRNAs that is critical for the function of T reg cells. Interestingly, like many other markers of T reg cells such as CD25, cytotoxic T lymphocyte antigen 4, and glucocorticoid-induced TNFR, a T reg cell miRNA signature is also induced in naive CD4+ T cells after TCR activation (28).
Although spontaneous inflammatory disease appeared to be related to a loss of T reg cell function in both DroshaF/Δ Foxp3Cre/Cre(Y) and DroshaF/Δ CD4-cre mice, only in the former was there massive lymphoproliferative disease, whereas lymphopenia occurred in the latter. The striking difference between the lifespans of DroshaF/Δ Foxp3Cre/Cre(Y) and DroshaF/Δ CD4-cre mice suggests that different mechanisms may be involved in the two diseases. It is clear that Drosha-dependent pathways are required for T reg cell function. However, the finding that the conventional T cells in DroshaF/Δ CD4-cre mice did not proliferate inappropriately to cause lymphadenopathy and early onset disease like the Drosha-sufficient conventional T cells in DroshaF/Δ Foxp3Cre/Cre(Y) mice suggests that Drosha-deficient T cells are also defective in some way, even though they are able to polarize into inflammatory effector cells both in vitro and in vivo. One possible defect may be the proliferative potential of Drosha-deficient T cells, as suggested by the lymphopenia rather than lymphoproliferation in Drosha CD4-cre mice. Indeed, both Drosha-deficient CD4+ and CD8+ T cells displayed reduced proliferation in vitro in response to anti-CD3 stimulation, but this was overcome when anti-CD28 co-stimulation was provided (Fig. S9 A, available at http://www.jem.org/cgi/content/full/jem.20081219/DC1). Furthermore, these cells also proliferated marginally slower than wild-type cells when adoptively cotransferred into the same lymphopenic Rag-deficient hosts (Fig. S9 B).
miRNAs may be involved in regulating TCR signaling and/or proliferation in conventional T cells. One miRNA, miR-181a, has previously been shown to modulate TCR sensitivity (38). Increasing the levels of miR-181a in T cells was found to lower the threshold of signaling. Thus, loss of miR-181a may render T cells less sensitive to TCR engagement. In DroshaF/Δ Foxp3Cre/Cre(Y) and DicerF/F Foxp3Cre/Cre(Y) mice, there was a dramatic expansion of all leukocytes, as has been previously shown in mice with mutations in Foxp3 or lacking Foxp3+ cells 2–4. However, in moribund DroshaF/Δ CD4-cre mice, there were only increases in Gr-1+ and γδT cells without lymphoproliferation, despite only T cells being defective for miRNA biogenesis. This suggests that the generalized lymphoproliferation and lymphadenopathy in DroshaF/Δ Foxp3Cre/Cre(Y), DicerF/F Foxp3Cre/Cre(Y), or Foxp3-deficient mice is entirely secondary to the activation of T cells.
Another difference was the appearance of IFN-γ+– and IL-17A+– but not IL-4–producing CD4+ T cells in moribund DroshaF/Δ CD4-cre mice, whereas IFN-γ– and IL-4–producing cells were dominant in DroshaF/Δ Foxp3Cre/Cre(Y) mice. The differentiation of IL-17A+ cells in DroshaF/Δ CD4-cre mice may be related to the inability of Drosha-deficient conventional T cells to rapidly proliferate and up-regulate IFN-γ and IL-4 in the perinatal period, as occurs in DroshaF/Δ Foxp3Cre/Cre(Y) mice. The massive expansion of Th1 and Th2 cells in the latter case may inhibit the differentiation of Th17 cells.
This study has clearly demonstrated a critical requirement for miRNA-dependent regulation in the prevention of spontaneous inflammation/autoimmunity. Might polymorphisms or mutations in miRNA machinery components or specific miRNA genes contribute to inflammatory or autoimmune disease in humans? Although mutations in this pathway have not yet been identified in humans with autoimmune disease, it has been shown that miR-17-92 overexpression in mice leads to spontaneous autoimmunity (39). A link to human disease, however, has been made for Drosha. Expression of Drosha, but not Dicer or DGCR8, is up-regulated in cervical squamous cell carcinomas and esophageal tumors (40, 41). Furthermore, the oncogenic ALL-1 fusion proteins have been shown to recruit Drosha to specific miRNA loci in leukemic cells lines (42), which may underlie the overexpression of certain miRNAs in leukemia. There is now a significant body of evidence correlating mutations or misexpression of miRNAs with numerous human cancers (43). Thus, continued efforts to gain a better understanding of miRNA biogenesis and function will undoubtedly provide significant insight into human disease.
MATERIALS AND METHODS
Mice.
A targeting construct containing exon 9 of Drosha flanked by LoxP sites and a LopP-flanked neomycin selection cassette (Neo) was assembled from genomic fragments subcloned from the bacterial artificial chromosome clone RP23-354M9. A LoxP-exon9-LoxP-Neo-LoxP configuration was used such that cre-mediated recombination between the 5′ and middle LoxP sites would yield a conditional allele, whereas recombination between the 5′ and 3′ LoxP sites would yield a null allele. EcoRI and SacI sites were inserted at the 5′ and 3′ LoxP sites, respectively, to allow for screening by Southern blotting. The construct was targeted into E14 129Sv-derived embryonic stem cells, from which mice were derived. These were bred with EIIA-cre mice, which transiently express cre in the early embryo, to obtain progeny harboring either the conditional or the null allele, and were then backcrossed onto the C57BL/6 genetic background for four more generations.
DicerF/F (provided by M. McManus, University of California, San Francisco, San Francisco, CA) (44), CD4-cre (25), and Foxp3Cre (31) mice have previously been described. EIIA-cre, Rosa26CreER, and Cd45.1 mice were purchased from Jackson ImmunoResearch Laboratories, and Rag2−/− mice were purchased from Taconic. The mice were housed in specific pathogen-free conditions at the animal facility of the Skirball Institute. All animal experiments were performed in accordance with approved protocols for the New York University Institutional Animal Care and Usage Committee.
Organ preparation and cell purifications.
Lymphocytes were prepared from thymi, spleens, and lymph nodes by grinding the organs through a 100-μm mesh. Red blood cells were removed from splenic cell suspensions by lysis with ACK buffer (BioWhittaker). Lymphoid populations were enriched by MACS magnetic bead purification (Miltenyi Biotec) and sorted to high purity on a FACSAria (BD Biosciences). Organs for histological analyses were fixed in 10% buffered formalin, dehydrated, and paraffin embedded before sectioning and staining with hematoxylin and eosin (H&E).
Southern and Northern blotting.
Genomic DNA was prepared from cells by standard techniques and digested overnight with restriction enzymes. DNA fragments were resolved on a 0.7% agarose gel before transfer onto a membrane (Hybond-XL; GE Healthcare), and were probed with radiolabeled DNA probes.
Total RNA was purified using TRIzol (Invitrogen) according to the manufacturer's protocol, with one modification: RNAs were precipitated from the aqueous phase using 2× volumes of ethanol at −80°C for 1 h before centrifugation. Detection of small RNA species was performed by resolving 10 μg of total RNA on a 15% polyacrylamide gel and Northern blotting with radiolabeled oligonucleotide probes, as previously described (45). For the detection of long RNA species, 20 μg of total RNA was resolved on a 1% formaldehyde agarose gel before Northern blotting with a radiolabeled oligonucleotide probe. Northern blotting was performed with the following probes: 5′-TAACCGATTTCAGATGGTGCTA-3′ (miR-29a), 5′-AGTAGTGCTTTCTACTTTATG-3′ (miR-142-5p), 5′-TCCATAAAGTAGGAAACACTACA-3′ (miR-142-3p), and 5′-CACTGGTACAAGGGTTGGGAGA-3′ (miR-150).
RT-PCRs.
First-strand RT was performed on total RNA using Superscript II (Invitrogen). The following forward primers were used to detect specific exon boundaries in the Drosha cDNA: 5′-GGCTGGGCTTTGATGGCGGCGACCCCGGGA-3′ (exon 1 forward), 5′-AACCTACATCGTGAGCGGAAGATGTAGCCT-3′ (exon 3 forward), 5′-GACCCCAAAACCTGCGACTTCTTCATC-3′ (exon 4), 5′-GCAGATCACCGTCTCTAGAAA-3′ (exon 5), 5′-GCACACCAGAGTTGCCTGGGGAGATGATTA-3′ (exon 6), and 5′-AAGCAGGCTCCGTGATTTGTATGAC-3′ (exon 8), with the reverse primer 5′-CTCTTCTCCGGGATAAATGCTGTGG-3′ (exon 10).
The following primer pairs were used to detect Drosha exon boundaries by quantitative PCR: 5′-TGCAAGGCAATACGTGTCAT-3′ and 5′-GGCCTGAGAGTTCGAGTAGG for exon 3/4; and 5′-GACGACGACAGCACCTGTT-3′ and 5′-GATAAATGCTGTGGCGGATT-3′ for exon 9/10. 5′-CACAGCTTCTTTGCAGCTCCTT-3′ and 5′-CGTCATCCATGGCGAACTG-3′ were used to amplify β-actin as a control.
Western blotting.
Cells were lysed in 50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, and protease inhibitors, and the insoluble components were removed by centrifugation. The equivalent of 2 × 106 cells was resolved on a 7% polyacrylamide gel and immunoblotted with a rabbit polyclonal antibody against the N terminus of Drosha (Millipore) or a mouse monoclonal antibody against β-tubulin (Invitrogen).
FACS analyses.
Analyses were performed on an LSRII (BD Biosciences). All antibodies were purchased from eBioscience or BD Biosciences. For the detection of intracellular cytokine expression, total cells were restimulated in vitro with 50 ng/ml PMA and 1 μM ionomycin for 3 h in the presence of GolgiStop (BD Biosciences). After staining for cell surface markers, the cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) and stained for cytokine expression. Detection of intranuclear Foxp3 expression was performed by fixation and permeabilization with Foxp3 fixation/permeabilization buffer (eBioscience) before staining for Foxp3.
In vitro T cell culture.
FACS-purified T cells were activated in vitro with 5 μg/ml of plate-bound anti-CD3 and 1 μg/ml of soluble anti-CD28 antibodies in RPMI supplemented with 10% FCS, 5 mM β-mercaptoethanol, and antibiotics. Cells were plated at a density of 5 × 105/ml at day 0 of culture. Various cytokines and neutralizing antibodies were added at day 0 or 1 of culture. All cytokines and antibodies were purchased from PeproTech or eBioscience.
For in vitro suppression assays, naive (CD62Lhi44lo25−) CD4+ T cells (effectors) were FACS purified from Cd45.1 mice and loaded with 5 μM CFSE (Invitrogen), and mixed with FACS-purified CD25+CD4+ T reg cells at various ratios. Total splenocytes from C57BL/6 mice inactivated with 50 μg/ml mitomycin C (Sigma-Aldrich) for 45 min were used as APCs. A total of 4 × 105 CD4+ cells (CFSE-loaded CD25− plus CD25+) was mixed with 105 APCs plus 1 μg/ml anti-CD3 antibody per well of a 96-well round-bottom plate. Proliferation of the Cd45.1 effector cells was analyzed by CFSE dilution.
T cell transfers.
CD45.1 and CD45.2 congenically marked cells were mixed at a 1:1 ratio and loaded with CFSE. The cells were resuspended in PBS at 108 cells/ml, and a total of 107 cells were transferred into Rag2−/− recipients by injection into the retroorbital plexus.
Online supplemental material.
Fig. S1 shows that multiple Drosha splice isoforms are expressed in mouse tissues. Fig. S2 depicts the generation of the conditional Drosha allele. Fig. S3 shows the accumulation of inflammatory cytokine-producing CD4+ T cells in the spleens of aging DroshaF/Δ CD4-cre mice. Fig. S4 depicts moribund DroshaF/Δ CD4-cre mice that do not develop splenomegaly or lymphadenopathy. Fig. S5 shows that defective TGF-β–induced Foxp3 expression in the absence of Drosha is not due to thymocyte selection defects. Fig. S6 shows that enhanced IFN-γ expression in Drosha-deficient CD4+ cells is not due to thymocyte selection defects. Fig. S7 depicts the induction of IFN-γ expression in Drosha-deficient CD8+ cells. Fig. S8 shows that Foxp3 is not coexpressed with IFN-γ or IL-17A in moribund DroshaF/Δ CD4-cre or DroshaF/Δ Foxp3Cre/Cre(Y) mice. Fig. S9 shows that Drosha-deficient T cells display a partial proliferation defect in response to TCR stimulation or lymphopenia. Table S1 provides a tissue survey of moribund DroshaF/Δ CD4-cre mice. Table S2 provides a tissue survey of moribund DroshaF/Δ Foxp3Cre/Cre(Y) mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081219/DC1.
Acknowledgments
We wish to thank Michael McManus for providing the Dicer conditional mice.
M.M.W. Chong was a recipient of a postdoctoral fellowship from the Cancer Research Institute and is currently funded by a Helen L. and Martin S. Kimmel Center for Stem Cell Biology Postdoctoral Award. This work was supported by the Howard Hughes Medical Institute (D.R. Littman and A.Y. Rudensky).
The authors have no conflicting financial interests.
Abbreviations used: DP, double positive; dsRNA, double-stranded RNA; H&E, hematoxylin and eosin; miRNA, microRNA; pri-miRNA, primary miRNA; RA, retinoic acid; rRNA, ribosomal RNA; siRNA, small inhibitory RNA.
References
- 1.Bennett, C.L., M.E. Brunkow, F. Ramsdell, K.C. O'Briant, Q. Zhu, R.L. Fuleihan, A.O. Shigeoka, H.D. Ochs, and P.F. Chance. 2001. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics. 53:435–439. [DOI] [PubMed] [Google Scholar]
- 2.Brunkow, M.E., E.W. Jeffery, K.A. Hjerrild, B. Paeper, L.B. Clark, S.A. Yasayko, J.E. Wilkinson, D. Galas, S.F. Ziegler, and F. Ramsdell. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73. [DOI] [PubMed] [Google Scholar]
- 3.Kim, J.M., J.P. Rasmussen, and A.Y. Rudensky. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. [DOI] [PubMed] [Google Scholar]
- 4.Wildin, R.S., F. Ramsdell, J. Peake, F. Faravelli, J.L. Casanova, N. Buist, E. Levy-Lahad, M. Mazzella, O. Goulet, L. Perroni, et al. 2001. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27:18–20. [DOI] [PubMed] [Google Scholar]
- 5.Hughes, P.D., G.T. Belz, K.A. Fortner, R.C. Budd, A. Strasser, and P. Bouillet. 2008. Apoptosis regulators Fas and Bim cooperate in shutdown of chronic immune responses and prevention of autoimmunity. Immunity. 28:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutcheson, J., J.C. Scatizzi, A.M. Siddiqui, G.K. Haines III, T. Wu, Q.Z. Li, L.S. Davis, C. Mohan, and H. Perlman. 2008. Combined deficiency of proapoptotic regulators Bim and Fas results in the early onset of systemic autoimmunity. Immunity. 28:206–217. [DOI] [PubMed] [Google Scholar]
- 7.Weant, A.E., R.D. Michalek, I.U. Khan, B.C. Holbrook, M.C. Willingham, and J.M. Grayson. 2008. Apoptosis regulators Bim and Fas function concurrently to control autoimmunity and CD8+ T cell contraction. Immunity. 28:218–230. [DOI] [PubMed] [Google Scholar]
- 8.Alexander, W.S., R. Starr, J.E. Fenner, C.L. Scott, E. Handman, N.S. Sprigg, J.E. Corbin, A.L. Cornish, R. Darwiche, C.M. Owczarek, et al. 1999. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 98:597–608. [DOI] [PubMed] [Google Scholar]
- 9.Morita, Y., T. Naka, Y. Kawazoe, M. Fujimoto, M. Narazaki, R. Nakagawa, H. Fukuyama, S. Nagata, and T. Kishimoto. 2000. Signals transducers and activators of transcription (STAT)-induced STAT inhibitor-1 (SSI-1)/suppressor of cytokine signaling-1 (SOCS-1) suppresses tumor necrosis factor alpha-induced cell death in fibroblasts. Proc. Natl. Acad. Sci. USA. 97:5405–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornish, A.L., G.M. Davey, D. Metcalf, J.F. Purton, J.E. Corbin, C.J. Greenhalgh, R. Darwiche, L. Wu, N.A. Nicola, D.I. Godfrey, et al. 2003. Suppressor of cytokine signaling-1 has IFN-gamma-independent actions in T cell homeostasis. J. Immunol. 170:878–886. [DOI] [PubMed] [Google Scholar]
- 11.Fontenot, J.D., J.P. Rasmussen, L.M. Williams, J.L. Dooley, A.G. Farr, and A.Y. Rudensky. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. [DOI] [PubMed] [Google Scholar]
- 12.Chen, W., W. Jin, N. Hardegen, K.J. Lei, L. Li, N. Marinos, G. McGrady, and S.M. Wahl. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rana, T.M. 2007. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 8:23–36. [DOI] [PubMed] [Google Scholar]
- 14.Han, J., Y. Lee, K.H. Yeom, J.W. Nam, I. Heo, J.K. Rhee, S.Y. Sohn, Y. Cho, B.T. Zhang, and V.N. Kim. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 125:887–901. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V.N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature. 425:415–419. [DOI] [PubMed] [Google Scholar]
- 16.Chendrimada, T.P., R.I. Gregory, E. Kumaraswamy, J. Norman, N. Cooch, K. Nishikura, and R. Shiekhattar. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight, S.W., and B.L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 293:2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein, E., S.Y. Kim, M.A. Carmell, E.P. Murchison, H. Alcorn, M.Z. Li, A.A. Mills, S.J. Elledge, K.V. Anderson, and G.J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 35:215–217. [DOI] [PubMed] [Google Scholar]
- 19.O'Rourke, J.R., S.A. Georges, H.R. Seay, S.J. Tapscott, M.T. McManus, D.J. Goldhamer, M.S. Swanson, and B.D. Harfe. 2007. Essential role for Dicer during skeletal muscle development. Dev. Biol. 311:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynn, F.C., P. Skewes-Cox, Y. Kosaka, M.T. McManus, B.D. Harfe, and M.S. German. 2007. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 56:2938–2945. [DOI] [PubMed] [Google Scholar]
- 21.Koralov, S.B., S.A. Muljo, G.R. Galler, A. Krek, T. Chakraborty, C. Kanellopoulou, K. Jensen, B.S. Cobb, M. Merkenschlager, N. Rajewsky, and K. Rajewsky. 2008. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 132:860–874. [DOI] [PubMed] [Google Scholar]
- 22.Tam, O.H., A.A. Aravin, P. Stein, A. Girard, E.P. Murchison, S. Cheloufi, E. Hodges, M. Anger, R. Sachidanandam, R.M. Schultz, and G.J. Hannon. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 453:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe, T., Y. Totoki, A. Toyoda, M. Kaneda, S. Kuramochi-Miyagawa, Y. Obata, H. Chiba, Y. Kohara, T. Kono, T. Nakano, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 453:539–543. [DOI] [PubMed] [Google Scholar]
- 24.Wu, H., H. Xu, L.J. Miraglia, and S.T. Crooke. 2000. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275:36957–36965. [DOI] [PubMed] [Google Scholar]
- 25.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov, I.I., B.S. McKenzie, L. Zhou, C.E. Tadokoro, A. Lepelley, J.J. Lafaille, D.J. Cua, and D.R. Littman. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang, W., J.K. Kolls, and Y. Zheng. 2008. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 28:454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobb, B.S., A. Hertweck, J. Smith, E. O'Connor, D. Graf, T. Cook, S.T. Smale, S. Sakaguchi, F.J. Livesey, A.G. Fisher, and M. Merkenschlager. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson, M.J., K. Pino-Lagos, M. Rosemblatt, and R.J. Noelle. 2007. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204:1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mucida, D., Y. Park, G. Kim, O. Turovskaya, I. Scott, M. Kronenberg, and H. Cheroutre. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 317:256–260. [DOI] [PubMed] [Google Scholar]
- 31.Rubtsov, Y.P., J.P. Rasmussen, E.Y. Chi, J. Fontenot, L. Castelli, X. Ye, P. Treuting, L. Siewe, A. Roers, W.R. Henderson Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh, C.S., Y. Liang, A.J. Tyznik, S.G. Self, D. Liggitt, and A.Y. Rudensky. 2004. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 21:267–277. [DOI] [PubMed] [Google Scholar]
- 33.Pacholczyk, R., J. Kern, N. Singh, M. Iwashima, P. Kraj, and L. Ignatowicz. 2007. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 27:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuellar, T.L., T.H. Davis, P.T. Nelson, G.B. Loeb, B.D. Harfe, E. Ullian, and M.T. McManus. 2008. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. USA. 105:5614–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer, A., D. O'Carroll, C.L. Tan, D. Hillman, M. Sugimori, R. Llinas, and P. Greengard. 2007. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 204:1553–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyara, M., and S. Sakaguchi. 2007. Natural regulatory T cells: mechanisms of suppression. Trends Mol. Med. 13:108–116. [DOI] [PubMed] [Google Scholar]
- 37.Hori, S., T. Nomura, and S. Sakaguchi. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061. [DOI] [PubMed] [Google Scholar]
- 38.Li, Q.J., J. Chau, P.J. Ebert, G. Sylvester, H. Min, G. Liu, R. Braich, M. Manoharan, J. Soutschek, P. Skare, et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 129:147–161. [DOI] [PubMed] [Google Scholar]
- 39.Xiao, C., L. Srinivasan, D.P. Calado, H.C. Patterson, B. Zhang, J. Wang, J.M. Henderson, J.L. Kutok, and K. Rajewsky. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muralidhar, B., L.D. Goldstein, G. Ng, D.M. Winder, R.D. Palmer, E.L. Gooding, N.L. Barbosa-Morais, G. Mukherjee, N.P. Thorne, I. Roberts, et al. 2007. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J. Pathol. 212:368–377. [DOI] [PubMed] [Google Scholar]
- 41.Sugito, N., H. Ishiguro, Y. Kuwabara, M. Kimura, A. Mitsui, H. Kurehara, T. Ando, R. Mori, N. Takashima, R. Ogawa, and Y. Fujii. 2006. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin. Cancer Res. 12:7322–7328. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, T., E. Canaani, and C.M. Croce. 2007. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc. Natl. Acad. Sci. USA. 104:10980–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esquela-Kerscher, A., and F.J. Slack. 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6:259–269. [DOI] [PubMed] [Google Scholar]
- 44.Harfe, B.D., M.T. McManus, J.H. Mansfield, E. Hornstein, and C.J. Tabin. 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. USA. 102:10898–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau, N.C., L.P. Lim, E.G. Weinstein, and D.P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 294:858–862. [DOI] [PubMed] [Google Scholar]